Abstract
This study assesses the feasibility of using a sample-efficient model to investigate radiomics changes over time for predicting progression-free survival in rare diseases. Eighteen high-grade glioma patients underwent two L-3,4-dihydroxy-6-[18F]-fluoro-phenylalanine positron emission tomography (PET) dynamic scans: the first during treatment and the second at temozolomide chemotherapy discontinuation. Radiomics features from static/dynamic parametric images, alongside conventional features, were extracted. After excluding highly correlated features, 16 different models were trained by combining various feature selection methods and time-to-event survival algorithms. Performance was assessed using cross-validation. To evaluate model robustness, an additional dataset including 35 patients with a single PET scan at therapy discontinuation was used. Model performance was compared with a strategy extracting informative features from the set of 35 patients and applying them to the 18 patients with 2 PET scans. Delta-absolute radiomics achieved the highest performance when the pipeline was directly applied to the 18-patient subset (support vector machine (SVM) and recursive feature elimination (RFE): C-index = 0.783 [0.744–0.818]). This result remained consistent when transferring informative features from 35 patients (SVM + RFE: C-index = 0.751 [0.716–0.784], p = 0.06). In addition, it significantly outperformed delta-absolute conventional (C-index = 0.584 [0.548–0.620], p < 0.001) and single-time-point radiomics features (C-index = 0.546 [0.512–0.580], p < 0.001), highlighting the considerable potential of delta radiomics in rare cancer cohorts.
Similar content being viewed by others
Introduction
Radiomics analysis of positron emission tomography (PET) imaging is of great interest, given its ability to characterize tumor heterogeneity and its prognostic value1. In neuro-oncology, radiomics studies have primarily focused on amino-acid PET radiotracers for glioma grading at the initial diagnosis2,3 and for predicting the presence of key molecular characteristics4,5,6,7,8,9 in accordance with the latest World Health Organization (WHO) classifications10. The potential benefits of radiomics for prognosis11,12,13, detecting recurrences14,15,16, and identifying early progression after glioma chemoradiation17 have also been evaluated. More specifically, studies have examined radiomics derived from both static and dynamic acquisitions at the initial diagnosis18,19, with the aim of assessing prognosis20 or identifying glioma recurrences21. Neuro-oncological applications of radiomics face a unique challenge, due to the relatively low occurrence of brain tumors worldwide, and therefore require the use of dedicated modeling workflows and/or data augmentation techniques22,23. Although gliomas account for almost 80% of newly diagnosed malignant primary brain tumors and the vast majority are high-grade glioblastomas24, gliomas only represent 2% of all adult cancers. Indeed, the worldwide age-standardized annual incidence of primary malignant brain tumors is approximately 3 per 100,00025. Despite the standard treatment, which involves surgical resection and adjuvant chemoradiation therapy26,27, our understanding of the prognosis for these tumors remains limited. The use of amino-acid PET radiotracers28, specifically L-3,4-dihydroxy-6-[18F]-fluoro-phenylalanine (18F-FDOPA), in conjunction with magnetic resonance imaging (MRI), is currently recommended29 as a non-invasive diagnostic approach for assessing glioma progression30,31,32. The application of amino-acid PET imaging by integrating dynamic33 and radiomics34 analyses, has had a significant impact in the field of neuro-oncology and has improved the diagnostic performance.
The traditional radiomics model utilizes single-time-point medical images to evaluate or predict patient outcomes, disregarding changes in tumor characteristics during treatment or influenced by external factors such as chemotherapy or radiotherapy. An alternative approach, known as delta radiomics, introduces a time component by extracting quantitative features from images acquired at different treatment and follow-up time points. The delta radiomics concept, which employs the change in radiomic features during or after treatment to guide clinical decisions, may potentially be more suitable for evaluating tumor response to treatment. Using machine-learning algorithms, delta radiomics analysis has shown a superior capacity to predict clinical outcomes compared to single-time-point radiomics features. Although a number of studies have focused on the application of delta radiomics in glioma patients, these have been restricted to MRI imaging and classification tasks35,36,37. Moreover, changes in radiomic features derived from PET/computed tomography (CT) imaging have also been assessed to predict survival outcomes in non-rare glioma cancers such as non-small cell lung cancer38,39 or head and neck carcinoma40 with effectiveness over at least 45 patients.
To the best of our knowledge, no study has to date explored the potential of amino-acid PET-based delta radiomics on time-to-event survival data in a rare disease such as glioma. The aim of our current study is therefore to evaluate the feasibility of using a sample-efficient model investigating radiomics changes over time to predict progression-free survival (PFS) in high-grade glioma (HGG). The objectives of this paper also include assessing the robustness of the model and comparing the delta radiomics results to both delta conventional features and single-time-point radiomics features.
Material and methods
Patient cohort
The clinical trial consisted in the retrospective analysis of images and medical records with the inclusion of 18 HGG patients with two consecutive PET scans, the first one during treatment (\({PET}_{0}\)) and the second at the time of adjuvant temozolomide (TMZ) chemotherapy discontinuation (\({PET}_{1}\)), allowing to assess changes in extracted features over time. This population of 18 patients, was selected as a subset from a total of 53 patients who underwent dynamic 18F-FDOPA PET scans, after following a Stupp protocol, in the Nuclear Medicine Department of the Regional University Hospital (CHRU) of Nancy, between January 2018 and May 2022. Minimum follow-up was 1 year. The decision to treat with the Stupp protocol and initiate or discontinue adjuvant TMZ therapy was determined individually for each patient during the multidisciplinary neuro-oncology tumor board meetings. Tumors were classified as HGG based on the surgical sample or biopsies, in accordance with the WHO 2021 classification guidelines10. Following the RANO41 criteria, the clinical endpoint of PFS was defined as the time in months from the initial treatment to either the date of progression or censoring. Informed consent was obtained from all patients. The institutional ethics committee (Comité d'Éthique du CHRU de Nancy) approved the study, on 26 August 2020. The trial was registered at ClinicalTrials.gov (NCT04469244) and complied with the principles of the Helsinki declaration.
Data acquisition
Prior to undergoing 18F-FDOPA PET scans, all patients fasted for a minimum of 4 h. Several patients received Carbidopa 1 h prior to the examination to enhance tracer uptake in the brain42. This institutional procedure was implemented from February 2018 to September 2020. PET acquisitions were performed with a digital PET/CT device (Vereos, Philips Healthcare®, Eindhoven, The Netherlands). Each patient initially underwent a CT scan, then, following the injection of 2MBq of 18F-FDOPA per kg of body weight, a 30-min dynamic PET acquisition was performed. Using an OSEM 3D algorithm, a static image was reconstructed based on the last 20 min of the acquisition (2 iterations, 10 subsets, 256 × 256 × 164 voxels of 1 × 1 × 1 mm3). Dynamic images were reconstructed with 30 frames, each lasting 1 min (3 iterations, 15 subsets, 128 × 128 × 82 voxels of 2 × 2 × 2 mm3). During this process, images were corrected for attenuation, dead time as well as random and scattered coincidences. Point-spread function corrections were only applied to static images43.
Image pre-processing
An experienced nuclear physician (A.Z.) manually delineated, healthy brain, striatum, and tumor volumes of interest (VOIs) on static images with the LifeX software44 (lifexsoft.org). For patients who underwent 2 PET scans, different segmented VOIs from both \({PET}_{0}\) and \({PET}_{1}\) were considered. The healthy brain VOI was selected as a crescent-shaped region on three consecutive slices of the semi-oval center from the unaffected hemisphere29. Semi-automatic segmentation was performed using a 70% threshold of the maximum standardized uptake value (SUVmax) for the striatum VOI, and a threshold of 1.6 times the mean SUV (SUVmean) of healthy brain for the tumor VOI, to yield the metabolic tumor volume (MTV)29. Dynamic images were registered on the CT to correct for patient movement. Considering our focus on the voxel-based analysis and the need to reduce the impact of noise on voxel time activity curves (TACs), dynamic images underwent a denoising process45. As recommended elsewhere46, the impact of Carbidopa on SUV measurements was reduced by normalizing static images to the SUVmean of healthy brain, which provided static tumor-to-background-ratio (TBR) parametric images. For dynamic images, region/voxel-based TACs were initially fitted and subsequently normalized to the fitted mean brain TACs. Time-to-peak (TTP) values, representing the time interval between the start of acquisition and maximum uptake, were extracted. This procedure generated dynamic TTP parametric images for the voxel-based analysis.
Radiomics analysis
Feature extraction
The feature extraction step applied two distinct approaches i.e., the region- and voxel-based analysis. For the region-based analysis, conventional features such as mean, maximum and peak of TBR, and tumor-to-striatum ratios were extracted from the tumor VOIs. We also obtained the MTV, region-based dynamic TTP, and calculated the slope of the linear regression for the data acquired between the 10th and 30th min4, which provided 9 conventional features. For the voxel-based analysis, we initially resampled the dynamic TTP parametric images with a linear interpolation, to achieve a common resolution of 1 × 1 × 1 mm3 with the static TBR parametric images, as described by the Image Biomarker Standardization Initiative47. First-order statistics, morphological, intensity histogram, and textural features were extracted using the pyradiomics package48.
The previously described in-house software4 was used to identify local intensity features (https://github.com/TimZaragori/Sklearn_NestedCV/blob/master/Radiomics_gliomas_article/local_intensity_features.py) that were not available in pyradiomics. Discretization of the parametric images was performed using a fixed bin width of 0.1 for the static TBR and 1 min for dynamic TTP parametric images, resulting in 199 extracted radiomics features (94 static TBR radiomics features, 94 dynamic TTP radiomics features and 11 morphological features).
Feature sets
Four methods were used to compute changes in radiomics and conventional features. These included delta-absolute (\({F}_{{PET}_{1}}- {F}_{{PET}_{0}}\), with ΔAR and ΔAC defined as delta-absolute radiomics and conventional features, respectively) and delta-relative (\(\left({F}_{{PET}_{1}}- {F}_{{PET}_{0}}\right)/{F}_{{PET}_{0}}\), where ΔRR and ΔRC represent delta-relative radiomics and conventional features, respectively).
In addition, we also evaluated weighted delta-absolute (\(\left({F}_{{PET}_{1}}- {F}_{{PET}_{0}}\right)/\left({t}_{{PET}_{1}}- {t}_{{PET}_{0}}\right)\), by considering WΔAR and WΔAC for weighted delta-absolute radiomics and conventional features, respectively) and weighted delta-relative (\(\left({F}_{{PET}_{1}}- {F}_{{PET}_{0}}\right)/{F}_{{PET}_{0}}\left({t}_{{PET}_{1}}- {t}_{{PET}_{0}}\right)\), with WΔRR and WΔRC referring to weighted delta-relative radiomics and conventional features, respectively), which normalized the delta features by the time interval between the two collected PET scans. The analysis also included the single-time-point feature (\({F}_{{PET}_{1}}\), where STPR refers to the single-time-point radiomics feature). Here, \({F}_{{PET}_{0}}\), \({F}_{{PET}_{1}}\) represent the extracted features and \({t}_{{PET}_{0}}\) and \({t}_{{PET}_{1}}\) indicate the acquisition dates of the PET scans collected during treatment and after the last treatment sessions respectively.
Feature selection methods and machine learning algorithms
To reduce the risk of overfitting in this small patient cohort, several steps were considered in the applied pipeline. For a given training and test set, all transformations and algorithms were initially fitted exclusively to the training set and then applied to the test set for output prediction. After the feature extraction, variables with zero variance were eliminated, while the other variables underwent Z-score normalization. To address the problem of highly correlated features, we used hierarchical clustering based on the absolute Spearman correlation coefficient as a similarity metric, with a threshold of 0.9 to form the clusters49. The selected feature was the medoid of each cluster, which represents the most centrally located feature in terms of maximizing the similarity to all other features within the same cluster. Due to the limited number of samples, where the algorithm is sensitive to small perturbations, the consensus clustering50 technique was implemented to enhance the stability and robustness of the clustering process. This involved performing the clustering on each of the 500 bootstrap samples extracted from each training fold, resulting in 500 clustering outcomes, each representing a different arrangement of features. The final clusters were then determined by comparing the cluster assignments using a co-association matrix, based on a 0.5 threshold. This matrix represents the frequency or probability of features being assigned to the same cluster across multiple clustering runs.
In order to obtain non-redundant features after the clustering step, feature selection was performed and we compared several approaches including univariate concordance index (C-index), mutual information (MI), LASSO Cox and recursive feature elimination (RFE) methods to identify the most informative features. For the filter-based feature selection methods, i.e. univariate C-index and MI, the process was repeated 500 times using bootstrap samples from each training fold with features ranked according to the score in each bootstrap sample and overall rank computed with the importance score49. To model time-to-event survival data, Cox proportional hazards (CoxPH), ElasticNet Cox, support vector machine (SVM), and gradient boosting with component-wise least squares as base learner (GB-Linear) algorithms51 were employed, resulting in a total of 16 different models being trained (4 feature selections multiplied by 4 regressors).
Model training strategy
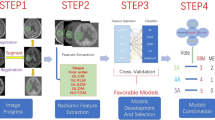
Two different strategies (Fig. 1) were explored for predicting PFS based on feature changes using the described pipeline.
In the main strategy, referred to as the default strategy, the pipeline was directly applied to the subset of 18 samples with 2 PET scans. Due to the small number of samples, the model hyperparameters were fixed, without undergoing further optimization (Supplementary Table S1). In this strategy, the feature selection methods were restricted to select consecutively 1, 2, and 3 single-time-point and delta features for comparison purpose. Model performance was evaluated using a 4-fold cross-validation (CV) with 25 repetitions.
The second strategy, serving as a validation of the first, is termed the fine-tuned strategy and involves a two-stage process. Initially, optimized hyperparameters and informative features were obtained from a separate and larger sample set (stage 1) and subsequently transferred to a smaller cohort of interest (stage 2). Focusing on single-time-point features in stage 1, information was extracted from all the patient with a single PET scan only (n = 35). This information was then used to build the model for stage 2, specifically aimed at evaluating the delta features extracted from the 18 patients with 2 PET scans. During stage 1, a 7-fold CV (ensuring the presence of at least 5 samples in the validation set) was repeated 20 times. The model hyperparameters were optimized (Supplementary Table S1) through a Bayesian search using Gaussian processes (https://github.com/scikit-optimize/scikit-optimize) over 100 iterations, with a focus on maximizing the C-index. Feature selection methods were limited to selecting at most 3 single-time-point radiomics and conventional features. In stage 1, the importance of features was calculated for each model across all folds using SHapley Additive exPlanations52 (SHAP) values, and subsequently, single-time-point features were ranked based on their importance. In stage 2, the analysis of single-time-point and delta features focused on the previously optimized hyperparameters in stage 1 and compared models by consecutively inputting 1, 2, and 3 top-ranked features identified in stage 1. The final model performance was assessed during stage 2 through a repeated CV process (4 folds, 25 repetitions) on the population of 18 patients with 2 PET scans.
The results were finally compared to a CoxPH model applied to conventional features only, which served as the baseline model (Baseline). The CoxPH model was also used to evaluate both absolute and relative changes of TBRmax and TBRmean for a univariate comparison.
Model robustness evaluation
Firstly, to address the limited number of samples with 2 PET scans, a fine-tuned strategy was developed (Fig. 1), transferring information from a larger set (n = 35) with single-time-point features to assess delta features within the smaller set (n = 18), serving as a validation of the default strategy.
Secondly, the results of different models employed for the default strategy were compared in terms of performance.
Thirdly, the applied pipeline integrated the bootstrap-based consensus clustering technique to improve cluster stability in each fold. The adjusted rand index53 (ARI), used as a similarity measure between two clustering results, was computed between the clustering results of all possible CV fold pairs and presented as the average of these pairs. Cluster similarity was determined by a negative ARI for discordant clustering, an ARI close to 0 for random labeling, and an ARI close to 1 when the clustering results were identical. ARI was also used to assess the similarity between the clusters of the first fold, arbitrary considered as the reference fold, and those derived from subsequent folds. For each cluster of the first fold, the most analogous cluster and the corresponding medoid features were identified, allowing us to evaluate the stability of these medoid features throughout CV folds.
Lastly, feature importance was calculated through SHAP values to assess whether the same features were selected across various models of the default strategy.
Statistical analysis
Categorical variables are expressed as counts and percentages and continuous variables as medians with interquartile ranges (IQR). The model performance on the test set was evaluated using the C-index, which assesses the probability that the sample with the highest predicted risk experiences an event before the sample with the lowest predicted risk among all the possible pairs. The integrated time-dependent area under the curve (iAUC) was calculated as a complementary metric to address potential limitations of the C-index. The iAUC estimates how effectively a predictive model can differentiate individuals who will experience an event within a specific time period from those who will not. The final result was obtained from one thousand bootstrap iterations of the CV performance distribution and summarized as the mean and standard error values with a 95% confidence interval (CI). To evaluate whether delta radiomics are superior to both delta conventional features and single-time-point radiomics features, one-sided Mann–Whitney U tests were conducted on the C-indices. To account for multiple comparisons, the p-values were corrected using the Benjamini-Hochberg54 method. A p-value < 0.05 was considered statistically significant. All analyses were performed in Python 3.8, using the scikit-survival51 library. Important features were identified using SHAP values for both the default strategy model and the second stage of the fine-tuned strategy.
Results
Patient characteristics
Among the 53 HGG patients identified, 18 patients with the following characteristics had undergone 2 PET scans (Supplementary Table S2). The median age was 62 years (IQR, 45–69) and the patients included 6 (33%) women. The median PFS was 11 months (IQR, 8–21), with a total of 16 (89%) patients relapsing. Carbidopa premedication was administered to 5 (28%) patients prior to 18F-FDOPA PET acquisition. The patients underwent either surgery (78%) or biopsy (22%) with tumors classified at the initial diagnosis according to the WHO 2021 glioma classification10. There were 3 (17%) IDH-mutant and 1p/19q non-codeleted astrocytomas (67% grade III and 33% grade IV) and 15 (83%) IDH-wildtype glioblastomas.
Survival analysis
Results obtained by selecting 1, 2, and 3 features were largely consistent (Fig. 2). The highest prediction performance was obtained with the SVM model in combination with RFE for delta-absolute radiomics (ΔAR: C-index = 0.783 [0.744, 0.818]) and with C-index feature selection for delta-relative radiomics (ΔRR: C-index = 0.740 [0.700, 0.778]), both based on a single selected feature (Fig. 2). Interestingly, GLCM Information Correlation 2 from dynamic TTP parametric images emerged as the most important radiomics feature in both models. The previously mentioned SVM + RFE model for delta-absolute and the SVM + C-index model for delta-relative features demonstrated significantly better performance for delta radiomics compared to single-time-point radiomics (Table 1), highlighting the positive influence of delta radiomics features on model decision-making (STPR of SVM + RFE: C-index = 0.546 [0.512, 0.580], p < 0.001 and STPR of SVM + C-index: C-index = 0.555 [0.514, 0.594], p < 0.001). These models also displayed a significantly better result for the delta radiomics compared to the delta conventional features (ΔAC of SVM + RFE: C-index = 0.584 [0.548, 0.620], p < 0.001 and ΔRC of SVM + C-index: C-index = 0.552 [0.513, 0.589], p < 0.001, when comparing the results of delta-absolute/relative radiomics features with their corresponding pairs of conventional features). Weighted delta radiomics results (WΔAR of SVM + RFE: C-index = 0.721 [0.682, 0.759] and WΔRR of SVM + C-index: C-index = 0.719 [0.682, 0.757], Supplementary Table S3) indicated a similar trend, with a better performance compared to single-time-point radiomics (p < 0.001 for both WΔAR and WΔRR) and weighted delta conventional features (WΔAC of SVM + RFE: C-index = 0.533 [0.492, 0.577], p < 0.001 and WΔRC of SVM + C-index: C-index = 0.451 [0.408, 0.495], p < 0.001).
Heatmap representing the C-indices of the default strategy based on the machine-learning algorithms (y-axis) and feature selection methods (x-axis) of the test set. The bold values indicate a significant p-value when comparing the radiomics model with its respective Baseline. GB: gradient boosting; SVM: support vector machine; MI: mutual information; RFE: recursive feature elimination.
Model robustness evaluation
Firstly, the fine-tuned strategy demonstrated optimal performance using the same combination SVM + RFE model as those used with the default strategy but with 2 selected features (Supplementary Fig. S1), revealing the delta-absolute and relative GLCM Cluster Prominence from dynamic TTP parametric image as the most informative radiomics features in stage 2 (ΔAR: C-index = 0.751 [0.716, 0.784] and ΔRR: C-index = 0.779 [0.742, 0.812], Table 2). Aligned with the default strategy, delta radiomics in the fine-tuned strategy also performed better than those obtained with single-time-point radiomics (p < 0.001 for both delta-absolute/relative radiomics) and delta conventional features (p < 0.001 for both delta-absolute/relative radiomics). Results from weighted delta radiomics reinforced this observation (see Supplementary Table S4), outperforming both single-time-point radiomics (p < 0.001 for both weighted delta-absolute/relative radiomics) and weighted delta conventional features (p = 0.006 for weighted delta-absolute, and p = 0.007 for weighted delta-relative radiomics). In both strategies, delta radiomics derived from the dynamic TTP parametric image and notably belonging to the same second-order radiomics family (GLCM) emerged as the most informative attribute (Fig. 3).
SHAP values indicate the feature importance in (a) the default strategy based on a single selected delta-absolute radiomics, and in (b) the second stage of the fine-tuned strategy based on two selected delta-absolute radiomics employing the SVM + RFE model in both strategies. The presence of TTP at the end of a feature name indicates that the corresponding feature is extracted from dynamic TTP parametric images.
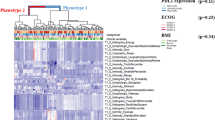
Secondly, in the default strategy, the methods employed to calculate changes in radiomics features did not significantly influence the results (the C-index range of 0.688 to 0.783 for delta-absolute and 0.619 to 0.740 for delta-relative, each based on a single selected feature), indicating a reliable extraction of relevant information from the input features by the model (Fig. 1). Similarly, the majority of radiomics models in the default strategy outperformed their conventional pairs (Fig. 1). Implementing bootstrap-based consensus clustering efficiently reduced the dimension of highly correlated features, resulting in a mean ARI value of 0.59 across CV folds for the SVM + RFE and a single selected feature within this strategy. Despite moderate cluster stability (ARI = 0.59), 70% of the medoid features assigned to the clusters were consistently observed in at least 80% of CV folds (Fig. 4). Furthermore, subsequent feature selection demonstrated consistency, with the GLCM Information correlation 2 extracted from dynamic TTP parametric images provided as the highest rank feature in terms of importance across all model combinations (Fig. 5).
The consistency of medoid features assigned to clusters across CV folds of the default strategy, based on a single selected feature and using the SVM + RFE model. The right side of the figure exhibits the medoid feature observed in the majority of folds, along with its corresponding percentage. The presence of TTP at the end of a feature name indicates that the corresponding feature is extracted from dynamic TTP parametric images. CV: cross-validation; GLRLM: gray level run length matrix; GLCM: gray level co-occurrence matrix; GLSZM: gray level size zone matrix; NGTDM: neighboring gray tone difference matrix; NGLDM: neighboring gray level dependence matrix.
SHAP value distributions for the top 10 ranked features across all 16 model combinations based on 1, 2, and 3 selected features in each CV fold within the default strategy. The presence of TTP at the end of a feature name indicates that the corresponding feature is extracted from dynamic TTP parametric images. CV: cross-validation; GLCM: gray level co-occurrence matrix; GLSZM: gray level size zone matrix; NGTDM: neighboring gray tone difference matrix; NGLDM: neighboring gray level dependence matrix.
Discussion
This study evaluated the prognostic value of delta radiomics features extracted from 18F-FDOPA PET scans acquired during HGG treatment and after the final treatment session and how these correlated with time-to-event survival data. Our results demonstrate that delta radiomics can be effectively performed and evaluated in rare cancer cohorts and that they outperform delta conventional features and single-time-point radiomics features. Notably, the delta radiomics model applied to rare cohorts in this study has demonstrated some criteria of robustness.
To the best of our knowledge, this study is the first to investigate the potential of PET-based radiomics changes over time to predict PFS in a relatively small cohort, typical of rare disease patient cohorts such as glioma. The aim of this methodological approach was to assess whether delta radiomics features were amenable to small patient numbers and how were their performances compared to both delta conventional features and single-time-point radiomics features. In addition to extracting single-time-point features from the PET scan conducted after the final treatment session, we also considered the absolute and relative changes of the extracted features, as well as their normalized versions, taking into account the time intervals between the 2 PET scans. We developed a robust pipeline incorporating key steps to enhance the reliability of the analysis, and thereby address the limitations of examining a relatively small number of cases. The pipeline specifically included feature normalization to ensure fair comparisons, bootstrap-based consensus clustering to obtain stable uncorrelated features, diverse feature selection methods for informative feature identification, and time-to-event survival algorithms to explore associations with event outcomes. Based on a CV estimation of the performance, a PFS prediction was conducted by directly applying the pipeline to the cohort of 18 HGG patients with 2 PET scans, referred to as the default strategy.
For the default strategy, delta radiomics, whether assessing absolute or relative changes, outperformed delta conventional features and single-time-point radiomics features (Table 1). Similar results were found for the weighted delta radiomics (Supplementary Table S3). These findings suggest that delta radiomics may capture more pertinent information for the PFS prediction task, irrespective of the specific time interval between the two collected PET scans. In general, models from the default strategy yielded superior performance when evaluated with a single selected feature. This may be attributed to the small number of longitudinal samples, as fewer features made it easier to identify meaningful patterns and extract relevant information.
To address the limited number of samples with 2 PET scans, several robustness criteria were assessed to validate the default strategy. First of all, we developed the fine-tuned strategy applied to a larger cohort of 35 patients with a single PET scan, and subsequently, the information obtained was used to evaluate delta features within the smaller cohort of 18 patients. Interestingly, the fine-tuned strategy also exhibited a similar trend than the default one, highlighting the efficiency of delta radiomics, particularly when integrating the SVM algorithm with the RFE method for feature selection (Table 2). In both strategies, the delta radiomics extracted from dynamic TTP parametric images, along with the same second-order radiomics family (GLCM), were identified as the most informative features, reinforcing the prognostic value of dynamic analysis33. Notably, the GLCM Information Correlation 2 feature played a significant role in the default strategy, while the GLCM Cluster Prominence feature was crucial for the fine-tuned strategy (Fig. 3).
Dealing with the common challenge of limited sample size in rare cancer cohorts, it is essential to implement a robust pipeline, as attempted in this study. Additional criteria for ensuring the robustness of the default strategy included the following considerations. The methods used for computing changes in radiomics features did not show a notable impact on the outcomes, suggesting that the model reliably extracted pertinent information from the input features. Likewise, most radiomics models demonstrated superior performance compared to their conventional pairs, highlighting the potential efficacy of the default strategy (Fig. 2). In addition, integrating bootstrap-based consensus clustering into the dimensionality reduction step of the pipeline resulted in an average ARI value of 0.59 across CV folds for the best model within the default strategy, and subsequent evaluation of medoid features demonstrated consistency (Fig. 4). Interestingly, the GLCM Information Correlation 2 attribute emerged as the highest rank feature across all 16 model combinations (Fig. 5).
Some studies have explored the effectiveness of the delta SUVmax feature in predicting PFS and overall survival in diffuse large B-cell lymphoma55,56,57. However, in the present study, the analysis of the delta feature demonstrated that, for both strategies, delta radiomics performed better than delta TBRmax, the normalized version of SUVmax using the mean of the brain (Tables 1 and 2, p < 0.001 when comparing delta-absolute of radiomics with TBRmax for both strategies, and p = 0.003 and p < 0.001 when comparing delta-relative of radiomics with TBRmax for default and fine-tuned strategies respectively) and delta TBRmean, the normalized version of SUVmean using the mean of the brain (Tables 1 and 2, p < 0.001 when comparing the results of delta-absolute/relative radiomics features with their corresponding pairs of TBRmean feature for both default and fine-tuned strategies) showcasing the superior performance of delta radiomics. This finding emphasizes the importance of further investigation within established guidelines such as PERCIST58. It is crucial to note that to comprehensively understand the implications of radiomics features, conducting additional evaluations over a larger cohort with more diverse samples is essential. Several studies have investigated deep learning and multitask learning approaches for analyzing time-to-event survival data39,59,60,61. While these algorithms demonstrate efficiency at capturing non-linear temporal patterns and managing temporal relationships across multiple time points, they also introduce complexity and require a larger sample size for generalization. These methods could nevertheless be a matter of future research. Conversely, working explicitly with delta radiomics provides several advantages, including reducing feature dimensionality and directly capturing temporal changes for enhanced interpretability and computational efficiency, which is adapted to the small number of samples. The proposed pipeline in this work can effectively be applied to explore the potential of radiomics changes over time in other rare patient cohorts.
The current study has several limitations. Firstly, as a retrospective study, the data was collected from a preselected patient population, thereby restricting patient inclusion. Given that our study was a single-center analysis, it would be useful to consider including samples from other centers or collecting data from new patients in our center for external validation to provide further verification in terms of the robustness and generalizability of the models developed. Admittedly, due to the retrospective nature of our study and the unavailability of raw data, we did not perform the repeatability of radiomics features in the current analysis. A feature repeatability assessment is, nevertheless worth considering in future studies to ensure reliable performance of the generated radiomics models22,23. Furthermore, although the study involved a limited number of patients who underwent 2 PET scans, the sample size corresponds to the typical range of longitudinal case numbers observed in rare diseases such as glioma35,36,37, and therefore reflects real life conditions. It is important to highlight that the primary objective of this study was to investigate the potential additive value of radiomics changes over time for predicting PFS in a small cohort of rare cancer patients. To address this, we implemented a robust pipeline and employed two distinct strategies, both of which yielded similar results, confirming the efficacy of the default strategy for delta radiomics.
Conclusion
This study highlights the considerable potential of delta radiomics analysis when applied to a relatively small cohort of patients with rare HGG disease, and demonstrates its superior performance compared to both delta conventional features and single-point radiomics features. The proposed pipeline, assessed through robustness criteria and adapted to small case numbers, needs to be evaluated in other cancer indications to confirm the effectiveness of delta radiomics. Such analyses may lead to the development of novel prognostic biomarkers for patients with rare cancers.
Data availability
Available in supplementary information files.
References
Hatt, M. et al. Joint EANM/SNMMI guideline on radiomics in nuclear medicine: Jointly supported by the EANM Physics Committee and the SNMMI Physics, Instrumentation and Data Sciences Council. Eur. J. Nucl. Med. Mol. Imaging 50, 352–375 (2023).
Pyka, T. et al. Textural analysis of pre-therapeutic [18F]-FET-PET and its correlation with tumor grade and patient survival in high-grade gliomas. Eur. J. Nucl. Med. Mol. Imaging 43, 133–141 (2015).
Russo, G. et al. Feasibility on the use of radiomics features of 11[C]-MET PET/CT in central nervous system tumours: Preliminary results on potential grading discrimination using a machine learning model. Curr. Oncol. 28, 5318–5331 (2021).
Zaragori, T. et al. 18F-FDOPA PET for the non-invasive prediction of glioma molecular parameters: A radiomics study. J. Nucl. Med. https://doi.org/10.2967/jnumed.120.261545 (2021).
Lohmann, P. et al. Predicting IDH genotype in gliomas using FET PET radiomics. Sci. Rep. 8, 13328 (2018).
Papp, L. et al. Sex-specific radiomic features of L-[S-methyl-11C] methionine PET in patients with newly-diagnosed gliomas in relation to IDH1 predictability. Front. Oncol. https://doi.org/10.3389/fonc.2023.986788 (2023).
Zhou, W. et al. Integrated CT radiomics features could enhance the efficacy of 18F-FET PET for non-invasive isocitrate dehydrogenase genotype prediction in adult untreated gliomas: A retrospective cohort study. Front. Oncol. https://doi.org/10.3389/fonc.2021.772703 (2021).
Qian, J. et al. Prediction of MGMT status for glioblastoma patients using radiomics feature extraction from 18F-DOPA-PET imaging. Int. J. Radiat. Oncol. Biol. Phys. 108, 1339–1346 (2020).
Haubold, J. et al. Non-invasive tumor decoding and phenotyping of cerebral gliomas utilizing multiparametric 18F-FET PET-MRI and MR fingerprinting. Eur. J. Nucl. Med. Mol. Imaging 47, 1435–1445 (2020).
Louis, D. N. et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro Oncol. 23, 1231–1251 (2021).
Papp, L. et al. Glioma survival prediction with combined analysis of in vivo 11C-MET PET features, ex vivo features, and patient features by supervised machine learning. J. Nucl. Med. 59, 892–899 (2018).
Carles, M. et al. FET-PET radiomics in recurrent glioblastoma: Prognostic value for outcome after re-irradiation?. Radiat. Oncol. 16, 46 (2021).
Wiltgen, T. et al. 18F-FET PET radiomics-based survival prediction in glioblastoma patients receiving radio(chemo)therapy. Radiat. Oncol. 17, 198 (2022).
Wang, K. et al. Individualized discrimination of tumor recurrence from radiation necrosis in glioma patients using an integrated radiomics-based model. Eur. J. Nucl. Med. Mol. Imaging 47, 1400–1411 (2020).
Hotta, M., Minamimoto, R. & Miwa, K. 11C-methionine-PET for differentiating recurrent brain tumor from radiation necrosis: Radiomics approach with random forest classifier. Sci. Rep. 9, 15666 (2019).
Müller, M. et al. Static FET PET radiomics for the differentiation of treatment-related changes from glioma progression. J. Neurooncol. 159, 519–529 (2022).
Lohmann, P. et al. FET PET radiomics for differentiating pseudoprogression from early tumor progression in glioma patients post-chemoradiation. Cancers 12, 3835 (2020).
Li, Z. et al. Prediction of TERTp-mutation status in IDH-wildtype high-grade gliomas using pre-treatment dynamic [18F]FET PET radiomics. Eur. J. Nucl. Med. Mol. Imaging 48, 4415–4425 (2021).
von Rohr, K. et al. Can radiomics provide additional information in [18F]FET-negative gliomas?. Cancers 14, 4860 (2022).
Li, Z. et al. Combination of pre-treatment dynamic [18F]FET PET radiomics and conventional clinical parameters for the survival stratification in patients with IDH-wildtype glioblastoma. Eur. J. Nucl. Med. Mol. Imaging 50, 535–545 (2023).
Ahrari, S. et al. Relevance of dynamic 18F-DOPA PET radiomics for differentiation of high-grade glioma progression from treatment-related changes. Biomedicines 9, 1924 (2021).
Gutsche, R. et al. Evaluation of FET PET radiomics feature repeatability in glioma patients. Cancers 13, 647 (2021).
Zounek, A. J. et al. Feasibility of radiomic feature harmonization for pooling of [18F]FET or [18F]GE-180 PET images of gliomas. Zeitschrift für Medizinische Physik 33, 91–102 (2023).
Ostrom, Q. T. et al. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2008–2012. Neuro-Oncology 17, 1–62 (2015).
Bondy, M. L. et al. Brain tumor epidemiology: Consensus from the Brain Tumor Epidemiology Consortium. Cancer 113, 1953–1968 (2008).
Sanai, N. & Berger, M. S. Surgical oncology for gliomas: The state of the art. Nat. Rev. Clin. Oncol. 15, 112–125 (2018).
Stupp, R. et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 10, 459–466 (2009).
Verger, A., Arbizu, J. & Law, I. Role of amino acid PET in HGGs: Limitations and perspectives. Q. J. Nucl. Med. Mol. Imaging https://doi.org/10.23736/S1824-4785.18.03092-3 (2018).
Law, I. et al. Joint EANM/EANO/RANO practice guidelines/SNMMI procedure standards for imaging of gliomas using PET with radiolabelled amino acids and [18F]FDG: version 1.0. Eur. J. Nucl. Med. Mol. Imaging 46, 540–557 (2019).
Karunanithi, S. et al. 18F-FDOPA PET/CT for detection of recurrence in patients with glioma: Prospective comparison with 18F-FDG PET/CT. Eur. J. Nucl. Med. Mol. Imaging 40, 1025–1035 (2013).
Zaragori, T. et al. Use of static and dynamic [18F]-F-DOPA PET parameters for detecting patients with glioma recurrence or progression. EJNMMI Res. 10, 56 (2020).
Prather, K. Y. et al. A systematic review of amino acid PET in assessing treatment response to temozolomide in glioma. Neuro-Oncol. Adv. 4, vdac008 (2022).
Verger, A., Imbert, L. & Zaragori, T. Dynamic amino-acid PET in neuro-oncology: A prognostic tool becomes essential. Eur. J. Nucl. Med. Mol. Imaging 48, 4129–4132 (2021).
Lohmann, P. et al. Radiomics in neuro-oncology: Basics, workflow, and applications. Methods 188, 112–121 (2021).
Chang, Y. et al. An investigation of machine learning methods in delta-radiomics feature analysis. PLoS ONE 14, e0226348 (2019).
Siakallis, L. et al. Longitudinal structural and perfusion MRI enhanced by machine learning outperforms standalone modalities and radiological expertise in high-grade glioma surveillance. Neuroradiology 63, 2047–2056 (2021).
Jeong, J. et al. Machine-learning based classification of glioblastoma using delta-radiomic features derived from dynamic susceptibility contrast enhanced magnetic resonance images. Quant. Imaging Med. Surg. 9, 1201–1213 (2019).
Fave, X. et al. Delta-radiomics features for the prediction of patient outcomes in non-small cell lung cancer. Sci. Rep. 7, 588 (2017).
Forouzannezhad, P. et al. Multitask learning radiomics on longitudinal imaging to predict survival outcomes following risk-adaptive chemoradiation for non-small cell lung cancer. Cancers 14, 1228 (2022).
Šedienė, S. et al. Development of a model based on delta-radiomic features for the optimization of head and neck squamous cell carcinoma patient treatment. Medicina 59, 1173 (2023).
Leao, D. J., Craig, P. G., Godoy, L. F., Leite, C. C. & Policeni, B. Response assessment in neuro-oncology criteria for gliomas: Practical approach using conventional and advanced techniques. Am. J. Neuroradiol. 41, 10–20 (2020).
Schiepers, C., Chen, W., Cloughesy, T., Dahlbom, M. & Huang, S.-C. 18F-FDOPA kinetics in brain tumors. J. Nucl. Med. 48, 1651–1661 (2007).
Ahrari, S. et al. Implementing the point spread function deconvolution for better molecular characterization of newly diagnosed gliomas: A dynamic 18F-FDOPA PET radiomics study. Cancers 14, 5765 (2022).
Nioche, C. et al. LIFEx: A freeware for radiomic feature calculation in multimodality imaging to accelerate advances in the characterization of tumor heterogeneity. Cancer Res. 78, 4786–4789 (2018).
Floberg, J. M. et al. Improved kinetic analysis of dynamic PET data with optimized HYPR-LR. Med. Phys. 39, 3319–3331 (2012).
Bros, M. et al. Effects of carbidopa premedication on 18F-FDOPA PET imaging of glioma: A multiparametric analysis. Cancers 13, 5340 (2021).
Zwanenburg, A. et al. The image biomarker standardization initiative: Standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology 295, 328–338 (2020).
van Griethuysen, J. J. M. et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 77, e104–e107 (2017).
Leger, S. et al. A comparative study of machine learning methods for time-to-event survival data for radiomics risk modelling. Sci. Rep. 7, 13206 (2017).
Monti, S., Tamayo, P., Mesirov, J. & Golub, T. Consensus clustering: A resampling-based method for class discovery and visualization of gene expression microarray data. Mach. Learn. 52, 91–118 (2003).
Pölsterl, S. scikit-survival: A Library for Time-to-Event Analysis Built on Top of scikit-learn.
Lundberg, S. M. & Lee, S.-I. A Unified Approach to Interpreting Model Predictions. In Advances in Neural Information Processing Systems Vol. 30 (Curran Associates, Inc., 2017).
Steinley, D. Properties of the Hubert-Arabie adjusted Rand index. Psychol. Methods 9, 386–396 (2004).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 57, 289–300 (1995).
Feres, C. et al. PET-CT quantitative parameter delta suvmax as a predictor of early relapse in patients with difuse large B-cell lymphoma. Blood 138, 4580 (2021).
Rekowski, J. et al. Interim PET evaluation in diffuse large B-cell lymphoma using published recommendations: Comparison of the deauville 5-point scale and the ΔSUVmax method. J. Nucl. Med. 62, 37–42 (2021).
Wolsztynski, E., O’Sullivan, F., O’Sullivan, J. & Eary, J. F. Statistical assessment of treatment response in a cancer patient based on pre-therapy and post-therapy FDG-PET scans. Stat. Med. 36, 1172–1200 (2017).
Pinker, K., Riedl, C. & Weber, W. A. Evaluating tumor response with FDG PET: Updates on PERCIST, comparison with EORTC criteria and clues to future developments. Eur. J. Nucl. Med. Mol. Imaging 44, 55–66 (2017).
Thammasorn, P. et al. Regularizing the deepsurv network using projection loss for medical risk assessment. IEEE Access 10, 8005–8020 (2022).
Han, J. et al. MS-ResNet: Disease-specific survival prediction using longitudinal CT images and clinical data. Int. J. CARS 17, 1049–1057 (2022).
Farina, B. et al. Integration of longitudinal deep-radiomics and clinical data improves the prediction of durable benefits to anti-PD-1/PD-L1 immunotherapy in advanced NSCLC patients. J. Transl. Med. 21, 174 (2023).
Acknowledgements
The authors thank Petra Neufing for the English editing of the manuscript.
Author information
Authors and Affiliations
Contributions
The authors participated in the (1) study conception and design or analysis and interpretation of the data, or both: S.A., T.Z., A.Z., and A.V.; (2) drafting of the manuscript or revising it critically for important intellectual content: S.A., T.Z., A.Z., J.O., L.I., and A.V.; (3) final approval of the submitted manuscript: T.Z., J.O., L.I., and A.V. All authors reviewed and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ahrari, S., Zaragori, T., Zinsz, A. et al. Application of PET imaging delta radiomics for predicting progression-free survival in rare high-grade glioma. Sci Rep 14, 3256 (2024). https://doi.org/10.1038/s41598-024-53693-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-53693-x
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.