Abstract
At the molecular level, triple-negative breast cancer (TNBC) is frequently categorized as PAM50 basal-like subtype, but despite the advances in molecular analyses, the clinical outcome for these subtypes is uncertain. Long non-coding RNAs (lncRNAs) are master regulators of genes involved in hallmarks of cancer, which makes them suitable biomarkers for breast cancer (BRCA) diagnosis and prognosis. Here, we evaluated the regulatory role of lncRNA SOX9-AS1 in these subtypes. Using the BRCA-TCGA cohort, we observed that SOX9-AS1 was significantly overexpressed in basal-like and TNBC in comparison with other BRCA subtypes. Survival analyzes showed that SOX9-AS1 overexpression was associated with a favorable prognosis in TNBC and basal-like patients. To study the functions of SOX9-AS1, we determined the expression levels in a panel of nine BRCA cell lines finding increased levels in MDA-MB-468 and HCC1187 TNBC. Using subcellular fractionation in these cell lines, we ascertained that SOX9-AS1 was located in the cytoplasmic compartment. In addition, we performed SOX9-AS1 gene silencing using two short-harping constructs, which were transfected in both cell models and performed a genome-wide RNA-seq analysis. Data showed that 351 lncRNAs and 740 mRNAs were differentially expressed in MDA-MB-468 while 56 lncRNAs and 100 mRNAs were modulated in HCC1187 cells (Log2FC < - 1.5 and > 1.5, p.adj value < 0.05). Pathway analysis revealed that the protein-encoding genes potentially regulate lipid metabolic reprogramming, and epithelial–mesenchymal transition (EMT). Expression of lipid metabolic-related genes LIPE, REEP6, GABRE, FBP1, SCD1, UGT2B11, APOC1 was confirmed by RT-qPCR. Functional analysis demonstrated that the knockdown of SOX9-AS1 increases the triglyceride synthesis, cell migration and invasion in both two TNBC cell lines. In conclusion, high SOX9-AS1 expression predicts an improved clinical course in patients, while the loss of SOX9-AS1 expression enhances the aggressiveness of TNBC cells.
Similar content being viewed by others
Introduction
Triple-negative breast cancer (TNBC) is a heterogeneous BRCA subtype characterized by the lack of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) expression, as determined by immunohistochemical staining1. At the molecular level, subtyping using the PAM50 gene signature often classifies BRCA into four intrinsic subtypes: luminal A, luminal B, HER2-enriched and basal-like2,3,4,5,6. As high as 60–80% of basal-like tumors are categorized as TNBC, nevertheless, the remaining basal-like tumors (expressing receptors) are grouped into other molecular subtypes7,8. The heterogeneity of TNBC and lack of molecular targets results in a diversity of treatment approaches unguided by tumor biology9. Therefore, a deeper characterization of the hallmarks of TNBC is necessary for identifying the events that drive oncogenesis. A pathogenic outcome can be accomplished by affecting metabolic reprogramming and other processes in TNBC cells10. However, more exhaustive oncogenomic analyses of TNBCs are required to understand the heterogeneity of the disease11.
LncRNAs are non-protein coding transcripts involved in diverse biological phenomena, and their specific patterns of expression coordinate different normal and disease processes12,13. In cancer, lncRNAs play a significant role in the regulation of gene expression and are involved in multiple hallmarks, suggesting that these molecules might be used as diagnostic or prognostic biomarkers14,15. However, given the vast number of lncRNAs identified using transcriptomic studies, it is critical to develop effective strategies for studying their roles and mechanisms in cancer16,17. In this regard, our research group previously evaluated lncRNA expression profiles in 156 TNBC biopsies through microarray expression. In this study, 710 lncRNAs showed differential expression between TNBC molecular subtypes, and most of these lncRNAs have not been characterized. One of these deregulated lncRNAs was SOX9-AS118. In another study including 74 tumors classified by PAM50, we found that SOX9-AS1 was significantly overexpressed in basal-like tumors19. However, the biological role of SOX9-AS1 in this BRCA subtype is poorly understood.
SOX9-AS1 is a human lncRNA, transcribed from cytogenetic band 17q24.3 and measuring 3033 bp20. It has been established that SOX9-AS1 promotes hepatocellular carcinoma (HCC) progression, through its sponge function of miR-5590-3p, causing SOX9 to be overexpressed. In turn, SOX9 activates SOX9-AS1 expression21. A meta-analysis identified SOX9-AS1 as part of a signature deregulated lncRNA-miRNA-mRNA network for TNBC. Taken together, this evidence suggests that SOX9-AS1 might have an important biological and even prognostic role in basal-like and TNBC. However, there is a need for comprehensive examination of this lncRNA expression profile to characterize its expression, molecular functions, and roles in TNBC22.
In this study, we analyzed SOX9-AS1 expression in a BRCA-TCGA cohort and determined its prognostic value in TNBC and the PAM50 basal-like subtype. Additionally, we elucidate its biological role through differential gene expression (DGE) analysis, KEGG pathways, Gene Ontology and GSEA in these subtypes from the same cohort. Moreover, transcriptome analysis using RNA-Seq of SOX9-AS1 deficient-cells indicate that silencing of this lncRNA regulates a plethora of protein-encoding genes that increases progression of two different TNBC cell lines. Therefore, we propose an onco-suppressor biological and clinical role for this novel lncRNA in TNBC and basal-like BRCA subtypes.
Results
SOX9-AS1 exhibits different expression patterns in multiple tissue types
Few studies have reported SOX9-AS1 expression levels in tumors21,23. In this regard, we determined its expression pattern across 31 cancer types using the TCGA database and the GEPIA2 platform. We found that SOX9-AS1 was overexpressed in 13 different normal tissues versus samples from diverse tumors (BRCA, TGCT, SKCM, PAAD, SARC, KICH, THCA, STAD, PRAD, HNSC, LUAD, LUSC, and PCPG), while it was underexpressed in 17 different normal tissues versus tumors (DLBC, THYM, BLCA, CESC, ESCA, OV, UCEC, READ, UCS, COAD, LIHC, KIRC, KIRP, CHOL, GBM, and LGG). Additionally, we found one tumor type with the same expression level (ACC), and one non-solid tumor without expression (LAML) (Fig. 1A). As shown in Fig. 1B, SOX9-AS1 expression was significantly downregulated in BRCA samples (p value = 0.0001) compared to normal tissues. These results suggest that SOX9-AS1 could have a relevant role in cancer biology.
SOX9-AS1 expression is deregulated in different human tumors. (A) SOX9-AS1 expression profile across all tumor samples and normal tissues (bar plot). The height of bar represents the median expression of tumor (red bars) and normal tissue (black bars) (data obtained from GEPIA2 platform). (B) SOX9-AS1 expression in normal breast tissue samples versus BRCA tissue determined by RNA-seq from the BRCA-TCGA cohort. Statistical differences between groups were compared using the Mann–Whitney test, ****p < 0.0001.
High SOX9-AS1 expression in TNBC and basal-like is associated with better prognosis
Next, we determined the expression level of SOX9-AS1 among tumor subtypes classified by IHC and the molecular PAM50 classifier. Notably, the expression of SOX9-AS1 was higher in the TNBC and basal-like molecular subtypes compared to receptor-positive BRCA (p value = 0.0001) (Fig. 2A,B). As shown in Supplementary Table 1, there was a significant association between SOX9-AS1 expression and ER (p value = 5.53e−21) and PR (p value = 4.94e−45) status, being higher in hormone receptor-negative samples. Furthermore, when using PAM50 to discriminate molecular subtypes (p value = 5.449e−50) there was a clear enrichment of SOX9-AS1 in basal-like tissue. Our results suggest that the high expression of SOX9-AS1 is a possible biomarker for TNBC and basal-like subtypes.
High SOX9-AS1 expression is associated with better prognosis in basal-like and TNBC. SOX9-AS1 expression between (A) receptor status (IHC), and (B) molecular subtypes classified by PAM50. The horizontal lines in the box plots represent the medians, the boxes represent the interquartile ranges, and the whiskers represent the 5th and 95th percentiles. RNA-seq data from the BRCA-TCGA cohort. Dunn's multiple comparisons and Kruskal–Wallis tests, ****p < 0.0001. (C) Kaplan–Meier curve of TNBC patients classified into high- and low-risk groups using the upper and lower quartiles of SOX9-AS1 expression for disease-specific survival (the analysis was conducted on 144 patients). Log-rank test between patients with low fold-change ( < -1.5) and high fold-change (> 1.5) and p value < 0.05. (D) Overall Survival in TNBC patients (the analysis was conducted on 144 patients), and (E) disease-free survival in basal-like patients, using Kaplan–Meier curves (the analysis was conducted on 442 patients), determined by Kaplan–Meier Plotter (breast). In red: patients with expression of SOX9-AS1 above the automatic cutoff calculation. In black, patients with expressions of SOX9-AS1 below automatic cutoff calculation.
To investigate the clinical value of SOX9-AS1 expression in TNBC and basal-like patients, Kaplan–Meier survival analyses were performed. As shown in Fig. 2C, TCGA TNBC patients have an increased disease-specific survival (DSS) time in the high SOX9-AS1 expression group, compared with those expressing low levels (Log-rank test, p value = 0.039). To discern if the prognostic value of SOX9-AS1 was replicated in independent cohorts, we used the Kaplan–Meier plotter service to analyze the SOX9-AS1 expression data24,25. We found that high expression of SOX9-AS1 was associated with an increased overall survival (OS) in TNBC patients (HR = 0.41, 95% CI (0.18–0.93); Log-rank p value = 0.028) (Fig. 2D). Overexpression of SOX9-AS1 was also related to a significant increase in disease-free survival (DFS) in basal-like patients (HR = 0.55, 95% CI (0.4–0.76); Log-rank test, p value = 0.00021) (Fig. 2E). These data indicate that high SOX9-AS1 expression predicts an increased DSS, OS, and DFS in TNBC and basal-like patients.
SOX9-AS1 potentially regulates processes involved in metabolism in TNBC and Basal-like samples from TCGA cohort
To clarify the role of SOX9-AS1 in gene regulation and biology of basal-like and TNBC, we performed a DGE analysis between the SOX9-AS1 high expression group and the SOX9-AS1 low expression group (see “Materials and methods” for details). We identified 1,173 differentially expressed genes (DEGs) in TNBC, of which 791 (67.4%) were overexpressed and 382 (32.6%) were underexpressed (Fig. 3A). In basal-like samples we identified 619 DEGs of which 352 (56.9%) were overexpressed and 267 (43.1%) underexpressed (Fig. 3B) (Supplementary Table 2). Figure 3C,D shows the top 5 DEGs in TNBC and basal-like, respectively. 428 genes were shared between TNBC and basal-like subtypes (Supplementary Fig. 1A; Supplementary Table 3). In addition, we identified 697 differentially expressed lncRNAs of which 481 (69%) lncRNAs were overexpressed and 216 (31%) were underexpressed in TNBC (Supplementary Fig. 1B,C). In contrast, the basal-like subtype only presented 343 differentially expressed lncRNAs, 189 (55.1%) lncRNAs overexpressed and 154 (44.9%) lncRNAs underexpressed (Supplementary Fig. 1D,E; Supplementary Table 4).
GSEA analyses of DEGs in TNBC and basal-like samples of the BRCA-TCGA cohort. The Volcano plots represent the distribution of DEGs. Blue genes are downregulated, red genes are upregulated, and gray genes are not differentially expressed in (A) TNBC-TCGA, and (B) basal-like-TCGA samples. Up- and downregulated genes were defined as Log2FC < - 1.5 and > 1.5 with adjusted p value < 0.05. Heatmap of the DGEs contributing to top 5 in (C) TNBC and (D) basal-like samples. Green: low expression, red: high expression according to Log2 fold change (visualized on the color bar above the heatmap). (E) Using the Hallmark gene set, genes involved in xenobiotic metabolism, peroxisome pathway, oxidative phosphorylation, reactive oxygen species pathway, and adipogenesis were significantly enriched in TNBC samples with FDR q value < 0.25. The NOM p value and FDR q value are displayed.
To elucidate the cellular processes and pathways of DEGs regulated by SOX9-AS1 in TNBC and basal-like subtypes, we performed GO and KEGG pathway analyses. We observed that similar processes and functions were shared across both TNBC and basal-like subtypes (Supplementary Fig. 2A,B). GO analysis showed numerous genes associated with regulation of hydrolase activity, skin development and differentiation, extracellular matrix structural organization, and hormone/xenobiotic metabolic process were significatively enriched (Supplementary Table 5). KEGG pathway enrichment analysis revealed that DEGs were significantly enriched in neuroactive ligand-receptor interaction, calcium signaling pathway, metabolism of organic compounds, protein absorption and digestion, and secretion of saliva and gastric acid (Supplementary Fig. 2C,D; Supplementary Table 6). Altogether, these results indicate that SOX9-AS1 could regulate a plethora of genes mostly involved in the metabolism of organic and chemical compounds in TNBC and basal-like BRCA samples.
In order to reduce variation and redundancy and to provide a narrower view of the biological role of SOX9-AS1, we performed a GSEA analysis using the Hallmark gene set26 with the same DEGs in TNBC and basal-like samples from the BRCA-TCGA cohort. The results with a NOM (p value < 0.05) and FDR (q value < 0.25) showed expression patterns associated with xenobiotic metabolism (i.e., BLVRB, CYP4F2, TMBIM6), peroxisome pathway (i.e., SULT2B1, DHCR24, IDH2), oxidative phosphorylation (i.e., PDHB, UQCRQ, POR), reactive oxygen species pathway (i.e., HMOX2, G6PD, PTPA), and adipogenesis (i.e., REEP6, CMBL, TKT) in TNBC samples (Fig. 3E). However, in basal-like samples, we did not identify significantly enriched Hallmark gene sets, although the same Hallmarks gene sets reported in TNBC were observed (Supplementary Table 7). Collectively, this suggests that SOX9-AS1 might regulate genes directly related to metabolic pathways in TNBC samples.
Relationship between SOX9-AS1 expression and protein-coding genes of TNBC and Basal-like samples
Given that predicting functions of lncRNAs remains a substantial challenge27, we attempted to predict functions of SOX9-AS1 by means of a correlation analysis based on RNA-seq data between the expression levels of SOX9-AS1 and genes in TNBC and basal-like samples from TCGA. The expression of SOX9-AS1 in TNBC samples was positively correlated with 5944 and negatively correlated with 723 genes (Spearman rank, p value < 0.05). While in basal-like samples, the expression of SOX9-AS1 was positively correlated with 4,503 and negatively correlated with 711 genes (Spearman rank, p value < 0.05) (Supplementary Table 8). The top 5 genes (positive- and negative-correlation) in TNBC and basal-like are shown in Fig. 4A,B, respectively. In TNBC, a strong positive correlation was observed between SOX9-AS1 and MIA (r = 0.63, p value ≤ 0.0001) and MIA-RAB4B (r = 0.65, p value ≤ 0.0001) (Fig. 4C). Furthermore, these genes showed significantly higher expression levels in the basal-like molecular subtype (Fig. 4E). In contrast, a moderate positive correlation with the ST3GAL6 (r = 0.49, p value ≤ 0.0001) and CDH19 (r = 0.47, p value ≤ 0.0001) genes was observed in basal-like BRCA (Fig. 4D).
Correlation of SOX9-AS1 expression with genes in TNBC and basal-like samples from BRCA-TCGA cohort. Heatmap showing the top 5 genes that show the greatest negative and positive correlation with the SOX9-AS1 expression in (A) TNBC and (B) basal-like samples. (C) Correlation plots of SOX9-AS1 expression with MIA, and MIA-RAB4B in TNBC. (D) Correlation plots of SOX9-AS1 expression with ST3GAL6, CDH19 in basal-like samples. Spearman's rank correlation critical value was used for the analysis. (E) Expression level of MIA, MIA-RAB4B, ST3GAL6, and CDH19 between molecular subtypes classified by PAM50 (determined by RNA-seq in the BRCA-TCGA cohort). Dunn's multiple comparisons and Kruskal–Wallis tests (****p < 0.0001). (F) Kaplan–Meier curve depicting the influence of SOX9-AS1, MIA and MIA-RAB4B mean expression on Overall Survival probability for basal-like patients (the analysis was conducted on 309 patients).
We then reasoned that SOX9-AS1 may exert a survival effect on patients through its related genes, i.e., the highly correlated genes MIA and MIA-RAB4B. Thus, we performed a Kaplan–Meier analysis in order to assess this hypothesis. As shown in Fig. 4F, high mean expression of SOX9-AS1, MIA and MIA-RAB4B is significantly associated with an increased OS rate in basal-like BRCA patients (HR = 0.41; CI [0.22–0.79], p value = 0.006). We then performed the same analysis including the moderately correlated genes ST3GAL6 and CDH19, which did not yield statistically significant results. Altogether, these data suggest that SOX9-AS1, MIA and MIA-RAB4B expression are not just statistically correlated, but indeed biologically associated or even co-regulated in this model.
SOX9-AS1 location is enriched in the cytoplasm of TNBC and Basal-like breast cancer cell lines
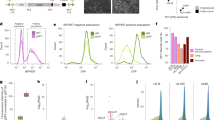
To further understand the functional role of SOX9-AS1 in TNBC cell lines, we examined its expression by RT-qPCR in various cell models of BRCA. Results showed that SOX9-AS1 was enriched in cell lines classified as TNBC or basal-like in comparison to luminal and HER2 subtypes (Fig. 5A,B). Through subcellular fractionation, we show that SOX9-AS1 is located mainly in the cytoplasm of MDA-MB-468 and HCC1187 cell lines, which correspond to the BL1 and IM subtypes of TNBC, respectively (Fig. 5C,D). We then predicted the subcellular localization of SOX9-AS1 in five cell lines through the LncAtlas website (http://lncatlas.crg.eu/). SOX9-AS1 was found to be distributed in the cytoplasm of both H1.Hesc (H1 human embryonic stem cell line) and NCI.H460 (non-small-cell lung cancer cell line), and in the nucleus of GM12878 (B-lymphoblastoid cell line), HepG2 (human hepatocellular carcinoma cell line), and SK.N.SH cells (human neuroblastoma cell line), but was not described in the MCF-7 luminal BRCA cell line (Fig. 5E), and LncLocator predictor showed that SOX9-AS1 was localized to the cytoplasm (Fig. 5F). These results show that SOX9-AS1 could have a biological role in cytoplasm.
Expression and subcellular localization of SOX9-AS1 in TNBC and basal-like cell lines. (A) Broad Institute Cancer Cell Line expression data of SOX9-AS1 across 46 distinct BRCA cell lines examined by RNA-seq. (B) SOX9-AS1 expression between BRCA cell lines examined by RT-qPCR. Mean ± SD. One-way ANOVA comparison test, ****p < 0.0001. (C) Subcellular localization of SOX9-AS1 in MDA-MB-468, and (D) HCC1187 cells was determined by by RT-qPCR after nuclear/cytoplasmic fractionation using MALAT1 as control for nucleus and GAPDH as control for cytoplasm. Analyses were performed three times in triplicate in vitro. (E) Subcellular localization of SOX9-AS1 in various cell lines predicted using the lncATLAS database. (F) Subcellular localizations of SOX9-AS1 determined by lncLocator predictor which includes RNA localization data from experimental evidence. ND: not detected.
Knockdown of SOX9-AS1 potentially regulates genes involved in metabolic and immunological processes and pathways
In order to elucidate the functional role of SOX9-AS1 in basal-like and TNBC biology, we performed gene silencing experiments using two shRNAs (see “Materials and methods”). SOX9-AS1 expression in SOX9-AS1-silenced cells was significantly reduced in MDA-MB-468 and HCC1187 (Fig. 6A,B). Moreover, after silencing, we assessed gene expression levels on a global scale using RNA-seq and evaluated genes that significantly changed their expression (Log2FC < − 1.5 to > 1.5, adjusted p-value < 0.05). In MDA-MB-468 cells, we found 740 DEGs. Of those, 44.7% (331 mRNAs) were upregulated, while 55.3% (409 mRNAs) were found to be downregulated when comparing the sh-control and the silenced SOX9-AS1-1 group (Fig. 6C). In the HCC1187 cell line, 100 coding genes were deregulated, of which 85 were upregulated and 15 were downregulated compared to control (Fig. 6D). 10 DEGs were shared between MDA-MB-468 and HCC1187 cells (Supplementary Table 9). Figure 6E,F show the top 5 DEGs in MDA-MB-468 and HCC1187, respectively. Additionally, we identified a total of 351 differentially expressed lncRNAs in MDA-MB-468 cells, of which 120 (34.2%) were upregulated and 231 (65.8%) downregulated when SOX9-AS1 expression was inhibited (Supplementary Fig. 3A,B). In the HCC1187 cell line, we identified 56 differentially expressed lncRNAs, 32 were upregulated and 24 were downregulated (Supplementary Fig. 3C,D; Supplementary Table 10).
Knockdown of SOX9-AS1 and DEGs involved in metabolic and immunological processes and pathways in TNBC cells in vitro. Analysis of SOX9-AS1 levels by RT-qPCR assays in (A) MDA-MB-468, and (B) HCC1187 cells transfected with two SOX9-AS1-targeting shRNAs and control shRNA (sh-Control). Student T test. *p < 0.05, ****p < 0.0001. The Volcano plots represent the distribution of DEGs. Blue genes are downregulated, red genes are upregulated, and gray genes are not differentially expressed in (C) MDA-MB-468, and (D) HCC1187 cells. Up- and downregulated genes were defined as Log2 Fold Change ≤ − 1.5 and > 1.5 with adjusted p values < 0.05. Heatmap of the DGEs contributing to the top 5 in (E) MDA-MB-468 and (F) HCC1187 cells. Green: low expression, red: high expression according to Log2 fold change. Bubble map for the top 20 enriched GO classifications in (G) MDA-MB-468, and (H) HCC1187 cells. The most enriched KEGG pathways of DEGs in (I) MDA-MB-468, and (J) HCC1187 cell models. The size of the q-value is represented by the point’s color.
Interestingly, according to GO analysis, genes enriched participate in lipid, steroid, alcohol and cholesterol metabolism, biosynthetic processes, and a myeloid leukocyte migration process in MDA-MB-468 cells (Fig. 6G). Leukocyte, granulocyte, neutrophil chemotaxis and migration processes were mainly enriched in HCC1187 under the effect of SOX9-AS1 silencing (Fig. 6H) (Supplementary Table 11). KEGG analysis ranked by enrichment score and p value < 0.05 in the MDA-MB-468 cell line showed pathways related to cancer and lipid metabolism, such as PI3K-Akt signaling and PPAR signaling, as well as steroid and fatty acid production (Fig. 6I). These findings are consistent with the biological processes and pathways identified in the DGE analysis in TNBC and basal-like patient samples (Fig. 2). Only two pathways were enriched in the HCC1187 cell line (Fig. 6J) (Supplementary Table 12). These results indicate a major biological role for SOX9-AS1 as a potential regulator of genes that participate in cancer metabolism-related processes and inflammatory pathways in TNBC and basal-like subtypes.
SOX9-AS1 regulates genes involved in metabolic processes and pathways in TNBC and Basal-like cells
We further performed GSEA analysis using the Hallmarks gene set. Considering nominal p value < 0.05 and FDR q value < 0.25. The most representative gene sets positively correlated with the silencing of SOX9-AS1 in the MDA-MB-468 cell line were: mTORC1 signaling, androgen response, cholesterol homeostasis, fatty acid metabolism, peroxisome signaling, and adipogenesis, while the most representative negatively correlated gene sets were Hedgehog signaling, TGF-beta signaling, WNT/beta catenin signaling (Fig. 7A), and several other cellular processes (Supplementary Table 13). GSEA analysis in the HCC1187 cell line showed that the silencing of SOX9-AS1 was only negatively correlated with hallmark gene sets, such as TGF-β signaling, pancreatic beta cells, Hedgehog signaling, estrogen response early, heme metabolism, and glycolysis (Fig. 7B). Furthermore, diverse mechanisms were fairly similar to those discovered in the MDA-MB-468 cell line (Supplementary Table 13). These results suggest that the biological role of SOX9-AS1 is directly related to metabolic processes and pathways in both TNBC and basal-like subtypes.
SOX9-AS1 potentially regulates a variety of signalling pathways and metabolic cellular processes in TNBC cells. GSEA for the hallmark gene sets after SOX9-AS1 silencing showed significant positive and negative enrichment (FDR q-value ≤ 0.25) in (A) MDA-MB-468, and (B) HCC1187. The NOM p-val and FDR q-value are displayed. (C) Seven genes involved in cellular metabolism were validated. (D) Distribution of candidate genes across PAM50-subtypes from BRCA-TCGA cohort. Data represent median expression value. Dunn's multiple comparisons and Kruskal–Wallis tests (****p < 0.0001). Validation of gene expression by RT-qPCR in (E) MDA-MB-468, and (F) HCC1187 cells transfected with two SOX9-AS1-targeting shRNA and sh-Control. Bar graph shows the quantified results with means ± SD of three biological replicates. Tukey’s test (*p < 0.05 and ****p < 0.0001 vs. sh-Control). ND: not detected.
We next validated some DEGs poorly studied in TNBC and basal-like cells, but involved in cellular metabolism (Fig. 7C). First, we used the BRCA-TCGA cohort to determined the expression of these genes in different BRCA subtypes (Fig. 7D) and observed that the expression level of LIPE, REEP6, FBP1, SCD1, UGT2B11 and APOC1, were significantly underexpressed (p value = 0.0001) while GABRE expression was significantly increased in basal-like tumors (p value = 0001) compared to receptor-positive tissues (Fig. 7D). Then, we used RT-qPCR in our SOX9-AS1-silenced cell lines and found an opposite effect on the relative quantification of these seven genes when compared to the sh-Control in both MDA-MB-468 and HCC1187 cell lines (Fig. 7E,F). Thus, we confirmed that these genes were potentially regulated by SOX9-AS1 in TNBC and basal-like subtypes.
Knockdown of SOX9-AS1 stimulates fatty acid synthesis and intake in TNBC and Basal-like cells
Pathway analysis showed that many DEGs were significantly enriched in lipid metabolism, both in samples and cell lines (Figs. 3, 6, 7). Therefore, we explored whether SOX9-AS1 participates in triglyceride metabolism (lipid compounds of fatty acids and glycerol) in TNBC cells. The levels of triglycerides in both MDA-MB-468 and HCC1187 cells after SOX9-AS1 knockdown were significantly increased compared with those in control cells (p value < 0.01) (Fig. 8A,B). Furthermore, we used lipoprotein lipase to convert triglycerides into glycerol, since triglycerides are highly insoluble and difficult to titrate in aqueous solution28. We found that there was a greater amount of glycerol in SOX9-AS1-silenced TNBC cells versus control cells (Fig. 8A,B). Additionally, we evaluated the capacity of SOX9-AS1 deficient-cells to accumulate intracellular lipids using the Oil Red O stain. However, the neutral lipids were not observed in SOX9-AS1-knockdown cells versus control cells (data not shown). These results showed that the loss of SOX9-AS1 increases intracellular triglyceride levels in TNBC cells.
SOX9-AS1-knockdown increases intracellular triglyceride levels in TNBC cells. Intracellular triglycerides were quantified as glycerol (in relative light units-RLU) following treatment with or without Lipase in (A) MDA-MB-468 and (B) HCC1187 cells transfected with sh-control, sh-SOX9-AS1-1 or sh-SOX9-AS1-2. Values represent mean ± SD for three biological replicates. One-way ANOVA analysis for each cell line. *p < 0.05, **p < 0.001, ***p < 000.1.
Knockdown of SOX9-AS1 increases migration, and invasion of MDA-MB-468 and HCC1187 cells
To further address and identify the molecular events induced by SOX9-AS1 in TNCB and basal-like subtype, we carried out a GSEA analysis that showed that SOX9-AS1 deregulated genes enriched in the process of epithelial-mesenchymal transition (EMT) in both MDA-MB-468 (NOM p value 0.04, FDR q value 0.14) and HCC1187 cells (NOM p value 0.02, FDR q value 0.08) (Fig. 9A,B). Given these observations, we performed migration and invasion assays. SOX9-AS1 silencing significantly increases the migration and invasion of MDA-MB-468 cells (p value ≤ 0.05 and < 0.01 for sh-SOX9-AS1-1 and sh-SOX9-AS1-2, respectively) (Fig. 9C) and HCC1187 cells (p value ≤ 0.05 for both sh-SOX9-AS1) (Fig. 9D) in comparison to control cells. Finally, these results confirm that loss of SOX9-AS1 expression results in a more aggressive phenotype for TNBC cells.
Knockdown of SOX9-AS1 increases cell migration and invasion in TNBC cells. The DEGs obtained by RNA-seq data from the SOX9-AS1-knockdown in (A) MDA-MB-468 and (B) HCC1187 cells was analyzed with GSEA enrichment plots for the HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION gene set. (C) Representative images of transwell migration and invasion assays after SOX9-AS1-knockdown in MDA-MB-468 cells and (D) HCC1187 cells and corresponding control cells. Bar graph shows the quantification of the results with means ± standard deviation. *p < 0.05 and **p < 0.01 vs. sh-Control.
Discussion
LncRNAs participate in the regulation of the Hallmarks of cancer, favoring or inhibiting oncogenesis29,30. In this context, we evaluated the clinical implication and the biological role of SOX9-AS1 in basal-like and TNBC subtypes using the BRCA-TCGA cohort and a panel of BRCA cell lines. First, we evidenced the expression levels of SOX9-AS1 across different cancer types compared to normal tissues. We found that there were higher expression levels of SOX9-AS1 in normal tissues versus cancer tissues, mainly those with secretory function (i.e., BRCA, TGCT, SKCM, PAAD). Additionally, we showed that some tumor types, like those of the nervous system (i.e., LGG, GMB), connective tissues (CHOL) and other excretory type cancer tissues (i.e., KIRP, KIRC) have higher expression levels of SOX9-AS1 compared to normal tissue. Additionally, tumors related to white blood cells presented null or low levels of expression of SOX9-AS1 (Fig. 1A). Due to these findings, we suggest that SOX9-AS1 is ubiquitously expressed but may have different tissue-specific functions. This has been shown to be the case for some lncRNAs that show different expression patterns between different types of normal and tumor tissues and therefore fulfill different functions31. An example is the FAM83H-AS1 lncRNA which is significantly overexpressed in 15/33 different tumor types, and is downregulated in LAML, suggesting that different mechanisms might exist for non-solid tumors32. LINC00460 was found to be overexpressed in five different types of cancer versus normal tissue (BRCA, COAD, HNSC, PAAD, and READ), while in two central nervous system cancers, LINC00460 showed low expression, compared to normal tissue counterparts (GBM and LGG), and a lack of expression in LAML33. This indicates that the biological role of lncRNAs is precise and closely related to spatial expression patterns between tissue types, and their dysfunction in cancer often influences disease development and progression34,35. Specifically, using BRCA-TCGA data, we revealed that the expression levels were significantly higher in TNBC and basal-like samples. These data strengthen our previous results using microarrays that show high expression of SOX9-AS1 in TNBC and basal-like samples18,19. Based on these results, we propose that SOX9-AS1 is a biomarker for these BRCA subtypes.
In addition, we found that SOX9-AS1 expression varies with age, being higher in patients under 50 years of age and lower expression in the group of women over 50 years (Supplementary Table 1). This may be due to the epidemiological finding that TNBC primarily affects young, premenopausal women between the ages of 40 and 46, who make up about 15–20% of all breast cancer patients36,37. Furthermore, we also observed a significant positive association of SOX9-AS1 expression with the basal-like subtype in the PAM50 molecular classification, being much higher that in the receptor-positive BRCA subtypes.
We believe that the strengths of our study are focused on the similarity of the processes and pathways identified in both samples and cell lines using the results from the DGE analysis of the TNBC and basal-like subtypes, which show genes enriched in hallmarks related to lipid metabolic reprogramming. Interestingly, we observed that SOX9-AS1 potentially regulates genes which are similar to metabolic reprogramming signatures described in TNBC by Gong et al.38 (see Supplementary Fig. 4; Supplementary Table 14). In particular, genes potentially regulated by SOX9-AS1 are enriched in the metabolic-pathway-based subtype 1 cluster (MSP1) which, in turn, is associated to the lipogenic subtype with upregulated lipid metabolism and with better clinical prognosis38. Collectively, SOX9-AS1-mediated modulation of these metabolic reprogramming-related genes can conceivably explain the effect of this lncRNA on the increased survival of TNBC patients (Fig. 2), since we observe that high SOX9-AS1 expression in patients displayed an increased OS, DSS, and DFS in TNBC and basal-like patients. In this context, we observe that the expression of SOX9-AS1 has a positive correlation with MIA and MIA-RAB4B genes in basal-like samples and that the increased expression of all three genes together show a better prognosis for patients with this molecular subtype (Fig. 4). We cannot discard, however, that some of the SOX9-AS1 mediated regulation of immune-related pathways identified in this study may play a role in TNBC and basal-like biology and clinical features. Further functional studies must address this issue.
Lipid metabolic reprogramming is an emerging hallmark of malignancy39, and any abnormal changes in fatty acid metabolism seem to provide advantages for the growth of cancer cells40. In this study, we found that SOX9-AS1 negatively regulates lipid levels in TNBC cells, since silencing this lncRNA increases triglyceride accumulation. This effect is probably regulated by thousands of genes modulated by this lncRNA. In this sense, we validated seven lipid metabolic-related genes (LIPE, REEP6, GABRE, FBP1, SCD1, UGT2B11 and APOC1), which are poorly studied in basal-like and TNBC. Interestingly, these genes, as well as many other DEGs found in our study, participate in lipid biosynthesis and fatty acid degradation in cancer (SCD1, SCD5, FASN, ACSL1, ACSL4, ACSS2, LPIN1), lipolysis (LIPE, PLIN5, PNPLA3), transport and uptake (CD36, RBP3) and lipid storage/lipid droplets (ACAT2) (see Supplementary Table 9)39,41,42,43. Additionally, we demonstrate that the activation of these genes by SOX9-AS1 silencing, increases intracellular triglyceride levels in MDA-MB-468 and HCC1187 cell lines compared to control cells (Fig. 8). However, we did not observe intracellular lipid droplets, presumably because of increased lipid utilization for cell growth, proliferation, migration, invasion and cell survival of cancer cells44,45. This could also explain the increased EMT of the TNBC cells.
A previous study reported that SOX9-AS1 expression is upregulated in hepatocellular carcinoma (HCC), as is SOX9, which regulates SOX9-AS1 expression. In turn, SOX9 activates the Wnt/beta-catenin pathway to promote EMT. Additionally, SOX9-AS1 maintains increased SOX9 expression by inhibiting miR-5590-3p, thereby promoting HCC progression and is therefore associated to poor outcome in HCC patients21. Another study showed that SOX9-AS1 was overexpressed in tissues and cell lines of intrahepatic cholangiocarcinoma (ICC) and its upregulation was related to shorter OS and recurrence-free survival. Additionally, SOX9-AS1 promotes growth, migration and invasion of ICC cells23. In contrast, our study shows that overexpression of SOX9-AS1 predicts an improved clinical course in TNBC and basal-like patients based on TCGA and GEO databases. Furthermore, using the DGE data from TCGA, we identified that SOX9-AS1 expression positively correlates with genes involved in the metabolic reprogramming of oxidative phosphorylation (OXPHOS), reactive oxygen species (ROS), and adipogenesis pathways in TNBC samples (Fig. 2), which are an important part of the metabolic phenotype of TNBC tumors10,46. Our experimental results show that SOX9-AS1 is located mainly in the cytoplasm where it can fulfill its functional role. Based on our RNA-seq data of SOX9-AS1-knockdown TNBC cells, this lncRNA positively correlates with a plethora of genes involved in metabolic reprogramming of glycolysis, fatty acid metabolism, peroxisome pathway, and adipogenesis (Fig. 6), a major metabolic program that could provide a survival advantage to TNBC cells10. Moreover, these processes are triggered by mTOR signaling and PI3K-Akt-mTOR signaling (Fig. 6), which are responsible for driving tumorigenesis and modulating metabolism in cancer cells47. Additionally, silenced SOX9-AS1 negatively correlates with genes involved in Hedgehog, TGF-β, and WNT signaling pathways (Fig. 7). These pathways trigger intracellular kinase cascades to induce transcription factors that activate the expression of EMT-associated genes48. In this sense, the in vitro results of the present study also demonstrate that SOX9-AS1 can negatively regulate EMT of MDA-MB-468 and HCC1187 cells, since cell migration and invasion were increased in SOX9-AS1-knockdown TNBC cells. Studies reveal that cancer-associated EMT requires complex and comprehensive metabolic reprogramming, including gene expression and epigenetic changes49. Therefore, transcription program switching in EMT is induced by different signaling pathways, such as transforming growth factor b (TGF-b) and bone morphogenetic protein (BMP), Wnt-β-catenin, Notch, Hedgehog, and receptor tyrosine kinases48. Although the number of studies involving SOX9-AS1 is limited and has more frequently been described as a predictive factor for poor prognosis, as shown for HCC, and ICC cells, we suggest that high expression of SOX9-AS1 might serve as a biomarker of good prognosis in TNCB tissue specific biological mechanisms in the cytoplasm of TNBC cells. Taken together, our results suggest that the loss of expression of SOX9-AS1 in TNBC cells promotes a more aggressive phenotype.
Conclusions
Our study demonstrates that SOX9-AS1 is a master regulator of a plethora of genes involved in metabolic reprogramming, cell migration and invasion in patient samples and cell models, and its biological function is influenced by the dynamic properties of EMT through different signaling pathways. Our results further our understanding of the important role of SOX9-AS1 in the molecular cell biology of TNBC and basal-like subtypes. In this context, we suggest that SOX9-AS1 could act as an onco-suppressor, since its elevated expression levels can result in a better clinical outcome for basal-like and TNBC.
Materials and methods
Analysis of SOX9-AS1 in the GEPIA2 (Gene Expression Profiling Interactive Analysis) database
SOX9-AS1 expression levels were screened in 31 TCGA adjacent tissue and tumor datasets and GTEx normal tissues (unrelated donors) using the GEPIA2 database (http://gepia2.cancer-pku.cn/#index)50. For our research, we screened 291 normal tissues, of which 179 were healthy normal from GTEx data and 112 adjacent tumor tissues from the TCGA-BRCA cohort, and 810 BRCA samples. The 31 tumors included in the analysis are enlisted in Supplementary Materials. Bar plots were generated to visualize the median expression levels transformed to Log2(TPM+1).
BRCA-TCGA dataset mining and stratification according to SOX9-AS1 expression
Clinical information from TNBC and its molecular classification by PAM50 were obtained from the TCGA database through cBioPortal (https://www.cbioportal.org/), and raw RNA-seq expression data was obtained from the GDC Data Portal (https://portal.gdc.cancer.gov/projects). Female patients who received neoadjuvant treatment or that lacked overall survival (OS), disease-specific survival (DSS), disease-free survival (DFS) and progression-free survival (PFS) data were excluded from this analysis. According to these criteria, 112 TNBC and 167 basal-like samples were included in this study. The expression data was normalized to Log2(TPM+1) and was used to stratify patients into groups with low and high expression of SOX9-AS1 (NCBI Reference Sequence: NR_103738.1), (Ensemble ID: ENSG00000234899.11), based on the lower (25%) and upper quartiles (75%), respectively.
Survival analyses
Kaplan–Meier survival analyses for OS, DSS, DFS and PFS were performed with the Log-rank test for BRCA-TCGA cohorts. p-values < 0.05 were considered statistically significant. Groups of patients with low and high expression of SOX9-AS1 were stratified based on the lower (25%) and upper quartiles (75%), respectively. The analyses were performed with survival (v. 3.1-8) and survminer (v. 0.4.8) R packages. To analyze OS and RFS of GEO-derived cohorts we used the Kaplan Meier Plotter online tool (https://kmplot.com/analysis/), using the automatic cutoff calculation and JetSet probes only51. The Breast RNA-seq Kaplan Meier Plotter tool was used for the three-gene (SOX9-AS1, MIA and MIA-RAB4B) combined survival analysis in basal-like patients, using the mean expression of the genes, and the auto-detection cutoff parameter.
SOX9-AS1-genes correlation analysis
To determine relationships between the SOX9-AS1 expression and gene expression, we performed Spearman correlation analysis. p-value < 0.05 was considered statistically significant. This analysis was performed in the R platform (v. 4.2.2).
Cell culture
The BRCA cell lines (MCF7, ZR-75-1, MDA-MB-361, SKBR3, MDA-MB-468, HCC1187, HS578T, MDA-MB-231, MDA-MB-453, BT-20) and non-tumorigenic epithelial cell line MCF10A, were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). Cell lines were routinely maintained in RPMI-1640 or DMEM medium (ATCC), supplemented with 10% fetal bovine serum (FBS) (ATCC), in a humidified incubator at 37 °C with 5% CO2 using the standard media and conditions established by the ATCC, except for MDA-MB-468 cell line, which was maintained in DMEM/High Glucose (Gibco, Grand Island, NY) with 10% FBS (ATCC).
RNA extraction and reverse transcription quantitative PCR (RT-qPCR) assay
Total RNA was extracted from SOX9-AS1-deficient BRCA cell lines using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Briefly, cDNA was generated with 100 ng of total RNA in a final reaction volume of 20 µl using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems™, Foster City, California, USA) following the manufacturer’s instructions. The expression level of SOX9-AS1 was detected using the TaqMan Noncoding RNA assays (Hs01107818_m1) and the following genes were validated using Taqman assays (Applied Biosystems): LIPE (Hs00943410_m1), REEP6 (Hs00922109_m1), GABRE (Hs00608332_m1), FBP1 (Hs00983323_m1), SCD (Hs01682761_m1), UGT2B11 (Hs01894900_gH), APCO1 (Hs001555790_m1) using qPCR analysis (Applied Biosystems, Foster City, USA) in a QuantStudio™ 5 Real-Time PCR System (Life Technologies). The qPCR reaction contained 1 µl of cDNA, 5 µl 2× TaqMan Universal Master Mix (Applied Biosystems, ThermoFisher™ Scientific, Waltham, Massachusetts, USA), 0.5 µl TaqMan probe and 3.5 µl of nuclease-free water. The thermocycling conditions were as follows: initial denaturation at 95 °C for 5 min, 40 denaturation cycles at 95 °C for 30 s, hybridization at 50 °C for 30 s, and extension at 72 °C for 30 s. The mean cycle threshold (Ct) value for GAPDH (Hs99999905) housekeeping gene was used to normalize the raw Ct data and calculate 2−ΔCt. The 2−ΔΔCt method was used to estimate the relative expression level of genes under treatment conditions52.
Subcellular fractionation of TNBC and basal-like cell lines
Cellular fractionation was performed using PARIS™ Kit (Invitrogen Inc., Carlsbad, CA, USA), according to the manufacturer’s instructions. RNA was isolated from the nucleus and cytoplasm compartments in MDA-MB-468 and HCC1187 cells. Subsequently, the SOX9-AS1 intracellular localization analysis, by RT-qPCR assay, was performed as described above. The data was normalized using the formula % of input = 100 × [2^(Ct total RNA − Ct RNA fraction). MALAT1 and GAPDH expression was used as nuclear and cytoplasmic controls, respectively.
Knockdown of lncRNAS SOX9-AS1 and cell transfection of shRNAs
The online tool BLOCK-iT™ RNAi Designer (Thermo Fisher Scientific) was used for the design of the short hairpin RNAs (shRNAs) targeting SOX9-AS1 and the LacZ control (sh-Control) was supplied in the kit. Post-transcriptional gene silencing of the canonical transcript (NR_103738.1) and one transcript variant (NR_103737.1) was performed with two shRNAs (sh-SOX9-AS1-1 and sh-SOX9-AS1-2). The sequences are shown in Supplementary Table 15. Briefly, the construction with two dsOligos capable of generating shRNAs in cells was carried out using the BLOCK-iT U6 RNAi Entry Vector Kit (Invitrogen, Carlsbad, CA, EE. UU.), following the manufacturer's instructions. Once the plasmids were generated, the transformation was carried out in the competent cells Escherichia coli One Shot TOP10, which were subsequently seeded in LB medium plates with 50 mg/ml kanamycin. After transformation, plasmids were isolated using the PureLink Hipure Plasmid Maxiprep kit (Invitrogen), according to the manufacturer's guidelines. To confirm the presence and correct orientations of dsOligo inserts into the plasmids, Sanger sequencing was performed. Plasmid transfection for the specific silencing of SOX9-AS1 was carried out in the HCC1187 and MDA-MB-468 cell lines using Xfect Transfection Reagent (Clontech, California, EE. UU.). Five µg of plasmid were used, and the silencing efficiency was determined at 48 h post-transfection by measuring SOX9-AS1 expression levels through RT-qPCR.
RNA-sequencing libraries preparation and data processing
We performed RNA-seq on 12 samples, three independent replicas for each silencing condition (sh-Control and sh-SOX9-AS1-1 for MDA-MB-468 and HCC1187 cells). Paired-end RNA-seq libraries with fragments of 76 bp were generated from total RNA with the TruSeq® Stranded mRNA Library Prep Kit (Illumina, CA, USA), following the manufacturer’s recommendations. Briefly, the protocol started with 1 μg of total RNA, which was purified and fragmented using poly-T oligo. Then, cDNA was synthesized, and adapter ligation was performed. The constructed libraries were amplified through PCR, and then were quantified with the Agilent 4200 TapeStation System using High Sensitivity D1000 ScreenTape (Agilent, CA, USA), according to the manufacturer's protocol. The sequencing of RNA-seq libraries was carried out on the NextSeq 500 platform (Illumina, USA) at an average depth of 23 million raw paired reads per sample. The general statistics are shown in Supplementary Table 15. Quality control of raw fastq files was determined using FastQC (v. 0.11.9) and MultiQC (v. 1.11). Adapter sequences, poor quality reads with Q values ≤ 22 and reads with fragment sizes ≤ 70 bp were filtered through Trimmomatic (v. 0.39). The alignment of raw reads to GRCh38 (Homo sapiens) reference genome and the generation of bam files were carried out with Hisat2 (v. 2.1.0) and samtools, respectively. Abundance estimation and mapped reads counting was performed with featureCounts (v. 2.0.0). All the analyses were carried out in the Unix-Bash environment of Linux platform.
Differential gene expression analysis and functional annotation
DGE analysis was determined using DESeq2 (v.1.26.0). We used two groups of TNBC and basal-like TCGA data (SOX9-AS1 low vs high expression) and cell lines (sh-LacZ vs sh-SOX9-AS1-1). Before DGE analysis with cell line data, we excluded a technical replica due to an unsuccessful silencing in the MDA-MB-468 cell line. Then, RNA-seq expression data with raw counts less than 10 were filtered. Up- and downregulated genes were defined as Log2FoldChange (LogFC) < − 1.5 and > 1.5 with p adjusted values < 0.05. Volcano plots and heatmaps were generated with EnhancedVolcano (v. 1.4.0) and Pheatmap (v. 1.0.12). Subsequently, the lists of differentially expressed genes (DEGs) were used to perform GO over-representation analysis for biological processes and KEGG pathway analysis using clusterProfiler (v. 3.14.3). For these analyses, we used minGSSize = 10 and maxGSSize = 500 as default parameters. The results were represented as dot plots and adjusted p values < 0.05 were considered statistically significant. All analyses were executed in R (version 3.6.2).
Gene set enrichment analysis (GSEA)
Using the normalized RNA-seq expression data from groups of cell lines (sh-LacZ vs sh-SOX9-AS1-1) and BRCA-TCGA data (low vs high expression of SOX9-AS1) we performed GSEA, using the Hallmarks_all_v7.4 gene set (with min size = 10 and max size = 500 as default parameters). These analyses were performed with 1000 permutations. Gene sets with nominal p values < 0.05 and FDR < 0.25 were considered statistically significant53. The analyses were performed through GSEA software (version 4.1.0).
Triglyceride measurement
Intracellular triglyceride levels in HCC1187 and MDA-MB-468 cell lysates were detected by luminescence using Triglyceride-Glo Assay (Cat. J3160, Promega, USA), according to the manufacturer’s instructions. Briefly, 10,000 transfected cells were cultured 24 h before the assay on a 96-well plate. After 24 h (48 h post-transfection), cells were washed twice with PBS. Then, 25 µl of glycerol lysis solution with and without lipase was added and cells were incubated at 37 °C for 30 min. Then, 25 µl of glycerol detection reagent containing reductase substrate and kinetic enhancer reagent was added to the lysates. After 1 h, the luminescence value was detected using a plate luminometer Synergy/HTX (BioTek instrument, Inc. Winooski, VT). The relative light unit (RLU) values were expressed as mean ± standard deviation. Experiments were performed in triplicate and repeated three times.
Oil red O staining
For lipid staining in HCC1187 and MDA-MB-468 cells, 48 after transfection, cell monolayers were washed twice with PBS and fixed with 10% formalin (HT501128-4L Sigma-Aldrich, Sweden) for 1 h at room temperature. Cells were then stained with Oil Red O and incubated at room temperature for 15 min. Oil Red O solution was then removed and cells were washed four times with ddH2O. The cells were carefully visualized under the microscope.
Cell migration and invasion assays
Cell migration and invasion assays were performed using a 24‐well transwell chamber with a pore size of 8 μm (Corning, USA) in the absence and presence of Matrigel (Corning) for migration and invasion assays, respectively, following to the manufacturer’s protocol. MDA-MB-468 and HCC1187 cells (5 × 104 cells per well) were seeded in the upper of chamber. The medium containing 10% FBS was deposited in the lower chamber as a chemoattractant. 48 h post-transfection, the cells were fixed in 3.7% paraformaldehyde for 20 min and then stained with 0.5% crystal violet for 10 min. The cells in the upper chamber were removed with a cotton swab, and the images were captured under the microscope. The number of cells was counted by ImageJ software and the mean cell count of three independent membranes was defined as the migration or invasion index. Experiments were repeated three times.
Statistical analysis
Experimental results were reported as mean ± standard deviation (SD) of three independent experiments in triplicate. A two-tailed paired Student's t-test was used to analyze the distribution between independent groups and 2 × 2 Chi-squared tests were used to identify associations between clinicopathological variables and SOX9-AS1 expression levels, both statistical analyzes were performed using GraphPad Prism 8 software (GraphPad Software Inc., La Jolla, CA, USA). Multiple level Chi-squared tests (2xn), Bonferroni multiple correction and post-hoc analyses were performed using custom made functions (https://github.com/neuhofmo/chisq_test_wrapper) based on SciPy, Statsmodels and Itertools Python modules, using the pandas library54. The complete code is available at: https://github.com/mriosromero/Chi-square-tests-Cisneros-et-al-2023. p values < 0.05 were considered statistically significant.
Data availability
The data presented in this study are not publicly available due to privacy. However, the data can be requested from the corresponding authors.. To review BioProject-NCBI accession PRJNA964471. Go to (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA964471). Link from BioSample-NCBI (https://www.ncbi.nlm.nih.gov/biosample?Db=biosample&DbFrom=bioproject&Cmd=Link&LinkName=bioproject_biosample&LinkReadableName=BioSample&ordinalpos=1&IdsFromResult=964471).
References
Garrido-Castro, A. C., Lin, N. U. & Polyak, K. Insights into molecular classifications of triple-negative breast cancer: Improving patient selection for treatment. Cancer Discov. 9, 176–198. https://doi.org/10.1158/2159-8290.CD-18-1177 (2019).
Perou, C. M. et al. Molecular portraits of human breast tumours. Nature 406, 747–752. https://doi.org/10.1038/35021093 (2000).
Sorlie, T. et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 98, 10869–10874. https://doi.org/10.1073/pnas.191367098 (2001).
Hu, Z. et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genom. 7, 96. https://doi.org/10.1186/1471-2164-7-96 (2006).
Sorlie, T. et al. Distinct molecular mechanisms underlying clinically relevant subtypes of breast cancer: Gene expression analyses across three different platforms. BMC Genom. 7, 127. https://doi.org/10.1186/1471-2164-7-127 (2006).
Prat, A. et al. Clinical implications of the intrinsic molecular subtypes of breast cancer. Breast 24(Suppl 2), S26-35. https://doi.org/10.1016/j.breast.2015.07.008 (2015).
Perou, C. M. Molecular stratification of triple-negative breast cancers. Oncologist 16(Suppl 1), 61–70. https://doi.org/10.1634/theoncologist.2011-S1-61 (2011).
Prat, A. et al. Molecular characterization of basal-like and non-basal-like triple-negative breast cancer. Oncologist 18, 123–133. https://doi.org/10.1634/theoncologist.2012-0397 (2013).
Lyons, T. G. Targeted therapies for triple-negative breast cancer. Curr. Treat. Options Oncol. 20, 82. https://doi.org/10.1007/s11864-019-0682-x (2019).
Sun, X. et al. Metabolic reprogramming in triple-negative breast cancer. Front. Oncol. 10, 428. https://doi.org/10.3389/fonc.2020.00428 (2020).
Lehmann, B. D. et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Investig. 121, 2750–2767. https://doi.org/10.1172/JCI45014 (2011).
Derrien, T. et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 22, 1775–1789. https://doi.org/10.1101/gr.132159.111 (2012).
Quinn, J. J. & Chang, H. Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 17, 47–62. https://doi.org/10.1038/nrg.2015.10 (2016).
Beylerli, O., Gareev, I., Sufianov, A., Ilyasova, T. & Guang, Y. Long noncoding RNAs as promising biomarkers in cancer. Noncoding RNA Res. 7, 66–70. https://doi.org/10.1016/j.ncrna.2022.02.004 (2022).
Ahadi, A. Functional roles of lncRNAs in the pathogenesis and progression of cancer. Genes Dis. 8, 424–437. https://doi.org/10.1016/j.gendis.2020.04.009 (2021).
Zhao, L. et al. NONCODEV6: An updated database dedicated to long non-coding RNA annotation in both animals and plants. Nucleic Acids Res. 49, D165–D171. https://doi.org/10.1093/nar/gkaa1046 (2021).
SanchezCalle, A., Kawamura, Y., Yamamoto, Y., Takeshita, F. & Ochiya, T. Emerging roles of long non-coding RNA in cancer. Cancer Sci. 109, 2093–2100. https://doi.org/10.1111/cas.13642 (2018).
Beltran-Anaya, F. O. et al. Expression of long non-coding RNA ENSG00000226738 (LncKLHDC7B) is enriched in the immunomodulatory triple-negative breast cancer subtype and its alteration promotes cell migration, invasion, and resistance to cell death. Mol. Oncol. 13, 909–927. https://doi.org/10.1002/1878-0261.12446 (2019).
Cedro-Tanda, A. et al. A lncRNA landscape in breast cancer reveals a potential role for AC0092831 in proliferation and apoptosis in HER2-enriched subtype. Sci. Rep. 10, 13146. https://doi.org/10.1038/s41598-020-69905-z (2020).
GeneCards. SOX9-AS1 Gene - SOX9 Antisense RNA 1. https://www.genecards.org/cgi-bin/carddisp.pl?gene=SOX9-AS1 (2023).
Zhang, W. et al. A SOX9-AS1/miR-5590-3p/SOX9 positive feedback loop drives tumor growth and metastasis in hepatocellular carcinoma through the Wnt/beta-catenin pathway. Mol. Oncol. 13, 2194–2210. https://doi.org/10.1002/1878-0261.12560 (2019).
Naorem, L. D., Prakash, V. S., Muthaiyan, M. & Venkatesan, A. Comprehensive analysis of dysregulated lncRNAs and their competing endogenous RNA network in triple-negative breast cancer. Int. J. Biol. Macromol. 145, 429–436. https://doi.org/10.1016/j.ijbiomac.2019.12.196 (2020).
Wu, L. et al. Impact of lncRNA SOX9-AS1 overexpression on the prognosis and progression of intrahepatic cholangiocarcinoma. Clin. Res. Hepatol. Gastroenterol. 46, 101999. https://doi.org/10.1016/j.clinre.2022.101999 (2022).
Gyorffy, B. et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 123, 725–731. https://doi.org/10.1007/s10549-009-0674-9 (2010).
Gyorffy, B. Survival analysis across the entire transcriptome identifies biomarkers with the highest prognostic power in breast cancer. Comput. Struct. Biotechnol. J. 19, 4101–4109. https://doi.org/10.1016/j.csbj.2021.07.014 (2021).
Liberzon, A. et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425. https://doi.org/10.1016/j.cels.2015.12.004 (2015).
Xiao, Y. et al. Predicting the functions of long noncoding RNAs using RNA-seq based on Bayesian network. Biomed. Res. Int. 2015, 839590. https://doi.org/10.1155/2015/839590 (2015).
Griffon, N. et al. Substrate specificity of lipoprotein lipase and endothelial lipase: Studies of lid chimeras. J. Lipid Res. 47, 1803–1811. https://doi.org/10.1194/jlr.M500552-JLR200 (2006).
Gutschner, T. & Diederichs, S. The hallmarks of cancer: A long non-coding RNA point of view. RNA Biol. 9, 703–719. https://doi.org/10.4161/rna.20481 (2012).
Di Gesualdo, F., Capaccioli, S. & Lulli, M. A pathophysiological view of the long non-coding RNA world. Oncotarget 5, 10976–10996. https://doi.org/10.18632/oncotarget.2770 (2014).
Jiang, C. et al. Identifying and functionally characterizing tissue-specific and ubiquitously expressed human lncRNAs. Oncotarget 7, 7120–7133. https://doi.org/10.18632/oncotarget.6859 (2016).
Rios-Romero, M. et al. FAM83H-AS1 is a potential modulator of cancer driver genes across different tumors and a prognostic marker for ER/PR + BRCA patients. Sci. Rep. 10, 14145. https://doi.org/10.1038/s41598-020-71062-2 (2020).
Cisneros-Villanueva, M. et al. LINC00460 is a dual biomarker that acts as a predictor for increased prognosis in basal-like breast cancer and potentially regulates immunogenic and differentiation-related genes. Front. Oncol. 11, 628027. https://doi.org/10.3389/fonc.2021.628027 (2021).
Bao, Z. et al. LncRNADisease 2.0: An updated database of long non-coding RNA-associated diseases. Nucleic Acids Res. 47, D1034–D1037. https://doi.org/10.1093/nar/gky905 (2019).
Lv, D. et al. LncSpA: LncRNA spatial atlas of expression across normal and cancer tissues. Cancer Res. 80, 2067–2071. https://doi.org/10.1158/0008-5472.CAN-19-2687 (2020).
Carey, L. A. et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. Jama 295, 2492–2502. https://doi.org/10.1001/jama.295.21.2492 (2006).
Morris, G. J. et al. Differences in breast carcinoma characteristics in newly diagnosed African-American and Caucasian patients: A single-institution compilation compared with the National Cancer Institute’s Surveillance, Epidemiology, and End Results database. Cancer 110, 876–884. https://doi.org/10.1002/cncr.22836 (2007).
Gong, Y. et al. Metabolic-pathway-based subtyping of triple-negative breast cancer reveals potential therapeutic targets. Cell Metab. 33, 51-64 e59. https://doi.org/10.1016/j.cmet.2020.10.012 (2021).
Cheng, C., Geng, F., Cheng, X. & Guo, D. Lipid metabolism reprogramming and its potential targets in cancer. Cancer Commun. (Lond.) 38, 27. https://doi.org/10.1186/s40880-018-0301-4 (2018).
Menendez, J. A. & Lupu, R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat. Rev. Cancer 7, 763–777. https://doi.org/10.1038/nrc2222 (2007).
Marino, N. et al. Upregulation of lipid metabolism genes in the breast prior to cancer diagnosis. NPJ Breast Cancer 6, 50. https://doi.org/10.1038/s41523-020-00191-8 (2020).
Yousuf, U., Sofi, S., Makhdoomi, A. & Mir, M. A. Identification and analysis of dysregulated fatty acid metabolism genes in breast cancer subtypes. Med. Oncol. 39, 256. https://doi.org/10.1007/s12032-022-01861-2 (2022).
Jin, H. R. et al. Lipid metabolic reprogramming in tumor microenvironment: From mechanisms to therapeutics. J. Hematol. Oncol. 16, 103. https://doi.org/10.1186/s13045-023-01498-2 (2023).
Gao, X. et al. A novel lipid prognostic signature of ADCY2, LIPE, and OLR1 in head and neck squamous cell carcinoma. Front. Oncol. 11, 735993. https://doi.org/10.3389/fonc.2021.735993 (2021).
Lung, J. et al. Lipid droplets in lung cancers are crucial for the cell growth and starvation survival. Int. J. Mol. Sci. https://doi.org/10.3390/ijms232012533 (2022).
Wang, Z., Jiang, Q. & Dong, C. Metabolic reprogramming in triple-negative breast cancer. Cancer Biol. Med. 17, 44–59. https://doi.org/10.20892/j.issn.2095-3941.2019.0210 (2020).
Lien, E. C., Lyssiotis, C. A. & Cantley, L. C. Metabolic reprogramming by the PI3K-Akt-mTOR pathway in cancer. Recent Results Cancer Res. 207, 39–72. https://doi.org/10.1007/978-3-319-42118-6_3 (2016).
Gonzalez, D. M. & Medici, D. Signaling mechanisms of the epithelial-mesenchymal transition. Sci. Signal 7, 8. https://doi.org/10.1126/scisignal.2005189 (2014).
Khaled, N. & Bidet, Y. New insights into the implication of epigenetic alterations in the EMT of triple negative breast cancer. Cancers (Basel) https://doi.org/10.3390/cancers11040559 (2019).
Tang, Z., Kang, B., Li, C., Chen, T. & Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 47, W556–W560. https://doi.org/10.1093/nar/gkz430 (2019).
Lanczky, A. & Gyorffy, B. Web-based survival analysis tool tailored for medical research (KMplot): Development and implementation. J. Med. Internet Res. 23, e27633. https://doi.org/10.2196/27633 (2021).
Schmittgen, T. D. & Livak, K. J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108. https://doi.org/10.1038/nprot.2008.73 (2008).
Subramanian, A. et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550. https://doi.org/10.1073/pnas.0506580102 (2005).
McKinney, W. Python for data analysis: Data wrangling with pandas, NumPy, and IPython. Second edition. edn.
Acknowledgements
We acknowledge INMEGEN for great support during the doctoral stay of M.C.-V. We also especially acknowledge those who participated in this study M.A.F.-M, M.R.-R, C.L.-C, S.J.-M, E.L, A.S.R-R, A.C.-T, D.H.-S, and A.H.-M.
Funding
This research was funded by Consejo Nacional de Ciencia y Tecnología (CONACYT), Mexico, Grant Ciencia Basica (number 258936) and Fronteras de la Ciencia (Grant number 1285). Mireya Cisneros-Villanueva received scholarships number 466600, from CONACYT, Mexico.
Author information
Authors and Affiliations
Contributions
M.C.-V., D.H.-S., and A.H.-M., and conceived and designed the study. M.C.-V. performed the methodology and investigation. M.A.F.-M., and M.R.-R. performed the formal analysis. M.C.-V., M.R.-R., E.L., and C.L.-C. performed writing—original draft. E.L., M.A.F.-M., M.R.-R., M.C.-V., S.J.-M., A.C.-T., and C.L.-C. performed writing – review and editing. M.A.F.-M., M.C.-V. M.R.-R. performed all the in-silico validation cohort-related analysis and the in silico prediction. M.A.F.-M., M.R.-R., and M.C.-V. performed statistical analysis, survival analysis, differential expression analysis, and risk analysis. A.S.R.-R., C.L.-C., and M.C.-V. performed additional experiments. A.H.-M. acquired the financing of the project. All authors interpreted and discussed the data. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Cisneros-Villanueva, M., Fonseca-Montaño, M.A., Ríos-Romero, M. et al. LncRNA SOX9-AS1 triggers a transcriptional program involved in lipid metabolic reprogramming, cell migration and invasion in triple-negative breast cancer. Sci Rep 14, 1483 (2024). https://doi.org/10.1038/s41598-024-51947-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-51947-2
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.