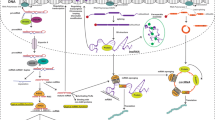
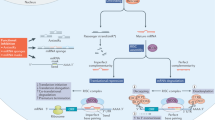
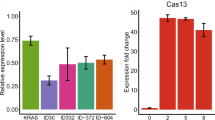
Abstract
RNA therapeutics (RNATx) aim to treat diseases, including cancer, by targeting or employing RNA molecules for therapeutic purposes. Amongst the most promising targets are long non-coding RNAs (lncRNAs), which regulate oncogenic molecular networks in a cell type-restricted manner. lncRNAs are distinct from protein-coding genes in important ways that increase their therapeutic potential yet also present hurdles to conventional clinical development. Advances in genome editing, oligonucleotide chemistry, multi-omics and RNA engineering are paving the way for efficient and cost-effective lncRNA-focused drug discovery pipelines. In this Review, we present the emerging field of lncRNA therapeutics for oncology, with emphasis on the unique strengths and challenges of lncRNAs within the broader RNATx framework. We outline the necessary steps for lncRNA therapeutics to deliver effective, durable, tolerable and personalized treatments for cancer.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Sung, H. et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021).
de Langen, A. J. et al. Sotorasib versus docetaxel for previously treated non-small-cell lung cancer with KRASG12C mutation: a randomised, open-label, phase 3 trial. Lancet 401, 733–746 (2023).
Amaral, P. P., Dinger, M. E., Mercer, T. R. & Mattick, J. S. The eukaryotic genome as an RNA machine. Science 319, 1787–1789 (2008).
Berber, B. et al. Gene editing and RNAi approaches for COVID-19 diagnostics and therapeutics. Gene Ther. 28, 290–305 (2021).
Kariko, K., Buckstein, M., Ni, H. & Weissman, D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23, 165–175 (2005).
Dykstra, P. B., Kaplan, M. & Smolke, C. D. Engineering synthetic RNA devices for cell control. Nat. Rev. Genet. 23, 215–228 (2022).
Sparmann, A. & Vogel, J. RNA-based medicine: from molecular mechanisms to therapy. EMBO J. 42, e114760 (2023).
Adams, D. et al. Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N. Engl. J. Med. 379, 11–21 (2018).
Chiriboga, C. A. et al. Results from a phase 1 study of nusinersen (ISIS-SMNRx) in children with spinal muscular atrophy. Neurology 86, 890–897 (2016).
Mattick, J. S. et al. Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 24, 430–447 (2023). This recent consensus statement from the lncRNA field synthesizes our present understanding of lncRNA biology and makes updated recommendations on nomenclature and definition.
Frankish, A. et al. Gencode 2021. Nucleic Acids Res. 49, D916–D923 (2021).
Hon, C. C. et al. An atlas of human long non-coding RNAs with accurate 5′ ends. Nature 543, 199–204 (2017).
Zhao, L. et al. NONCODEV6: an updated database dedicated to long non-coding RNA annotation in both animals and plants. Nucleic Acids Res. 49, D165–D171 (2021).
Uszczynska-Ratajczak, B., Lagarde, J., Frankish, A., Guigo, R. & Johnson, R. Towards a complete map of the human long non-coding RNA transcriptome. Nat. Rev. Genet. 19, 535–548 (2018).
Volders, P. J. et al. LNCipedia 5: towards a reference set of human long non-coding RNAs. Nucleic Acids Res. 47, D135–D139 (2019).
Ma, L. et al. LncBook: a curated knowledgebase of human long non-coding RNAs. Nucleic Acids Res. 47, D128–D134 (2019).
Mercer, T. R., Dinger, M. E. & Mattick, J. S. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 10, 155–159 (2009).
Slack, F. J. & Chinnaiyan, A. M. The role of non-coding RNAs in oncology. Cell 179, 1033–1055 (2019).
Winkle, M., El-Daly, S. M., Fabbri, M. & Calin, G. A. Noncoding RNA therapeutics — challenges and potential solutions. Nat. Rev. Drug Discov. 20, 629–651 (2021).
Kornienko, A. E. et al. Long non-coding RNAs display higher natural expression variation than protein-coding genes in healthy humans. Genome Biol. 17, 14 (2016).
Unfried, J. P. & Ulitsky, I. Substoichiometric action of long noncoding RNAs. Nat. Cell Biol. 24, 608–615 (2022).
Esposito, R. et al. Multi-hallmark long noncoding RNA maps reveal non-small cell lung cancer vulnerabilities. Cell Genom. 2, 100171 (2022). This study demonstrates how CRISPR screens can be integrated across multiple cancer hallmarks and cell backgrounds to prioritize high-promise therapeutic lncRNA targets.
Huarte, M. The emerging role of lncRNAs in cancer. Nat. Med. 21, 1253–1261 (2015).
Balas, M. M. & Johnson, A. M. Exploring the mechanisms behind long noncoding RNAs and cancer. Noncoding RNA Res. 3, 108–117 (2018).
Winkler, L. & Dimitrova, N. A mechanistic view of long noncoding RNAs in cancer. Wiley Interdiscip. Rev. RNA 13, e1699 (2022).
Gutschner, T. & Diederichs, S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 9, 703–719 (2012).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Vancura, A. et al. Is evolutionary conservation a useful predictor for cancer long noncoding RNAs? Insights from the cancer lncRNA census 3. Noncoding RNA 8 https://doi.org/10.3390/ncrna8060082 (2022).
Yang, M. et al. lncRNAfunc: a knowledgebase of lncRNA function in human cancer. Nucleic Acids Res. 50, D1295–D1306 (2022).
Wang, Z. et al. lncRNA epigenetic landscape analysis identifies EPIC1 as an oncogenic lncRNA that interacts with MYC and promotes cell-cycle progression in cancer. Cancer Cell 33, 706–720.e9 (2018).
Zhao, H. et al. Comprehensive landscape of epigenetic-dysregulated lncRNAs reveals a profound role of enhancers in carcinogenesis in BC subtypes. Mol. Ther. Nucleic Acids 23, 667–681 (2021).
Esposito, R. et al. Tumour mutations in long noncoding RNAs enhance cell fitness. Nat. Commun. 14, 3342 (2023). This study demonstrates that somatic mutations in lncRNAs can promote tumour cell fitness, and may be used to identify cancer-promoting lncRNAs.
Yan, X. et al. Comprehensive genomic characterization of long non-coding RNAs across human cancers. Cancer Cell 28, 529–540 (2015).
Sasaki, Y. T., Ideue, T., Sano, M., Mituyama, T. & Hirose, T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc. Natl Acad. Sci USA 106, 2525–2530 (2009).
Naveed, A. et al. NEAT1 polyA-modulating antisense oligonucleotides reveal opposing functions for both long non-coding RNA isoforms in neuroblastoma. Cell Mol. Life Sci. 78, 2213–2230 (2021).
Fu, J. W., Kong, Y. & Sun, X. Long noncoding RNA NEAT1 is an unfavorable prognostic factor and regulates migration and invasion in gastric cancer. J. Cancer Res. Clin. Oncol. 142, 1571–1579 (2016).
Hirose, T. et al. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol. Biol. Cell 25, 169–183 (2014).
Choudhry, H. et al. Tumor hypoxia induces nuclear paraspeckle formation through HIF-2α dependent transcriptional activation of NEAT1 leading to cancer cell survival. Oncogene 34, 4482–4490 (2015).
Prasanth, K. V. et al. Regulating gene expression through RNA nuclear retention. Cell 123, 249–263 (2005).
Gupta, R. A. et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464, 1071–1076 (2010).
Kogo, R. et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 71, 6320–6326 (2011).
Geng, Y. J., Xie, S. L., Li, Q., Ma, J. & Wang, G. Y. Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression. J. Int. Med. Res. 39, 2119–2128 (2011).
Liu, X. H. et al. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer 13, 464 (2013).
Somarowthu, S. et al. HOTAIR forms an intricate and modular secondary structure. Mol. Cell 58, 353–361 (2015).
Ji, P. et al. MALAT-1, a novel noncoding RNA, and thymosin β4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 22, 8031–8041 (2003).
Zhu, N., Ahmed, M., Li, Y., Liao, J. C. & Wong, P. K. Long noncoding RNA MALAT1 is dynamically regulated in leader cells during collective cancer invasion. Proc. Natl Acad. Sci. USA 120, e2305410120 (2023).
Tripathi, V. et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 39, 925–938 (2010).
Yang, L. et al. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell 147, 773–788 (2011).
Kim, J. et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat. Genet. 50, 1705–1715 (2018).
Zhou, Y., Zhang, X. & Klibanski, A. MEG3 noncoding RNA: a tumor suppressor. J. Mol. Endocrinol. 48, R45–R53 (2012).
Uroda, T. et al. Conserved pseudoknots in lncRNA MEG3 are essential for stimulation of the p53 pathway. Mol. Cell 75, 982–995.e9 (2019).
Zhou, Y. et al. Activation of p53 by MEG3 non-coding RNA. J. Biol. Chem. 282, 24731–24742 (2007).
Zheng, Q. et al. Long noncoding RNA MEG3 suppresses liver cancer cells growth through inhibiting β-catenin by activating PKM2 and inactivating PTEN. Cell Death Dis. 9, 253 (2018).
Marin-Bejar, O. et al. The human lncRNA LINC-PINT inhibits tumor cell invasion through a highly conserved sequence element. Genome Biol. 18, 202 (2017).
Marin-Bejar, O. et al. Pint lincRNA connects the p53 pathway with epigenetic silencing by the Polycomb repressive complex 2. Genome Biol. 14, R104 (2013).
Cabili, M. N. et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes. Dev. 25, 1915–1927 (2011).
Chen, J. et al. Evolutionary analysis across mammals reveals distinct classes of long non-coding RNAs. Genome Biol. 17, 19 (2016).
Hezroni, H. et al. Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Rep. 11, 1110–1122 (2015).
Washietl, S., Kellis, M. & Garber, M. Evolutionary dynamics and tissue specificity of human long noncoding RNAs in six mammals. Genome Res. 24, 616–628 (2014).
Necsulea, A. & Kaessmann, H. Evolutionary dynamics of coding and non-coding transcriptomes. Nat. Rev. Genet. 15, 734–748 (2014).
Yue, F. et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature 515, 355–364 (2014).
Andergassen, D. & Rinn, J. L. From genotype to phenotype: genetics of mammalian long non-coding RNAs in vivo. Nat. Rev. Genet. 23, 229–243 (2022).
Zhang, B. et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2, 111–123 (2012).
Ross, C. J. et al. Uncovering deeply conserved motif combinations in rapidly evolving noncoding sequences. Genome Biol. 22, 29 (2021).
Torarinsson, E., Sawera, M., Havgaard, J. H., Fredholm, M. & Gorodkin, J. Thousands of corresponding human and mouse genomic regions unalignable in primary sequence contain common RNA structure. Genome Res. 16, 885–889 (2006).
Seiler, J. et al. The lncRNA VELUCT strongly regulates viability of lung cancer cells despite its extremely low abundance. Nucleic Acids Res. 45, 5458–5469 (2017).
Liu, S. J. et al. Single-cell analysis of long non-coding RNAs in the developing human neocortex. Genome Biol. 17, 67 (2016).
Johnsson, P. et al. Transcriptional kinetics and molecular functions of long noncoding RNAs. Nat. Genet. 54, 306–317 (2022).
Pinkney, H. R., Black, M. A. & Diermeier, S. D. Single-cell RNA-seq reveals heterogeneous lncRNA expression in xenografted triple-negative breast cancer cells. Biology 10 https://doi.org/10.3390/biology10100987 (2021).
Cabili, M. N. et al. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. 16, 20 (2015).
Eissmann, M. et al. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 9, 1076–1087 (2012).
Gutschner, T. et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 73, 1180–1189 (2013). This seminal study demonstrates the practicality and efficacy of targeting oncogenic lncRNAs in vivo using ASOs.
Liu, S. J. et al. CRISPRi-based radiation modifier screen identifies long non-coding RNA therapeutic targets in glioma. Genome Biol. 21, 83 (2020). This work presents a CRISPRi screen to identify radio-sensitizing onco-lncRNAs in glioblastoma, making the important observation that cell-specific therapeutic targets do not necessarily have cell-specific expression.
Ramilowski, J. A. et al. Functional annotation of human long noncoding RNAs via molecular phenotyping. Genome Res. 30, 1060–1072 (2020). This work presents the most comprehensive data set of lncRNA-targeting ASOs to date, linking their inhibitory activity to molecular and cellular phenotypes, which represents a valuable resource for future ASO drug development.
Stojic, L. et al. Specificity of RNAi, LNA and CRISPRi as loss-of-function methods in transcriptional analysis. Nucleic Acids Res. 46, 5950–5966 (2018).
Guttman, M. & Rinn, J. L. Modular regulatory principles of large non-coding RNAs. Nature 482, 339–346 (2012).
Sun, D., Gao, W., Hu, H. & Zhou, S. Why 90% of clinical drug development fails and how to improve it? Acta Pharm. Sin. B 12, 3049–3062 (2022).
Muller, P. Y. & Milton, M. N. The determination and interpretation of the therapeutic index in drug development. Nat. Rev. Drug Discov. 11, 751–761 (2012).
Iyer, M. K. et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 47, 199–208 (2015). This work presents one of the first comprehensive surveys of dysregulated lncRNAs across diverse cancer types, representing a valuable resource for future discovery.
Diermeier, S. D. et al. Mammary tumor-associated RNAs impact tumor cell proliferation, invasion, and migration. Cell Rep. 17, 261–274 (2016).
Yu, A. T. et al. PHAROH lncRNA regulates Myc translation in hepatocellular carcinoma via sequestering TIAR. eLife 10 https://doi.org/10.7554/eLife.68263 (2021).
Hosono, Y. et al. Oncogenic role of THOR, a conserved cancer/testis long non-coding RNA. Cell 171, 1559–1572.e20 (2017).
Panzitt, K. et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology 132, 330–342 (2007).
Leucci, E. et al. Melanoma addiction to the long non-coding RNA SAMMSON. Nature 531, 518–522 (2016). These authors demonstrate how an exquisitely tumour-specific expressed lncRNA can be targeted for a potent disease-modifying activity with minimal effect on non-transformed cells.
Montes, M. et al. The lncRNA MIR31HG regulates p16INK4A expression to modulate senescence. Nat. Commun. 6, 6967 (2015).
Rheinbay, E. et al. Analyses of non-coding somatic drivers in 2,658 cancer whole genomes. Nature 578, 102–111 (2020).
Khalid, R., Naveed, H. & Khalid, Z. Computational prediction of disease related lncRNAs using machine learning. Sci. Rep. 13, 806 (2023).
de Goede, O. M. et al. Population-scale tissue transcriptomics maps long non-coding RNAs to complex disease. Cell 184, 2633–2648.e19 (2021).
Mitra, R., Adams, C. M. & Eischen, C. M. Systematic lncRNA mapping to genome-wide co-essential modules uncovers cancer dependency on uncharacterized lncRNAs. eLife 11 https://doi.org/10.7554/eLife.77357 (2022). This work presents a powerful, integrative approach combining diverse clinical evidence sources that demonstrates an in silico strategy for prioritizing therapeutic lncRNAs, which could be validated experimentally.
Villiers, W. et al. Multi-omics and machine learning reveal context-specific gene regulatory activities of PML::RARA in acute promyelocytic leukemia. Nat. Commun. 14, 724 (2023).
Zhang, Y. et al. Identifying cancer driver lncRNAs bridged by functional effectors through integrating multi-omics data in human cancers. Mol. Ther. Nucleic Acids 17, 362–373 (2019).
Delas, M. J. et al. lncRNA requirements for mouse acute myeloid leukemia and normal differentiation. eLife 6 https://doi.org/10.7554/eLife.25607 (2017).
Notzold, L. et al. The long non-coding RNA LINC00152 is essential for cell cycle progression through mitosis in HeLa cells. Sci. Rep. 7, 2265 (2017).
Tiessen, I. et al. A high-throughput screen identifies the long non-coding RNA DRAIC as a regulator of autophagy. Oncogene 38, 5127–5141 (2019).
Beermann, J. et al. A large shRNA library approach identifies lncRNA Ntep as an essential regulator of cell proliferation. Cell Death Differ. 25, 307–318 (2018).
Gutschner, T., Baas, M. & Diederichs, S. Noncoding RNA gene silencing through genomic integration of RNA destabilizing elements using zinc finger nucleases. Genome Res. 21, 1944–1954 (2011).
Lin, N. et al. An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol. Cell 53, 1005–1019 (2014).
Maamar, H., Cabili, M. N., Rinn, J. & Raj, A. linc-HOXA1 is a noncoding RNA that represses Hoxa1 transcription in cis. Genes Dev. 27, 1260–1271 (2013).
Lennox, K. A. & Behlke, M. A. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. 44, 863–877 (2016).
Gagnon, K. T., Li, L., Chu, Y., Janowski, B. A. & Corey, D. R. RNAi factors are present and active in human cell nuclei. Cell Rep. 6, 211–221 (2014).
Yip, C. W. et al. Antisense-oligonucleotide-mediated perturbation of long non-coding RNA reveals functional features in stem cells and across cell types. Cell Rep. 41, 111893 (2022).
Esposito, R. et al. Hacking the cancer genome: profiling therapeutically actionable long non-coding RNAs using CRISPR–Cas9 screening. Cancer Cell 35, 545–557 (2019).
Zhu, S. et al. Genome-scale deletion screening of human long non-coding RNAs using a paired-guide RNA CRISPR–Cas9 library. Nat. Biotechnol. 34, 1279–1286 (2016). This paper demonstrates the feasibility of high-throughput pooled CRISPR screening of lncRNAs, using a CRISPR-deletion strategy.
Liu, Y. et al. Genome-wide screening for functional long noncoding RNAs in human cells by Cas9 targeting of splice sites. Nat. Biotechnol. https://doi.org/10.1038/nbt.4283 (2018).
Brinkman, E. K. et al. Kinetics and fidelity of the repair of Cas9-induced double-strand DNA breaks. Mol. Cell 70, 801–813.e6 (2018).
Liu, S. J. et al. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 355 https://doi.org/10.1126/science.aah7111 (2017).
Koirala, P. et al. lncRNA AK023948 is a positive regulator of AKT. Nat. Commun. 8, 14422 (2017).
Goyal, A. et al. Challenges of CRISPR/Cas9 applications for long non-coding RNA genes. Nucleic Acids Res. 45, e12 (2017).
Cox, D. B. T. et al. RNA editing with CRISPR–Cas13. Science 358, 1019–1027 (2017).
Konermann, S. et al. Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell 173, 665–676.e14 (2018).
Abudayyeh, O. O. et al. RNA targeting with CRISPR–Cas13. Nature 550, 280–284 (2017).
Xu, D. et al. A CRISPR/Cas13-based approach demonstrates biological relevance of vlinc class of long non-coding RNAs in anticancer drug response. Sci. Rep. 10, 1794 (2020).
Wang, L. et al. CRISPR–Cas13d screens identify KILR, a breast cancer risk-associated lncRNA that regulates DNA replication and repair. Preprint at bioRxiv https://doi.org/10.1101/2023.11.16.567471 (2023).
Wong, L. S. et al. In vivo genome-wide CRISPR activation screening identifies functionally important long noncoding RNAs in hepatocellular carcinoma. Cell Mol. Gastroenterol. Hepatol. 14, 1053–1076 (2022). This work presents the first in vivo genome-wide CRISPRa screen to identify oncogenic lncRNAs in hepatocellular carcinoma.
Han, K. et al. CRISPR screens in cancer spheroids identify 3D growth-specific vulnerabilities. Nature 580, 136–141 (2020).
Shen, W. et al. Zebrafish xenograft model of human lung cancer for studying the function of LINC00152 in cell proliferation and invasion. Cancer Cell Int. 20, 376 (2020). This article demonstrates how, somewhat paradoxically, zebrafish can be used as a practical and faithful in vivo model of lung cancer, reducing the use of mammals in research.
Michels, B. E. et al. Pooled in vitro and in vivo CRISPR–Cas9 screening identifies tumor suppressors in human colon organoids. Cell Stem Cell 26, 782–792.e7 (2020).
Rinaldi, C. & Wood, M. J. A. Antisense oligonucleotides: the next frontier for treatment of neurological disorders. Nat. Rev. Neurol. 14, 9–21 (2018).
Crooke, S. T., Liang, X. H., Baker, B. F. & Crooke, R. M. Antisense technology: a review. J. Biol. Chem. 296, 100416 (2021).
Hong, D. et al. AZD9150, a next-generation antisense oligonucleotide inhibitor of STAT3 with early evidence of clinical activity in lymphoma and lung cancer. Sci. Transl. Med. 7, 314ra185 (2015). This study is one of the first to demonstrate disease-modifying activity of an ASO for a solid tumour in humans.
Crooke, S. T., Baker, B. F., Crooke, R. M. & Liang, X. H. Antisense technology: an overview and prospectus. Nat. Rev. Drug Discov. 20, 427–453 (2021).
Monia, B. P. et al. Evaluation of 2′-modified oligonucleotides containing 2′-deoxy gaps as antisense inhibitors of gene expression. J. Biol. Chem. 268, 14514–14522 (1993).
Havens, M. A. & Hastings, M. L. Splice-switching antisense oligonucleotides as therapeutic drugs. Nucleic Acids Res. 44, 6549–6563 (2016).
Latos, P. A. et al. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science 338, 1469–1472 (2012).
Schmidt, K. et al. Targeting the oncogenic long non-coding RNA SLNCR1 by blocking its sequence-specific binding to the androgen receptor. Cell Rep. 30, 541–554.e5 (2020).
Milazzo, C. et al. Antisense oligonucleotide treatment rescues UBE3A expression and multiple phenotypes of an Angelman syndrome mouse model. JCI Insight 6 https://doi.org/10.1172/jci.insight.145991 (2021).
Hsiao, J. et al. Upregulation of haploinsufficient gene expression in the brain by targeting a long non-coding RNA improves seizure phenotype in a model of Dravet syndrome. EBioMedicine 9, 257–277 (2016).
Modarresi, F. et al. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat. Biotechnol. 30, 453–459 (2012). This study shows how repressive lncRNAs can be targeted to restore healthy gene expression in vivo.
Tay, D. J. T. et al. Targeting RNA editing of antizyme inhibitor 1: a potential oligonucleotide-based antisense therapy for cancer. Mol. Ther. 29, 3258–3273 (2021).
Shin, M. et al. Intratracheally administered LNA gapmer antisense oligonucleotides induce robust gene silencing in mouse lung fibroblasts. Nucleic Acids Res. 50, 8418–8430 (2022).
Shah, P., Lalan, M. & Barve, K. Intranasal delivery: an attractive route for the administration of nucleic acid based therapeutics for CNS disorders. Front. Pharmacol. 13, 974666 (2022).
Mazur, C. et al. Brain pharmacology of intrathecal antisense oligonucleotides revealed through multimodal imaging. JCI Insight 4 https://doi.org/10.1172/jci.insight.129240 (2019).
Depreux, F. F. et al. Antisense oligonucleotides delivered to the amniotic cavity in utero modulate gene expression in the postnatal mouse. Nucleic Acids Res. 44, 9519–9529 (2016).
Calero, M. et al. Lipid nanoparticles for antisense oligonucleotide gene interference into brain border-associated macrophages. Front. Mol. Biosci. 9, 887678 (2022).
Bakowski, K. & Vogel, S. Evolution of complexity in non-viral oligonucleotide delivery systems: from gymnotic delivery through bioconjugates to biomimetic nanoparticles. RNA Biol. 19, 1256–1275 (2022).
Geary, R. S., Norris, D., Yu, R. & Bennett, C. F. Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv. Drug Deliv. Rev. 87, 46–51 (2015).
Scharner, J., Qi, S., Rigo, F., Bennett, C. F. & Krainer, A. R. Delivery of GalNAc-conjugated splice-switching ASOs to non-hepatic cells through ectopic expression of asialoglycoprotein receptor. Mol. Ther. Nucleic Acids 16, 313–325 (2019).
Yang, L. et al. Efficient delivery of antisense oligonucleotides using bioreducible lipid nanoparticles in vitro and in vivo. Mol. Ther. Nucleic Acids 19, 1357–1367 (2020).
Hammond, S. M. et al. Antibody–oligonucleotide conjugate achieves CNS delivery in animal models for spinal muscular atrophy. JCI Insight 7 https://doi.org/10.1172/jci.insight.154142 (2022).
Prakash, T. P. et al. Targeted delivery of antisense oligonucleotides to hepatocytes using triantennary N-acetyl galactosamine improves potency 10-fold in mice. Nucleic Acids Res. 42, 8796–8807 (2014).
Shen, W., Liang, X. H., Sun, H., De Hoyos, C. L. & Crooke, S. T. Depletion of NEAT1 lncRNA attenuates nucleolar stress by releasing sequestered P54nrb and PSF to facilitate c-Myc translation. PLoS ONE 12, e0173494 (2017).
Katsushima, K. et al. Targeting the Notch-regulated non-coding RNA TUG1 for glioma treatment. Nat. Commun. 7, 13616 (2016).
Carrette, L. L. G. et al. A mixed modality approach towards Xi reactivation for Rett syndrome and other X-linked disorders. Proc. Natl Acad. Sci. USA 115, E668–E675 (2018).
Gong, N., Teng, X., Li, J. & Liang, X. J. Antisense oligonucleotide-conjugated nanostructure-targeting lncRNA MALAT1 inhibits cancer metastasis. ACS Appl. Mater. Interfaces 11, 37–42 (2019).
Arun, G. et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 30, 34–51 (2016).
Lindow, M. et al. Assessing unintended hybridization-induced biological effects of oligonucleotides. Nat. Biotechnol. 30, 920–923 (2012).
Pollak, A. J. et al. Insights into innate immune activation via PS–ASO–protein–TLR9 interactions. Nucleic Acids Res. 50, 8107–8126 (2022).
Shen, W. et al. Chemical modification of PS–ASO therapeutics reduces cellular protein-binding and improves the therapeutic index. Nat. Biotechnol. 37, 640–650 (2019).
Dhuri, K. et al. Antisense oligonucleotides: an emerging area in drug discovery and development. J. Clin. Med. 9 https://doi.org/10.3390/jcm9062004 (2020).
Carlevaro-Fita, J. et al. Ancient exapted transposable elements promote nuclear enrichment of human long noncoding RNAs. Genome Res. 29, 208–222 (2019).
Doxtader Lacy, K. A., Liang, X. H., Zhang, L. & Crooke, S. T. RNA modifications can affect RNase H1-mediated PS–ASO activity. Mol. Ther. Nucleic Acids 28, 814–828 (2022).
Mouse Genome Sequencing Consortium et al. Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562 (2002).
Goyenvalle, A. et al. Considerations in the preclinical assessment of the safety of antisense oligonucleotides. Nucleic Acid Ther. 33, 1–16 (2023).
Ray, K. K. et al. Long-term efficacy and safety of inclisiran in patients with high cardiovascular risk and elevated LDL cholesterol (ORION-3): results from the 4-year open-label extension of the ORION-1 trial. Lancet Diabetes Endocrinol. 11, 109–119 (2023).
Chi, X., Gatti, P. & Papoian, T. Safety of antisense oligonucleotide and siRNA-based therapeutics. Drug Discov. Today 22, 823–833 (2017).
Chen, F., Li, Y., Feng, Y., He, X. & Wang, L. Evaluation of antimetastatic effect of lncRNA-ATB siRNA delivered using ultrasound-targeted microbubble destruction. DNA Cell Biol. 35, 393–397 (2016).
Ma, X. et al. Oncogenic role of lncRNA CRNDE in acute promyelocytic leukemia and NPM1-mutant acute myeloid leukemia. Cell Death Discov. 6, 121 (2020).
Tano, K. et al. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett. 584, 4575–4580 (2010).
Zhang, C., Wang, W., Lin, J., Xiao, J. & Tian, Y. lncRNA CCAT1 promotes bladder cancer cell proliferation, migration and invasion. Int. Braz. J. Urol. 45, 549–559 (2019).
Hu, B. et al. Therapeutic siRNA: state of the art. Signal Transduct. Target. Ther. 5, 101 (2020).
Wolfrum, C. et al. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat. Biotechnol. 25, 1149–1157 (2007).
Akinc, A. et al. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol. Ther. 18, 1357–1364 (2010).
Coelho, T. et al. Safety and efficacy of RNAi therapy for transthyretin amyloidosis. N. Engl. J. Med. 369, 819–829 (2013).
Juliano, R., Bauman, J., Kang, H. & Ming, X. Biological barriers to therapy with antisense and siRNA oligonucleotides. Mol. Pharm. 6, 686–695 (2009).
Hong, W., Zeng, J. & Xie, J. Antibiotic drugs targeting bacterial RNAs. Acta Pharm. Sin. B 4, 258–265 (2014).
Costales, M. G., Childs-Disney, J. L., Haniff, H. S. & Disney, M. D. How we think about targeting RNA with small molecules. J. Med. Chem. 63, 8880–8900 (2020).
Abulwerdi, F. A. et al. Selective small-molecule targeting of a triple helix encoded by the long noncoding RNA, MALAT1. ACS Chem. Biol. 14, 223–235 (2019).
Rakheja, I., Ansari, A. H., Ray, A., Chandra Joshi, D. & Maiti, S. Small molecule quercetin binds MALAT1 triplex and modulates its cellular function. Mol. Ther. Nucleic Acids 30, 241–256 (2022).
Brown, J. A. et al. Structural insights into the stabilization of MALAT1 noncoding RNA by a bipartite triple helix. Nat. Struct. Mol. Biol. 21, 633–640 (2014).
Aguilar, R. et al. Targeting Xist with compounds that disrupt RNA structure and X inactivation. Nature 604, 160–166 (2022).
Costales, M. G., Matsumoto, Y., Velagapudi, S. P. & Disney, M. D. Small molecule targeted recruitment of a nuclease to RNA. J. Am. Chem. Soc. 140, 6741–6744 (2018).
Yang, N. J. & Hinner, M. J. Getting across the cell membrane: an overview for small molecules, peptides, and proteins. Methods Mol. Biol. 1266, 29–53 (2015).
Kanasty, R. L., Whitehead, K. A., Vegas, A. J. & Anderson, D. G. Action and reaction: the biological response to siRNA and its delivery vehicles. Mol. Ther. 20, 513–524 (2012).
Takakusa, H. et al. Drug metabolism and pharmacokinetics of antisense oligonucleotide therapeutics: typical profiles, evaluation approaches, and points to consider compared with small molecule drugs. Nucleic Acid Ther. 33, 83–94 (2023).
Disney, M. D. Targeting RNA with small molecules to capture opportunities at the intersection of chemistry, biology, and medicine. J. Am. Chem. Soc. 141, 6776–6790 (2019).
Childs-Disney, J. L. et al. Targeting RNA structures with small molecules. Nat. Rev. Drug Discov. 21, 736–762 (2022).
Zhao, R., Fu, J., Zhu, L., Chen, Y. & Liu, B. Designing strategies of small-molecule compounds for modulating non-coding RNAs in cancer therapy. J. Hematol. Oncol. 15, 14 (2022).
Disney, M. D. et al. Inforna 2.0: a platform for the sequence-based design of small molecules targeting structured RNAs. ACS Chem. Biol. 11, 1720–1728 (2016).
Shi, Y. et al. Stabilization of lncRNA GAS5 by a small molecule and its implications in diabetic adipocytes. Cell Chem. Biol. 26, 319–330.e16 (2019).
Ward, M., Courtney, E. & Rivas, E. Fitness functions for RNA structure design. Nucleic Acids Res. 51, e40 (2023).
Jalali, S., Bhartiya, D., Lalwani, M. K., Sivasubbu, S. & Scaria, V. Systematic transcriptome wide analysis of lncRNA–miRNA interactions. PLoS ONE 8, e53823 (2013).
Zucchelli, S. et al. SINEUPs are modular antisense long non-coding RNAs that increase synthesis of target proteins in cells. Front. Cell Neurosci. 9, 174 (2015).
Ferre, F., Colantoni, A. & Helmer-Citterich, M. Revealing protein–lncRNA interaction. Brief. Bioinform. 17, 106–116 (2016).
Shaath, H. et al. Long non-coding RNA and RNA-binding protein interactions in cancer: experimental and machine learning approaches. Semin. Cancer Biol. 86, 325–345 (2022).
Wu, R. et al. The long noncoding RNA LUCAT1 promotes colorectal cancer cell proliferation by antagonizing Nucleolin to regulate MYC expression. Cell Death Dis. 11, 908 (2020).
Brockdorff, N., Bowness, J. S. & Wei, G. Progress toward understanding chromosome silencing by Xist RNA. Genes Dev. 34, 733–744 (2020).
He, S., Zhang, H., Liu, H. & Zhu, H. LongTarget: a tool to predict lncRNA DNA-binding motifs and binding sites via Hoogsteen base-pairing analysis. Bioinformatics 31, 178–186 (2015).
Nojima, T. et al. Deregulated expression of mammalian lncRNA through loss of SPT6 induces R-loop formation, replication stress, and cellular senescence. Mol. Cell 72, 970–984.e7 (2018).
Huang, D. et al. NKILA lncRNA promotes tumor immune evasion by sensitizing T cells to activation-induced cell death. Nat. Immunol. 19, 1112–1125 (2018).
Ma, Y., Zhang, J., Wen, L. & Lin, A. Membrane-lipid associated lncRNA: a new regulator in cancer signaling. Cancer Lett. 419, 27–29 (2018).
Wei, Z., Zhou, Y., Wang, R., Wang, J. & Chen, Z. Aptamers as smart ligands for targeted drug delivery in cancer therapy. Pharmaceutics 14 https://doi.org/10.3390/pharmaceutics14122561 (2022).
de Voogt, W. S., Tanenbaum, M. E. & Vader, P. Illuminating RNA trafficking and functional delivery by extracellular vesicles. Adv. Drug Deliv. Rev. 174, 250–264 (2021).
Kaseniit, K. E. et al. Modular, programmable RNA sensing using ADAR editing in living cells. Nat. Biotechnol. 41, 482–487 (2023).
Guo, S. et al. Ultra-thermostable RNA nanoparticles for solubilizing and high-yield loading of paclitaxel for breast cancer therapy. Nat. Commun. 11, 972 (2020).
Lin, C. et al. Importance of the long non-coding RNA (lncRNA) transcript HULC for the regulation of phenylalanine hydroxylase and treatment of phenylketonuria. Mol. Genet. Metab. 135, 171–178 (2022).
Li, Y. et al. A noncoding RNA modulator potentiates phenylalanine metabolism in mice. Science 373, 662–673 (2021). This work uses GalNAc-labelled HULC mimics as an in vivo therapeutic strategy to improve phenylalanine tolerance, demonstrating the promise of small lncRNA fragments as therapeutics.
Jin, F. et al. A functional motif of long noncoding RNA Nron against osteoporosis. Nat. Commun. 12, 3319 (2021). This work is another example of how a lncRNA fragment can be repurposed as a therapeutic oligonucleotide in vivo.
Sunwoo, H., Wu, J. Y. & Lee, J. T. The Xist RNA–PRC2 complex at 20-nm resolution reveals a low Xist stoichiometry and suggests a hit-and-run mechanism in mouse cells. Proc. Natl Acad. Sci. USA 112, E4216–E4225 (2015).
Dai, Y. et al. Programmable synthetic biomolecular condensates for cellular control. Nat. Chem. Biol. 19, 518–528 (2023).
Kirk, J. M. et al. Functional classification of long non-coding RNAs by k-mer content. Nat. Genet. 50, 1474–1482 (2018). This pioneering study demonstrates how lncRNA bioactivities can be modelled by combinations of short-sequence k-mers, offering a means of engineering RNAs with desired properties.
Wan, Y., Qu, K., Ouyang, Z. & Chang, H. Y. Genome-wide mapping of RNA structure using nuclease digestion and high-throughput sequencing. Nat. Protoc. 8, 849–869 (2013).
Van Nostrand, E. L. et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat. Methods 13, 508–514 (2016).
Lubelsky, Y. & Ulitsky, I. Sequences enriched in Alu repeats drive nuclear localization of long RNAs in human cells. Nature 555, 107–111 (2018).
Carrieri, C. et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 491, 454–457 (2012). This work is the archetypal example of lncRNA re-engineering for therapeutic gene regulation.
Toki, N., Takahashi, H., Zucchelli, S., Gustincich, S. & Carninci, P. Synthetic in vitro transcribed lncRNAs (SINEUPs) with chemical modifications enhance target mRNA translation. FEBS Lett. 594, 4357–4369 (2020).
Cao, C. et al. Enhancement of protein translation by CRISPR/dCasRx coupled with SINEB2 repeat of noncoding RNAs. Nucleic Acids Res. 51, e33 (2023).
Cao, Y. et al. RNA-based translation activators for targeted gene upregulation. Nat. Commun. 14, 6827 (2023).
Mercer, T. R. & Mattick, J. S. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 20, 300–307 (2013).
Gandhi, M., Caudron-Herger, M. & Diederichs, S. RNA motifs and combinatorial prediction of interactions, stability and localization of noncoding RNAs. Nat. Struct. Mol. Biol. 25, 1070–1076 (2018).
Roberts, T. C., Langer, R. & Wood, M. J. A. Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov. 19, 673–694 (2020).
Swayze, E. E. et al. Antisense oligonucleotides containing locked nucleic acid improve potency but cause significant hepatotoxicity in animals. Nucleic Acids Res. 35, 687–700 (2007).
Sugahara, K. N. et al. Tissue-penetrating delivery of compounds and nanoparticles into tumors. Cancer Cell 16, 510–520 (2009).
Guan, J. et al. iRGD-liposomes enhance tumor delivery and therapeutic efficacy of antisense oligonucleotide drugs against primary prostate cancer and bone metastasis. Adv. Funct. Mater. 31 https://doi.org/10.1002/adfm.202100478 (2021).
Moumne, L., Marie, A. C. & Crouvezier, N. Oligonucleotide therapeutics: from discovery and development to patentability. Pharmaceutics 14 https://doi.org/10.3390/pharmaceutics14020260 (2022).
Bauman, J. E. et al. Phase 1 study of EGFR-antisense DNA, cetuximab, and radiotherapy in head and neck cancer with preclinical correlatives. Cancer 124, 3881–3889 (2018).
Springate, C. M., Jackson, J. K., Gleave, M. E. & Burt, H. M. Efficacy of an intratumoral controlled release formulation of clusterin antisense oligonucleotide complexed with chitosan containing paclitaxel or docetaxel in prostate cancer xenograft models. Cancer Chemother. Pharmacol. 56, 239–247 (2005).
Boehnke, N. et al. Massively parallel pooled screening reveals genomic determinants of nanoparticle delivery. Science 377, eabm5551 (2022). This work shows that innovative genomic screening approaches are being brought to bear on the important problem of extrahepatic oligonucleotide delivery.
Joung, J. et al. Genome-scale activation screen identifies a lncRNA locus regulating a gene neighbourhood. Nature 548, 343–346 (2017).
Qu, L. et al. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell 29, 653–668 (2016).
Liu, R. et al. Long noncoding RNA expression signature to predict platinum-based chemotherapeutic sensitivity of ovarian cancer patients. Sci. Rep. 7, 18 (2017).
Seemann, S. E. et al. The identification and functional annotation of RNA structures conserved in vertebrates. Genome Res. 27, 1371–1383 (2017).
Diermeier, S. D. & Spector, D. L. Antisense oligonucleotide-mediated knockdown in mammary tumor organoids. Bio Protoc. 7 https://doi.org/10.21769/BioProtoc.2511 (2017).
Cimadamore, A. et al. Long non-coding RNAs in prostate cancer with emphasis on second chromosome locus associated with prostate-1 expression. Front. Oncol. 7, 305 (2017).
Adewunmi, O., Shen, Y., Zhang, X. H. & Rosen, J. M. Targeted inhibition of lncRNA Malat1 alters the tumor immune microenvironment in preclinical syngeneic mouse models of triple-negative breast cancer. Cancer Immunol. Res. 11, 1462–1479 (2023).
Amodio, N. et al. Drugging the lncRNA MALAT1 via LNA gapmeR ASO inhibits gene expression of proteasome subunits and triggers anti-multiple myeloma activity. Leukemia 32, 1948–1957 (2018).
Suzuki, M. M. et al. TUG1-mediated R-loop resolution at microsatellite loci as a prerequisite for cancer cell proliferation. Nat. Commun. 14, 4521 (2023).
Tasaki, Y. et al. Cancer-specific targeting of taurine-upregulated gene 1 enhances the effects of chemotherapy in pancreatic cancer. Cancer Res. 81, 1654–1666 (2021).
Taiana, E. et al. Long non-coding RNA NEAT1 targeting impairs the DNA repair machinery and triggers anti-tumor activity in multiple myeloma. Leukemia 34, 234–244 (2020).
Zhu, Z. et al. Knockdown long noncoding RNA nuclear paraspeckle assembly transcript 1 suppresses colorectal cancer through modulating miR-193a-3p/KRAS. Cancer Med. 8, 261–275 (2019).
Liang, H. & Peng, J. LncRNA HOTAIR promotes proliferation, invasion and migration in NSCLC cells via the CCL22 signaling pathway. PLoS One 17, e0263997 (2022).
Tian, L. I. et al. Targeting LncRNA LLNLR-299G3.1 with antisense oligonucleotide inhibits malignancy of esophageal squamous cell carcinoma cells in vitro and in vivo. Oncol. Res. 31, 463–479 (2023).
Li, M. et al. Antisense oligonucleotides targeting lncRNA AC104041.1 induces antitumor activity through Wnt2B/beta-catenin pathway in head and neck squamous cell carcinomas. Cell Death Dis. 11, 672 (2020).
Yang, L., Bai, H. S., Deng, Y. & Fan, L. High MALAT1 expression predicts a poor prognosis of cervical cancer and promotes cancer cell growth and invasion. Eur. Rev. Med. Pharmacol. Sci. 19, 3187–3193 (2015).
Guo, F., Li, Y., Liu, Y., Wang, J., Li, Y. & Li, G. Inhibition of metastasis-associated lung adenocarcinoma transcript 1 in CaSki human cervical cancer cells suppresses cell proliferation and invasion. Acta Biochim. Biophys. Sin. (Shanghai) 42, 224–229 (2010).
Yao, W. et al. Upregulation of MALAT-1 and its association with survival rate and the effect on cell cycle and migration in patients with esophageal squamous cell carcinoma. Tumour Biol. 37, 4305–4312 (2016).
Wang, W. et al. Long noncoding RNA MALAT1 promotes malignant development of esophageal squamous cell carcinoma by targeting beta-catenin via Ezh2. Oncotarget 7, 25668–25682 (2016).
Wang, X. et al. Silencing of long noncoding RNA MALAT1 by miR-101 and miR-217 inhibits proliferation, migration, and invasion of esophageal squamous cell carcinoma cells. J. Biol. Chem. 290, 3925–3935 (2015).
Li, P. et al. MALAT1 is associated with poor response to oxaliplatin-based chemotherapy in colorectal cancer patients and promotes chemoresistance through EZH2. Mol. Cancer Ther. 16, 739–751 (2017).
Liu, K. et al. MALAT1 promotes osteosarcoma development by regulation of HMGB1 via miR-142-3p and miR-129-5p. Cell Cycle 16, 578–587 (2017).
Zhao, H., Wang, Y., Hou, W., Ding, X. & Wang, W. Long non-coding RNA MALAT1 promotes cell proliferation, migration and invasion by targeting miR-590-3p in osteosarcoma. Exp. Ther. Med. 24, 672 (2022).
Kim, S. S., Harford, J. B., Moghe, M., Rait, A., Pirollo, K. F. & Chang, E. H. Targeted nanocomplex carrying siRNA against MALAT1 sensitizes glioblastoma to temozolomide. Nucleic Acids Res. 46, 1424–1440 (2018).
Wang, X., Sehgal, L., Jain, N., Khashab, T., Mathur, R. & Samaniego, F. LncRNA MALAT1 promotes development of mantle cell lymphoma by associating with EZH2. J. Transl. Med. 14, 346 (2016).
Amodio, N. et al. MALAT1: a druggable long non-coding RNA for targeted anti-cancer approaches. J. Hematol. Oncol. 11, 63 (2018).
Fang, H., Liu, H. M., Wu, W. H., Liu, H., Pan, Y. & Li, W. J. Upregulation of long noncoding RNA CCAT1-L promotes epithelial-mesenchymal transition in gastric adenocarcinoma. Onco. Targets Ther. 11, 5647–5655 (2018).
Kim, T. et al. Long-range interaction and correlation between MYC enhancer and oncogenic long noncoding RNA CARLo-5. Proc. Natl Acad. Sci. USA 111, 4173–4178 (2014).
Guttman, M. et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458, 223–227 (2009).
Faghihi, M. A. et al. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of β-secretase. Nat. Med. 14, 723–730 (2008).
Pandey, R. R. et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell 32, 232–246 (2008).
Engreitz, J. M. et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 539, 452–455 (2016).
Dimitrova, N. et al. LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Mol. Cell 54, 777–790 (2014).
Grelet, S. et al. A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nat. Cell Biol. 19, 1105–1115 (2017).
Guo, C. J. et al. Distinct processing of lncRNAs contributes to non-conserved functions in stem cells. Cell 181, 621–636.e2 (2020).
Lee, S. et al. Noncoding RNA NORAD regulates genomic stability by sequestering PUMILIO proteins. Cell 164, 69–80 (2016).
Somasundaram, K., Gupta, B., Jain, N. & Jana, S. lncRNAs divide and rule: the master regulators of phase separation. Front. Genet. 13, 930792 (2022).
Wu, M. et al. lncRNA SLERT controls phase separation of FC/DFCs to facilitate Pol I transcription. Science 373, 547–555 (2021).
Yoon, J. H. et al. LincRNA-p21 suppresses target mRNA translation. Mol. Cell 47, 648–655 (2012).
Acknowledgements
Work in the Johnson laboratory is funded by Science Foundation Ireland through Future Research Leaders award 18/FRL/6194 and by the Irish Research Council through Consolidator Laureate award (IRCLA/2022/2500). M.C. acknowledges the support of DevelopMed, which received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No 945425. The authors thank S. Ramnarayanan, T. Uroda and D. Harvey for helpful comments.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
R.J. is a paid consultant for NextRNA and inventor of the patent ‘Nucleic acid agents for treatment of non-small-cell lung cancer’ (WO2023061888A1). S.O. is CEO, founder and shareholder of HAYA Therapeutics SA. The other authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Genetics thanks Sarah Diermeier, Sven Diederichs and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Coan, M., Haefliger, S., Ounzain, S. et al. Targeting and engineering long non-coding RNAs for cancer therapy. Nat Rev Genet (2024). https://doi.org/10.1038/s41576-024-00693-2
Accepted:
Published:
DOI: https://doi.org/10.1038/s41576-024-00693-2