Abstract
The increasing prevalence of antimicrobial-resistant (AMR) pathogens has become a major global health concern. To address this challenge, innovative strategies such as bacteriophage therapy must be optimised. Genomic characterisation is a crucial step in identifying suitable phage candidates for combating AMR pathogens. The aim of this study was to characterise seven phages that infect the Escherichia coli O177 strain using a whole genome sequencing. The analysis of genome sequences revealed that these phages had linear dsDNA, with genome sizes spanning from 136, 483 to 166,791 bp and GC content varying from 35.39 to 43.63%. Taxonomically, the phages were classified under three different subfamilies (Stephanstirmvirinae, Tevenvirinae, and Vequintavirinae) and three genera (Phapecoctavirus, Tequatrovirus, and Vequintavirus) within the class Caudoviricetes. In silico PhageAI analysis predicted that all the phages were virulent, with confidence levels between 96.07 and 97.26%. The phage genomes contained between 66 and 82 ORFs, which encode hypothetical and putative functional proteins. In addition, the phage genomes contained core genes associated with molecular processes such as DNA replication, transcription modulation, nucleotide metabolism, phage structure (capsid and tail), and lysis. None of the genomes carried genes associated with undesirable traits such as integrase, antimicrobial resistance, virulence, and toxins. The study revealed high genome and proteome homology among E. coli O177 phages and other known Escherichia phages. The results suggest that the seven phages are new members of the genera Phapecoctavirus, Tequatrovirus, and Vequintavirus under the subfamilies Stephanstirmvirinae, Tevenvirinae, and Vequintavirinae, respectively.
Similar content being viewed by others
Introduction
Bacteriophages (phages), the viruses that infect and kill their bacterial host, are most abundant biological entity in the biosphere1,2. Phages are classified into temperate and lytic groups based on their reproductive strategies and how these affect their bacterial host3. The ability of lytic phages to lyse bacterial cell hosts has attracted the interest of pharmacologists and researchers in search of alternative antimicrobials . According to the International Committee of Taxonomy of Viruses (ICTV) and Bacterial and Archaeal Viruses Subcommittee, the most common lytic phages are linear double-stranded DNA (dsDNA) and tailed, belonging to the class Caudoviricetes4. The Caudoviricetes encompass several subfamilies including Stephanstirmvirinae, Tequatrovirinae, and Vequintavirinae4. Whole genome sequencing shows that phages have diverse genome sizes and levels of organisation5,6. Phages with genome sizes less than 200 kbp are classified as small or medium phages while those with genome sizes greater than 200 kbp but less than 500 kbp are referred to as jumbo phages5,7,8. There are over 22,000 complete phages in the NCBI database, with 21 419 classified as small or medium phages9. Most of the viruses with small or medium genomes belong to the Caudoviricetes class7.
Because of their diverse genome architecture, phages may infect the same and/or different bacteria species such as Acinetobacter baumannii, E. coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Vibrio alginolyticus species10,11,12,13,14. Despite this, phages that infect the same host may differ significantly in terms of their genome sequences6. Several studies have also reported that phage genomes contain a plethora of unique genes encoding hypothetical and putative functional proteins5,13,14. In addition, lytic phages possess genes responsible for genome replication and nucleotide metabolism, as well as DNA and RNA polymerases, which regulate gene expression5,15. Other small phages carry specialised genes encoding tRNAs and aminoacyl-tRNA synthetase10,15,16,17. Phage genomes may also carry anti-CRISPR (Acr) proteins, which interact with host CRISPR-Cas system18. In addition, lytic phages possess genes encoding proteins responsible for the formation of a phage pseudo-nucleus, which may provide phage protection against bacterial defence mechanisms such as clustered regularly interspaced short palindromic repeats (CRISPR-Cas) system and/or restriction modified enzymes19. The concerted action of these features does not only make lytic phages less dependent on the host replication machinery, but also enhance phage virulence and host range5,7. It is these attributes that make lytic phages ideal for biocontrol application7. Given that lytic phages harbour an array of genes with unknown function, there is a need to determine genetic diversity and understand the evolutionary strategies that they use to overcome host defence mechanisms. Therefore, to expand on our previous study20, the current study determines genomic and proteomic characteristics of phages infecting E. coli O177 strain.
Results
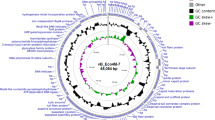
A summary of the genomic features of the seven phage genomes characterised in this study is shown in Table 1. All E. coli O177 phages were linear double-stranded DNA (dsDNA), varying in size from 136, 483 to 166,791 bp with a GC content between 35.39 and 43.63% (Table 1). Based on the BLASTn and PhageAI analysis, phages belonged to the class Caudoviricetes, under three different subfamilies (Stephanstirmvirinae, Tevenvirinae, and Vequintavirinae) and three genera (Phapecoctavirus, Tequatrovirus, and Vequintavirus) (Table 1). In silico PhageAI analysis predicted all the phages as virulent, with confidence levels between 96.07 and 97.26%. These high confidence levels suggest that these phages are likely to be lytic in nature. BLASTn analysis revealed that the E. coli O177 phage genomes had high sequence similarity (> 95%) to other Escherichia phages genomes from the NCBI database (Table S1). The total number of coding sequences (CDS) identified in phage genomes ranged from 220 to 284. Those CDS/genes encode structural proteins (major capsid, baseplate, and tail fiber), host lysis (endolysin and lysozyme), and other functions (DNA replication/transcription, repair/packaging proteins). No genes encoding undesirable (antimicrobial resistance, virulence, toxins, mobile genetic material determinants) features or anti-CRISPR proteins were detected in all the phage genomes.
Phage genomes encompassed between 66 and 82 ORFs, with 42 to 76% predicted as hypothetical proteins and 24 to 58% assigned to various putative functional proteins. The ORFs assigned to putative functional proteins were classified into four distinct modules, namely, DNA replication and regulation, DNA packaging, structural modules (major capsid and tail), and host lysis. As depicted in Fig. 1, 58% of ORFs encoding putative functional proteins were found in phagevB_EcoM_11B_SA_NWU. The tRNAscan-SE analysis showed that all the phage genomes contained tRNA features (Table 1 and Table S2A–G). The phage genomes harboured rho–independent transcription terminators sites (ranging from 37 to 48 sites) with stem-loop secondary structure (ΔG ≤ 9 kcal mol−1), (Table S3A–G).
Topology analysis showed that integral membrane protein found in phages vB_EcoM_3A1_SA_NWU, vB_EcoM_10C3_SA_NWU, and vB_EcoM_118_SANWU possessed 9 transmembrane domains (Fig. 2). However, no signal peptides were detected in the phage proteins. Table 2 indicates the physiochemical properties and secondary structure of lysin, and endolysin proteins. ExPASY ProtParam analysis indicated that lysin, and endolysin are large proteins with molecular weight of 17,423.00 and 19,779.16 kDa and Isoelectric point of 9.13 and 9.98, respectively. Both proteins were classified as stable, with the instability index (II) of 37.11 (lysozyme) and 27.11 (endolysin). Lysozyme and endolysin proteins from this study were found to be closely related (sharing ≥ 99% amino acids sequence identity) to 1AM7_A lysozyme and 5B2G_C endolysin proteins found in Enterobacteria phage lambda and Enterobacteria phage T4, respectively. MODELLER (v10.0) program was used to generate homology model of these three proteins. Figure 3 depicts 3D model of lysozyme, and endolysin containing α-helices (> 40%) random coils (> 35%), and lower frequencies of β-sheets. VERIFY_3D-1D and PROCHECK revealed reliability and quality (with > 80% of the residues averaged 3D-1D score greater than 0.2) of the predicted models. Ramachandran plot revealed that ≥ 88.1% residues of the models were placed within the favourable regions while ≤ 11.5% were within the additionally allowed region (Fgure S1). No residues were observed in the disallowed regions. PROSA Z-score of the lysozyme and endolysin were − 6.14 and − 4.08, respectively, which indicate that predicted models fall within a reasonable range of NMR and X-ray structures (Figure S2, 3). The global model quality estimate (GMQE) and Qmean values ranged from 0.6 to 0.91 and 0.46 ± 0. 06 to 0.82 ± 0.07, respectively. Overall, all the models were of high quality.
The VIRIDIC analysis (which uses a similar algorithm to that used by the International Committee on Taxonomy of Viruses (ICTV) and Bacterial and Archaeal Viruses Subcommittee) showed high (≥ 93.7%) intergenomic similarity between E. coli O177 phages and related Escherichia phages (Fig. 4). The intergenomic similarity of three phages from the current study (vB_EcoM_3A1_SA_NWU, vB_EcoM_10C3_SA_NWU, and vB_EcoM_118_SA_NWU) and Escherichia phage vB_EcoM-Ro121lw (accession no. MH160766.1) was 95.7%, indicating that these phages belong to the same species. VIRIDIC analysis also showed that four phages from the current study (vB_EcoM_10C2_SA_NWU, vB_EcoM_11B_SA_NWU, vB_EcoM_12A_SA_NWU, and vB_EcoM_366V_SA_NWU) had 93.7 to 93.9% intergenomic similarity to Escherichia phage ECP52 (accession no. ON782582.1) and Escherichia phage Rv5_ev158 (accession no. LR694611.1), suggesting that they belong to the same genus (default VIRIDIC similarity threshold of 70% for genus and 95% for species). Based on progressiveMauve alignment analysis, phages within the genera Phapecoctavirus and Vequintavirus contain ten locally collinear blocks (LCBs) while phages from Tequatrovirus had seven LCBs (Fig. 5). Escherichia coli O177 phages showed similar LCBs with closely related Escherichia phages. The phages within the Phapecoctavirus (vB_EcoM_3A1_SA_NWU, vB_EcoM_10C3_SA_NWU, and vB_EcoM_118_SA_NWU) and Vequintavirus (vB_EcoM_366V_SA_NWU) had all LCBs arranged in the forward orientation while Vequintavirus phages (vB_EcoM_10C2_SA_NWU, and vB_EcoM_12A_SA_NWU) showed unique rearrangement of homologous modules, with eight LCBs arranged in the reverse complement orientation. Phage vB_EcoM_11B_SA_NWU (Tequatrovirus) revealed five LCBs arranged in the reverse complement orientation, with two of the local LCBs arranged in the forward orientation (Fig. 5). The whole genome sequence phylogenetic tree revealed all phages belonging to the same genera are closely related (Fig. 6). Escherichia coli O177 phages cladded together with closely related phages from the NCBI database, which indicated that these phages shared a common evolutionary history. TBLASTX analysis showed that Phapecoctavirus, Tequatrovirus, and Vequintavirus (vB_EcoM_10C2_SA_NWU and vB_EcoM_12A_SA_NWU) phages shared ≥ 96% identity similarity (with ˃ 95% coverage) and genome organisation (Fig. 7A–C). Phage vB_EcoM_366V_SA_NWU had low genome homology with other phages within Vequintavirus (Fig. 7B).
The heatmap showing percentage intergenomic sequence similarities (upper right half) and alignment genome fraction and genome length ratios (lower left half) for E. coli O177 phages and their closely related four Escherichia phages computed using VIRIDIC. The horizontal and vertical coordinates depict the corresponding phages GenBank accession number and the E. coli O177 phages in this study are indicated by the red asterisk (*) symbol next to their accession numbers accession number.
ProgressiveMauve alignment of the complete genomes of ten Escherichia phages [seven E. coli O177 phages and three reference phages (Escherichia phage vB_EcoM Hdk5 (accession no: MK373780.1), Escherichia phage vB_EcoM_Schickermooser (accession no: NC_048196.1), and Escherichia phage vB_EcoM UFV10 (accession no. OP555981.1)] representing three genera (Phapecoctavirus, Tequatrovirus, and Vequintavirus). Genome similarity is indicated by a similarity plot within the coloured blocks with the height of the plot proportional to the average nucleotide identity. The fragments that are not aligned or specific to a particular genome are represented by white areas. The regions of homologous DNA shared among the genomes are defined as local collinear blocks (LCBs) presented by boxes with the identical colours. LCBs above the genome’s center line are in the forward orientation relative to the reference genome while LCBs below the genome’s center line are in the reverse complement orientation relative to the reference genome.
Comparison of the genome sequences of E. coli O177 phages with closely related members of Phapecoctavirus (A), Vequintavirus (B), and Tequatrovirus (C) genera, created using Easyfig. The grey colour between the genome maps indicates level of homology with the scales representing the percentage genome identity between the regions obtained through tBLASTx. The arrows represent the genes/CDSs. Genomes are drawn to scale; the scale bar indicates 10 Kbp.
ViPtree analysis showed high amino acid senteny among the Phapecoctavirus, Tequatrovirus, and Vequintavirus (Fig. 8A,B). Escherichia coli O177 phages clustered together with other phages (Escherichia phages) within the class Caudoviricetes, with the highest tBLASTx score (SG) level of > 0.99 (Fig. 8B). Phage vB_EcoM_11B_SA_NWU cladded separately from other E. coli O177 phages, which suggest a uniqueness at proteomic level. All the phages were classified into Pseudomonadota (synonym Proteobacteria) host group. Protein-to-protein network-based Phage cloud analysis linked E coli O177 phages with the phages belonging to the same genus with a distance range of 0.01–0.03 ratio (Fig. 9). The phylogenetic tree of the selected conserved protein sequences (major capsid, terminase large subunit (TerL), and tail fiber proteins) revealed various clustering patterns (Fig. 10A–C). As depicted in Fig. 10A, major capsid protein and TerL phylogenetic tree analysis showed that E coli O177 phages formed monophyletic clade together with their closely related Escherichia phages (from the representatives of Phapecoctavirus, Tequatrovirus, and Vequintavirus genera), which suggested that these proteins originated from a common ancestor. However, tail phylogenetic tree analysis generated different clustering patterns amongst the phages. Phages vB_EcoM_10C2_SA_NWU and vB_EcoM_11B_SA_ formed separate monophyletic group NWU (based on terminase large subunit and tail fiber protein sequences, respectively), which suggested a distinct relationship with other Escherichia phages from Phapecoctavirus, Tequatrovirus and Vequintavirus.
Viral phylogenetic tree (ViPTree) analysis based on genome-wide sequence similarities computed using tBLASTx. (A) ViPTree of 8 E. coli O177-infecting phage genomes and other 2480 phage genomes presented in the circular view. The coloured rings represent the virus families (inner ring) and host groups (outer ring) while the region marked with red asterisk represents the E. coli O177 phage genomes. (B) Rectangular phylogenetic tree (subset) of the phages generated using ViPTree, with the log scale on top representing the SG values. Red branches represent E. coli O177 phage genomes while black branches represent 26 linear dsDNA known phage genomes from ViPTree database. The right and left lines (green) represent the classification of the phages based on the host group and family level, respectively.
Phylogenetic neighbour-joining tree based on amino acid sequences of E. coli O177 phages and other Escherichia phages. (A) major capsid; (B) terminase large subunit (TerL), and (C) tail fiber protein. The trees were generated by MEGA 11 software using the neighbour-joining method and a bootstrap value of 1000 replicates.
Discussion
The emergence and transmission of AMR pathogens pose a significant threat to public health, especially considering limited novel antibiotic development21. Therefore, exploring alternative strategies like phage therapy is crucial. To develop phage therapy against AMR pathogens, it is essential to isolate and genomically characterise lytic phages. Accordingly, several studies have isolated and characterised phages that infect various pathogenic bacteria such as A. baumannii, E. coli, K. pneumoniae, P. aeruginosa, Salmonella, and Vibrio species12,17,20,22,23,24. Since lytic phage genome architecture harbours genes encoding several hypothetical proteins with unknown functions22, genomic analysis may help not only in the taxonomic classification of phages, but also identify good candidates for phage therapy.
The aim of this study was to characterise the complete genomes of seven Escherichia phages that infect multidrug resistant E. coli O177 strain isolated from cattle faeces. Genomic analysis showed that all phages contained linear dsDNA genomes spanning from 136,476 to 151,980 bp, with GC content that varied from 35.39 to 43.63%. Given that the genomes of E. coli O177 phages were smaller than 200 kbp, they were classified as small phages. These findings were consistent with previous studies, which have reported small phages infecting other pathogenic bacteria such as E. coli, K. pneumoniae, P. aeruginosa, and Salmonella species10,11,12,13,14,25. Although TEM analysis revealed that E. coli O177 phages had similar morphotype20, BLASTn and PhageAI analysis classified these phages under the subfamilies Stephanstirmvirinae, Tevenvirinae, and Vequintavirinae within the Caudoviricetes class. In addition, E. coli O177 phages were classified under three genera; Phapecoctavirus (vB_EcoM_3A1_SA_NWU, vB_EcoM_10C3_SA_NWU, and vB_EcoM_118_SA_NWU) Tequatrovirus, (vB_EcoM_11B_SA_NWU), and Vequintavirus (10C2_SA_NWU, vB_EcoM_12A_SA_NWU and vB_EcoM_366V_SA_NWU) as predicted by BLASTn and PhageAI tools. This suggests that phages presenting similar morphotype may belong to different families and/or genera. Interestingly, PhageAI results predicted all phages to be virulent, making them suitable candidates for phage therapy.
Escherichia coli O177 phages harboured a plethora of unique genes, which encode hypothetical and putative functional proteins. A substantial number of the CDSs and ORFs found in the phage genomes were predicted as hypothetical proteins with unknown functions. Similar observations have been reported in other phages that infect pathogenic bacteria species10,22,26,27. This indicates that phage genomes carry several genes whose functions are yet to be understood. Thus, research efforts must be directed at elucidating the true functions of these hypothetical proteins. Another interesting observation was that E. coli O177 phages contained genes coding for phage DNA replication, DNA synthesis and packaging, structural proteins (capsid and tail), and host lysis (lysozyme, and endolysin). In contrast with other phages, two phage genomes (vB_EcoM_3A1_SA_NWU, and vB_EcoM_366V_SA_NWU) contain genes encoding baseplate tail fiber, and tail spike proteins. Because tail fiber proteins are crucial for phage receptor recognition, a high abundance of baseplate, tail fiber, and tail spike proteins in a phage genome can enhance its infection capabilities and host range28. Phages also harboured tail tubular protein, whose enzymatic activity may hydrolyse polysaccharides and biofilm structures29. Indeed, tail tubular protein has been reported to play a role in bacteria cell lysis30, suggesting that E. coli O177 phages can also utilise tail tubular system to lyse the bacteria cell. Furthermore, E. coli O177 phage genomes carried the genes encoding DNA polymerase (DNAPs), RNA polymerase (RNAs), and RNA polymerase binding, which provide phages with a degree of autonomy when it comes to gene expression.
Genomes of the seven phages harboured genes encoding ribonucleoside-diphosphate reductase large subunit and ATP-dependent protease, which are involved in nucleotide and protein biosynthesis and metabolism. The ATPases have been reported to be a molecular motor, which catalyse the packaging process of the mature viral genome31. This makes the phages more self-sufficient with respect to their core replication machinery. Furthermore, the phages harboured genes coding for serine/threonine, activator middle promoter, Rha family transcriptional regulator, ADP-ribosyltransferase, and helix-turn-helix transcriptional regulator proteins. These proteins are involved in the transcription of viral early genes10. Moreover, our phage genomes harboured middle transcription regulatory protein motA and RNA polymerase proteins, which are responsible for activating middle and late prompters for the transcription of middle and later viral genes, respectively. It is worth noting that the ExPASY ProtParam analysis showed that lysozyme and endolysin proteins found in E. coli O177 phages are more stable and water soluble (GRAVY ranging between − 0.499 and − 0.261). The random helix predominated by α-helix contributes to the stability of secondary structures. Interestingly, in silico analysis showed that both proteins have a life span (in vivo) of ≥ 10 h in E. coli cell. Given that lysozyme and endolysin have antimicrobial activity, these attributes suggest that the proteins can be used as an antibacterial agent to combat AMR E. coli infections. Notably, no antibiotics resistance, virulence and/or lysogenic signatures were detected in any of the phage genomes.
While the exact roles of tRNAs in phages remain unclear, their presence in the phage genome is believed to enhance phage fitness and improve translational efficiency, potentially enabling independent translation from the host5,17. The tRNAs are frequently found in myoviruses with a large genome size32. Although tRNAs are commonly found in jumbo phages, some small/medium phage genomes may also carry tRNAs10,17. Similarly, tRNAscan-SE showed that our E. coli O177 phage genomes harboured 5–11 tRNA genes encoding various amino acids. Given that tRNAs play a key role in protein biosynthesis, these phages may have less tRNAs depended on the host translation mechanisms17. Interestingly, phages vB_EcoM_3A_SA_NWU and vB_EcoM_11B_SA_NWU possessed genes coding for other specialised tRNA enzymes (histidyl tRNA synthetase and valyl-tRNA synthetase), which may catalyse formation of tRNAs10. Histidyl tRNA synthetase, putative tRNA nucleotidyltransferase/poly(A) polymerase, and valyl-tRNA synthetase play a crucial role in protein synthesis by facilitating the attachment of amino acids to tRNA molecules33. Phage genomes also harboured genes coding for tRNA (Ile)-lysidine synthase that may play a role in tRNA maturation34. In addition, the tRNAs may contribute to high phage infectivity and virulence8,35,36.
Comparative genomic (VIRIDIC and ProgressiveMauve alignment) analysis revealed high sequence similarity between E. coli O177 phages and other Escherichia phages from Phapecoctavirus, Tequatrovirus, and Vequintavirus genera. These finding are consistent with the results obtained through BLASTn and PhageAI tools. Notably, VIRIDIC analysis showed that maximum intergenomic similarity scores ranged from 93.7 to 95.7% between E. coli O177 phages and other phages from the NCBI database. Based on 70% threshold, these findings suggest that E. coli O177 phages appear to be new members of Phapecoctavirus, Tequatrovirus, and Vequintavirus genera under Stephanstirmvirinae, Straboviridae (subfamily Tevenvirinae), and Vequintavirinae families4,37,38. In addition, three phages (vB_EcoM_3A1_SA_NWU, vB_EcoM_10C3_SA_NWU, and vB_EcoM_118_SA_NWU) and Escherichia phage vB_EcoM-Ro1211w had intergenomic similarity of 95.7%, which suggests that these phages belong to the same species37,38. The Easyfig analysis also showed that genome organisation of E. coli O177 phages was similar to that of closely related phages. These finding support the results obtained by VIRIDIC analysis. Furthermore, ViPtree analysis revealed that E. coli O177 phages clustered with previously described phages from Phapecoctavirus, Tequatrovirus, and Vequintavirus genera, confirming that these phages share from a common ancestor. Similarly, phylogenetic tree analysis based on the conserved genes (major capsid, terminase large subunits, and tail fiber) supported the above findings.
Conclusion
The current study provides insights into the genomic diversity of E. coli O177 phages. The results revealed that these phages harboured a vast array of hypothetical proteins with unknown function. In addition, these phages possessed genes encoding extra functions such as DNA replication, transcription, nucleotide metabolism, as well as lysis. The absence of antimicrobial resistance, lysogeny, and virulence signatures in E. coli O177 phages indicates that they are suitable candidates for biocontrol purpose. Genomic and proteomic analysis showed high similarity between E. coli O177 phages and other phages from the NCBI database. This suggests that our E. coli O177 phages are new members under the subfamilies Stephanstirmvirinae, Tevenvirinae, and Vequintavirinae.
Materials and methods
Propagation of phages
Seven bacteriophages, which were isolated from cattle faeces in our previous study20, were propagated using E. coli O177 strain. In brief, an aliquot of 100 µL of overnight culture of E. coli O177 was inoculated into 250 mL volumetric flasks containing 100 mL tryptic soya broth. A 200 µL aliquot of each phage lysate was added to the mixture. The flasks were incubated at 37 °C for 24 h in a shaking incubator (120 rpm). After incubation, the mixture was transferred into 50 mL falcon tubes and centrifuged at 10,000× g for 10 min. The supernatant was filter-sterilised using 0.22 µm pore-size acrodisc syringe filter. Phage stock was kept for the DNA extraction.
DNA extraction and phage genome sequence
Phage genomic DNA was extracted using the phenol–chloroform protocol39, with minor modifications. Briefly, 1.5 mL of phage lysate was transferred into a 2 mL Eppendorf tube. The samples were treated with 18 µL of DNase (10 mg/mL) and 8 µL of RNAse A (10 mg/mL). The samples were mixed and incubated at 37 °C for 30 min. After incubation, the samples were treated with 50 µL of SDS (10%), 18 µL proteinase K (20 mg/mL), and 40 µL of 0.5 M EDTA (pH 8.0), followed by incubation at 60 °C for 60 min. Subsequently, 500 µL phenol–chloroform–isoamyl alcohol (25:24:1) was added to the samples. The samples were inverted five times and then centrifuged at 10,000× g for 5 min. The aqueous layer was transferred into a new 2 mL Eppendorf tube and mixed with 500 µL chloroform–isoamyl alcohol (24:1), inverted five times, and centrifuged at 10,000× g for 5 min. The aqueous layer was transferred into a new 1.5 mL Eppendorf tube and mixed with 45 µL of 3 M sodium acetate (pH 7.5) and 500 µL isopropanol (100%). The samples were incubated at − 20 °C overnight. Subsequently, the samples were centrifuged at 14,800 × g for 30 min. The DNA pellet was washed with three times pre-chilled 70% alcohol. The DNA pellet was air-dried, resuspended in 70 µL of 1X TE buffer (10 mM Tris PH 8.0, 1 mM EDTA), and stored at − 20 °C.
Prior to genome sequencing, the DNA samples were purified using GeneJet PCR Purification Kit (ThermoFishre, Baltics UAB, Lithuania) following the protocol described by the manufacturer. Subsequently, the samples were transported to Inqaba Biotechnical Industries (Pty) Ltd for sequencing. The DNA libraries were prepared using the NEBNext® Ultra™ II DNA Library Preparation Kit for Illumina® (NEB, Ipswich, MA, USA) according to the manufacturer’s instructions. The libraries were quantified using Qubit 2.0 fluorometer (Thermo Fisher Scientific Inc., Waltham, MA, USA) and genome sequencing was performed using Illumina NextSeq sequencing platform. A total of 400 Mb data (2 × 150 bp paired-end reads) was generated per sample.
Bioinformatics analysis and phage genome annotation
The quality of the raw reads was assessed using FastQC (v0.11.9)40, and then the adapters, N bases, and low-quality reads were removed using Trimmomatic (v0.36)41. The reads with high quality score were assembled into contigs using Spades v3.15.3, with K-mer set at default42. The assembled contigs were annotated using the Rapid Annotation using System Technology Toolkit (RASTtk) pipeline43. The annotated proteins were manually curated and validated using UniProtKB (https://www.uniprot.org/, accessed on 20 February 2023). The complete genomes were compared with other phage genome sequences using BLASTn from the NCBI database (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 20 February 2023). Phage taxonomy and lifestyle were computationally predicted using BLASTn (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and PhageAI tools44. Potential open reading frames (ORFs) were predicted and annotated RAST (http://rast.nmpdr.org, accessed on 23 February 2023), GeneMark (http://opal.biology.gatech.edu/GeneMark/, accessed on 23 February 2023) and NCBI ORFfinder tool (https://www.ncbi.nlm.nih.gov/orffinder/, accessed on 23 February 2023). Predicted ORFs were manually validated and curated. Subsequently, homology searches for each identified ORF sequences were subjected to BLASTp against the non-redundant protein database (with parameters set at score of > 50, E-value of < 1.0 × 10−2)45. Codon usage frequencies in the phage genomes were computed using Codon Usage programme (https://www.bioinformatics.org/sms2/codon_usage.html, accessed on 4 March 2023). The sequences were used to search for the protein domains in InterProScan 5 (http://www.ebi.ac.uk/interpro/search/sequence/, accessed on 4 march 2023)46, and protein family (Pfam) database (http://pfam.xfam.org/, accessed on 4 March 2023)47.
The presence of tRNAs were predicted using ARAGORN and tRNAscan-SE tools48,49. The rho–independent transcription terminators were determined using ARNold (http://rssf.i2bc.paris-saclay.fr/toolbox/arnold/index.php, accessed on 10 March 2023) and Genome2D tools50. Putative promoters were searched using the phage promoter integrated in Galaxy platform v 0.1.0 (https://bit.ly/2Dfebfv, accessed on 10 March 2023) with the parameters set at: thresholds: 90%, phage family: Myoviridae (former), host bacteria genus: Escherichia coli, and phage type: virulent51. Possible anti-CRISPR genes were predicted using AcrDB tool (https://bcb.unl.edu/AcrFinder/, accessed on 4 March 2023)52. The presence of virulence determinants was screened using VICTORS53, and Virulence Factor Database (VFDB)54, while antimicrobial resistance, and temperate genetic signatures were determined using Comprehensive Antibiotic Research Database (CARD)55, and PhageLeads56.
Domain identification and prediction of E. coli O177 phages structural proteins by homology modelling
The domains on proteins were ascertained using HmmerWeb (v2.41.2)57, and the protein family database47. Topology of proteins was analysed for the presence of transmembrane and signal-arrest-release (SAR) domains as previously described58,59,60. Signal peptides were predicted using Phobius (v. 1.01)61, Topcons (v.1.0)62, SOSUI (v. 1.1)63, DeepTMHMM (v1.0.12) and SignalP-6.0 (v0.0.52)60. Physiochemical properties and secondary structure of lysozyme and endolysin proteins were predicted using the ExPASY ProtParam tool (https://web.expasy.org/protparam/, accessed on 12 April 2023) and SOPMA (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_sopma.html, accessed on 12 April 2023), respectively. Homology modelling was performed on lysozyme and endolysin proteins found in E. coli O177 phage genomes. Each protein sequence was used as a query to identify templates using HHPred server (https://toolkit.tuebingen.mpg.de/tools/hhpred, accessed on 20 April 2023)64. The templates with highest identity similarity of ≥ 95%, and E-value of ≤ 0 for each sequence (top hits) were selected to generate structural models using web-based MODELLER (v10.0) algorithm64,65. The predicted 3D structures were verified for accuracy by VERIFY_3D (https://saves.mbi.ucla.edu/, accessed on 20 April 2023), PROSA (https://prosa.services.came.sbg.ac.at/prosa.php, accessed on 20 April 2023), and PROCHECK (http://www.ebi.ac.uk/thornton-srv/software/PROCHECK/, accessed on 20 April 2023).
Comparative genomics and proteomics analysis of phages
The intergenomic nucleotide sequence similarity among E. coli O177 phages and other Escherichia phages from the GenBank database was determined using Virus Intergenomic Distance Calculator (VIRIDIC). The genomic similarity threshold was set at 70% for genus and 95% for species37. Whole genome comparison between E. coli O177 phages and closely related Escherichia phages was performed using progressiveMauve software (v2.3.1)66, and tBLASTx on the DiGAlign Dynamic Genomic Alignment sever (https://www.genome.jp/digalign, accessed on 10 June 2023)67. Linear genome map comparison of the phages belonging to the same genus were created using Easyfig (v2.2.5) (http://mjsull.github.io/Easyfig/, accessed on 20 June 2023), using tBLASTx68.
Viral proteomic tree (ViPTree) server (v3.1) (https://www.genome.jp/viptree/, accessed on 20 June 2023) was used to infer a tree of E. coli O177 phages and other known phage genome sequences based on genome-wide sequence similarities computed using tBLASTx67. In addition, Phage clouds analysis was used to visualise the genomic relationship between E. coli O177 phages and other phages in NCBI database based on their genomic distances (with the threshold of 0.2)69. The amino acid sequences of three conserved proteins sequences (major capsid, terminase large subunit (TerL), and tail fiber) were used to construct phylogenetic tree by the neighbour-joining method using MEGA 11 with 1000 bootstrap repeats70. The complete genome sequences were deposited into the NCBI database, and the accession numbers are stated in Table 1.
Data availability
Sequence data generated and presented in this study have been deposited into the NCBI database under the GenBank Accession numbers; https://www.ncbi.nlm.nih.gov/nuccore/OR062524; https://www.ncbi.nlm.nih.gov/nuccore/OR062525; https://www.ncbi.nlm.nih.gov/nuccore/OR062526; https://www.ncbi.nlm.nih.gov/nuccore/OR062527; https://www.ncbi.nlm.nih.gov/nuccore/OR062528; https://www.ncbi.nlm.nih.gov/nuccore/OR062529; https://www.ncbi.nlm.nih.gov/nuccore/OR062530.
References
Batinovic, S. et al. Bacteriophages in natural and artificial environments. Pathogens 8, 100. https://doi.org/10.3390/pathogens8030100 (2019).
Mushegian, A. R. Are there 10^31 virus particles on earth, or more, or fewer?. J. Bacteriol. 202(9), 2020. https://doi.org/10.1128/JB.00052-20 (2020).
Kutter, E. & Sulakvelidze, A. Bacteriophages: Biology and Applications Boca Raton (CRC Press, 2004).
Turner, D. et al. Abolishment of morphology-based taxa and change to binomial species names: 2022 taxonomy update of the ICTV bacterial viruses subcommittee. Arch. Virol. 168, 74. https://doi.org/10.1007/s00705-022-05694-2 (2023).
Yuan, Y. & Gao, M. Jumbo bacteriophages: An overview. Front. Microbiol. 8, 1–9. https://doi.org/10.3389/fmicb.2017.00403 (2017).
Zhu, Y., Shang, J., Peng, C. & Sun, Y. Phage family classification under caudoviricetes: A review of current tools using the latest ICTV classification framework. Front. Microbiol. 13, 1032186. https://doi.org/10.3389/fmicb.2022.1032186 (2022).
Guan, J. & Bondy-Denomy, J. Intracellular organization by jumbo bacteriophages. J. Bacteriol. 203, e00362-e420. https://doi.org/10.1128/jb.00362-20 (2020).
M Iyer, L., Anantharaman, V., Krishnan, A., Burroughs, A. M. & Aravind, L. Jumbo phages: A comparative genomic overview of core functions and adaptions for biological conflicts. Viruses 13, 63. https://doi.org/10.3390/v13010063 (2021).
Jo, D., Kim, H., Lee, Y., Kim, J. & Ryu, S. Characterization and genomic study of EJP2, a novel jumbo phage targeting antimicrobial resistant Escherichia coli. Front. Microbiol. https://doi.org/10.3389/fmicb.2023.1194435 (2023).
Kim, S. G. et al. Characterization of novel Erwinia amylovora jumbo bacteriophages from Eneladusvirus genus. Viruses 12, 1373. https://doi.org/10.3390/v12121373 (2020).
Lewis, R. et al. Isolation of a novel jumbo bacteriophage effective against Klebsiella aerogenes. Front. Med. 7, 67. https://doi.org/10.3389/fmed.2020.00067 (2020).
Lood, C. et al. Integrative omics analysis of Pseudomonas aeruginosa virus PA5oct highlights the molecular complexity of jumbo phages. Environ. Microbiol. 22, 2165–2181. https://doi.org/10.1111/1462-2920.14979 (2020).
Cucić, S., Kropinski, A. M., Lin, J., Khursigara, C. M. & Anany, H. Complete genome sequence of a jumbo bacteriophage, Escherichia phage vB_EcoM_EC001. Microbiol. Resour. Announc. 11, e00017-22. https://doi.org/10.1128/mra.00017-22 (2022).
Orozco-Ochoa, A. K. et al. Characterization and genome analysis of six novel Vibrio parahaemolyticus phages associated with acute hepatopancreatic necrosis disease (AHPND). Virus Res. 323, 198973. https://doi.org/10.1016/j.virusres.2022.198973 (2023).
Zhang, B., Xu, J., He, X., Tong, Y. & Ren, H. Interactions between jumbo phage SA1 and staphylococcus: a global transcriptomic analysis. Microorganisms 10, 1590. https://doi.org/10.3390/microorganisms10081590 (2022).
Yoshikawa, G. et al. Xanthomonas citri jumbo phage XacN1 exhibits a wide host range and high complement of tRNA genes. Sci. Rep. 8, 4486. https://doi.org/10.1038/s41598-018-22239-3 (2018).
Nazir, A., Ali, A., Qing, H. & Tong, Y. Emerging aspects of jumbo bacteriophages. Infect. Drug Resist. 14, 5041. https://doi.org/10.2147/IDR.S330560 (2021).
Li, Y. & Bondy-Denomy, J. Anti-CRISPRs go viral: The infection biology of CRISPR-Cas inhibitors. Cell Host Microb. 29, 704–714. https://doi.org/10.1016/j.chom.2020.12.007 (2021).
Chaikeeratisak, V. et al. The phage nucleus and tubulin spindle are conserved among large Pseudomonas phages. Cell Rep. 20, 1563–1571. https://doi.org/10.1016/j.celrep.2017.07.064 (2017).
Montso, P. K., Mlambo, V. & Ateba, C. N. Characterization of lytic bacteriophages infecting multidrug-resistant shiga toxigenic atypical Escherichia coli O177 strains isolated from cattle feces. Front. Public Heal. 7, 355. https://doi.org/10.3389/fpubh.2019.00355 (2019).
Gordillo Altamirano, F. L. & Barr, J. J. Phage therapy in the postantibiotic era. Clin. Microbiol. Rev. 32, e00066-18. https://doi.org/10.1128/cmr.00066-18 (2019).
Imam, M. et al. vB_PaeM_MIJ3, a novel jumbo phage infecting Pseudomonas aeruginosa, possesses unusual genomic features. Front. Microbiol. 10, 2772. https://doi.org/10.3389/fmicb.2019.02772 (2019).
Rai, P. et al. Characterisation of broad-spectrum phiKZ like jumbo phage and its utilisation in controlling multidrug-resistant Pseudomonas aeruginosa isolates. Microb. Pathog. 172, 105767. https://doi.org/10.1016/j.micpath.2022.105767 (2022).
Nicolas, M. et al. Isolation and characterization of a novel phage collection against avian-pathogenic Escherichia coli. Microbiol. Spectr. https://doi.org/10.1128/spectrum.04296-22 (2023).
Zaki, B. M., Fahmy, N. A., Aziz, R. K., Samir, R. & El-Shibiny, A. Characterization and comprehensive genome analysis of novel bacteriophage, vB_Kpn_ZCKp20p, with lytic and anti-biofilm potential against clinical multidrug-resistant Klebsiella pneumoniae. Front. Front. Cell. Infect. Microbiol. 13, 1077995. https://doi.org/10.3389/fcimb.2023.1077995 (2023).
Chinnadurai, L. et al. Draft genome sequence of Escherichia coli phage CMSTMSU, isolated from shrimp farm effluent water. Microbiol. Resour. Announc. 7, e01034-e1118. https://doi.org/10.1128/mra.01034-18 (2018).
Korn, A. M., Hillhouse, A. E., Sun, L. & Gill, J. J. Comparative genomics of three novel jumbo bacteriophages infecting Staphylococcus aureus. J. Virol. 95, e02391-e2420. https://doi.org/10.1128/jvi.02391-20 (2021).
Nobrega, F. L. et al. Targeting mechanisms of tailed bacteriophages. Nat. Rev. Microbiol. 16, 760–773. https://doi.org/10.1038/s41579-018-0070-8 (2018).
Brzozowska, E. et al. Hydrolytic activity determination of tail tubular protein A of Klebsiella pneumoniae bacteriophages towards saccharide substrates. Sci. Rep. 7, 18048. https://doi.org/10.1038/s41598-017-18096-1 (2017).
Pyra, A. et al. Tail tubular protein A: A dual-function tail protein of Klebsiella pneumoniae bacteriophage KP32. Sci. Rep. 7, 2223. https://doi.org/10.1038/s41598-017-02451-3 (2017).
Tajuddin, S. et al. Genomic analysis and biological characterization of a novel Schitoviridae phage infecting Vibrio alginolyticus. Appl. Microbiol. Biotechnol. 107, 749–768. https://doi.org/10.1007/s00253-022-12312-3 (2023).
Bailly-Bechet, M., Vergassola, M. & Rocha, E. Causes for the intriguing presence of tRNAs in phages. Genome Res. 17, 1486–1495. https://doi.org/10.1101/gr.6649807 (2007).
Waldron, A., Wilcox, C., Francklyn, C. & Ebert, A. Knock-down of histidyl-tRNA synthetase causes cell cycle arrest and apoptosis of neuronal progenitor cells in vivo. Front. Cell Dev. Biol. 7, 67. https://doi.org/10.3389/fcell.2019.00067 (2019).
Ahmad, A. A., Addy, H. S. & Huang, Q. Biological and molecular characterization of a jumbo bacteriophage infecting plant pathogenic Ralstonia solanacearum species complex strains. Front. Microbiol. https://doi.org/10.3389/fmicb.2021.741600 (2021).
Shahin, K. et al. Bio-control of O157: H7, and colistin-resistant MCR-1-positive Escherichia coli using a new designed broad host range phage cocktail. LWT 154, 112836. https://doi.org/10.1016/j.lwt.2021.112836 (2022).
de Almeida Kumlien, A. C. M., Pérez-Vega, C., González-Villalobos, E., Borrego, C. M. & Balcázar, J. L. Genome analysis of a new Escherichia phage vB_EcoM_C2–3 with lytic activity against multidrug-resistant Escherichia coli. Virus Res. 307, 198623. https://doi.org/10.1016/j.virusres.2021.198623r (2022).
Moraru, C., Varsani, A. & Kropinski, A. M. VIRIDIC—A novel tool to calculate the intergenomic similarities of prokaryote-infecting viruses. Viruses 12, 1268. https://doi.org/10.3390/v12111268 (2020).
Turner, D., Kropinski, A. M. & Adriaenssens, E. M. A roadmap for genome-based phage taxonomy. Viruses 13, g506. https://doi.org/10.3390/v13030506 (2021).
Zhao, F. et al. Characterization and genome analysis of a novel bacteriophage vB_SpuP_Spp16 that infects Salmonella enterica serovar pullorum. Virus Genes 55(4), 532–540. https://doi.org/10.1007/s11262-019-01664-0 (2019).
Arkin, A. P. et al. KBase: The United States department of energy systems biology knowledgebase. Nat. Biotechnol. 36, 566–569. https://doi.org/10.1038/nbt.4163 (2018).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. https://doi.org/10.1093/bioinformatics/btu170 (2014).
Bankevich, A. et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. https://doi.org/10.1089/cmb.2012.0021 (2012).
Aziz, R. K. et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 9, 1–15. https://doi.org/10.1186/1471-2164-9-75 (2008).
Tynecki, P., Guziński, A., Kazimierczak, J., Jadczuk, M., Dastych, J. & Onisko, A. PhageAI-bacteriophage life cycle recognition with machine learning and natural language processing. BioRxiv. https://doi.org/10.1101/2020.07.11.198606 (2020).
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. https://doi.org/10.1093/nar/25.17.3389 (1997).
Jones, P. et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 30, 1236–1240. https://doi.org/10.1093/bioinformatics/btu031 (2014).
Mistry, J. et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 49, D412–D419. https://doi.org/10.1093/nar/gkaa913 (2021).
Lowe, T. M. & Chan, P. P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44, W54–W57. https://doi.org/10.1093/nar/gkw413 (2016).
Tillich, M. et al. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45, W6–W11. https://doi.org/10.1093/nar/gkx391 (2017).
Baerends, R. J. et al. Genome2D: A visualization tool for the rapid analysis of bacterial transcriptome data. Genome Biol. 5, 37. https://doi.org/10.1186/gb-2004-5-5-r37 (2004).
Sampaio, M., Rocha, M., Oliveira, H. & Dias, O. Predicting promoters in phage genomes using PhagePromoter. Bioinformatics 35, 5301–5302. https://doi.org/10.1093/bioinformatics/btz580 (2019).
Yi, H. et al. AcrFinder: Genome mining anti-CRISPR operons in prokaryotes and their viruses. Nucleic Acids Res. 48, W358–W365. https://doi.org/10.1093/nar/gkaa351 (2020).
Sayers, S. et al. Victors: A web-based knowledge base of virulence factors in human and animal pathogens. Nucleic Acids Res. 47, D693–D700. https://doi.org/10.1093/nar/gky999 (2019).
Chen, L. et al. VFDB: A reference database for bacterial virulence factors. Nucleic Acids Res. 33, D325–D328. https://doi.org/10.1093/nar/gki008 (2005).
McArthur, A. G. et al. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 57, 3348–3357. https://doi.org/10.1128/aac.00419-13 (2013).
Yukgehnaish, K. et al. PhageLeads: Rapid assessment of phage therapeutic suitability using an ensemble machine learning approach. Viruses 14, 342. https://doi.org/10.3390/v14020342 (2022).
Potter, S. C. et al. HMMER web server: 2018 update. Nucleic Acids Res. 46, W200–W204. https://doi.org/10.1093/nar/gky448 (2018).
Gontijo, M. T. P., Vidigal, P. M. P., Lopez, M. E. S. & Brocchi, M. Bacteriophages that infect Gram-negative bacteria as source of signal-arrest-release motif lysins. Res. Microbiol. 172, 103794. https://doi.org/10.1016/j.resmic.2020.103794 (2021).
Gontijo, M. T. P., Teles, M. P., Vidigal, P. M. P. & Brocchi, M. Expanding the database of signal-anchor-release domain endolysins through metagenomics. Probiotics Antimicrob. Proteins 14, 603–612. https://doi.org/10.1007/s12602-022-09948-y (2022).
Hallgren, J., Tsirigos, K. D., Pedersen, M. D., Armenteros, J. J. A., Marcatili, P., Nielsen, H., Krogh, A. & Winther, O. DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. bioRxiv. https://doi.org/10.1101/2022.04.08.487609 (2022).
Käll, L., Krogh, A. & Sonnhammer, E. L. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338, 1027–1036. https://doi.org/10.1016/j.jmb.2004.03.016 (2004).
Tsirigos, K. D., Peters, C., Shu, N., Käll, L. & Elofsson, A. The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res. 43, W401–W407. https://doi.org/10.1093/nar/gkv485 (2015).
Hirokawa, T., Boon-Chieng, S. & Mitaku, S. SOSUI: Classification and secondary structure prediction system for membrane proteins. Bioinformatics 14, 378–379. https://doi.org/10.1093/bioinformatics/14.4.378 (1998).
Zimmermann, L. et al. A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 430, 2237–2243. https://doi.org/10.1016/j.jmb.2017.12.007 (2018).
Gabler, F. et al. Protein sequence analysis using the MPI bioinformatics toolkit. Curr. Protoc. Bioinform. 72, e108. https://doi.org/10.1002/cpbi.108 (2020).
Darling, A. E., Mau, B. & Perna, N. T. ProgressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. PloS ONE 5, e11147. https://doi.org/10.1371/journal.pone.0011147 (2010).
Nishimura, Y. et al. ViPTree: The viral proteomic tree server. Bioinformatics 33, 2379–2380. https://doi.org/10.1093/bioinformatics/btx157 (2017).
Sullivan, M. J., Petty, N. K. & Beatson, S. A. Easyfig: A genome comparison visualizer. Bioinformatics 27, 1009–1010. https://doi.org/10.1093/bioinformatics/btr039 (2011).
Rangel-Pineros, G. et al. From trees to clouds: PhageClouds for fast comparison of∼ 640,000 phage genomic sequences and host-centric visualization using genomic network graphs. Phage 2, 194–203. https://doi.org/10.1089/phage.2021.0008 (2021).
Tamura, K., Stecher, G. & Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evolut. 38, 3022–3027. https://doi.org/10.1093/molbev/msab120 (2021).
Acknowledgements
The CHPC Lengau Cluster (Account No. CBBI1482) was used for data analysis.
Funding
Open access funding provided by North-West University. This research was financially supported by the National Research Foundation, South Africa, (Grant No. 138545), Higher Degrees Committee of the Faculty of Natural and Agricultural Sciences, and Post-doctoral fellowship from the North-West University.
Author information
Authors and Affiliations
Contributions
P.K.M., C.N.A.; Conceptualization, P.K.M., A.M.K., F.M., R.E.P.; Data curation, P.K.M., A.M.K., F.M., R.E.P.; Formal Analysis, P.K.M., C.N.A.; Funding acquisition, P.K.M.; Investigation (Wet lab), () P.K.M., A.M.K., F.M., R.E.P.; Bioinformatics and sequences analysis, P.K.M.; Methodology, P.K.M., A.M.K., F.M., R.E.P.; Software, P.K.M., M.V.; Statistical analysis, P.K.M., A.M.K., F.M., R.E.P., M.V, C.N.A.; Validation, P.K.M., A.M.K., F.M., R.E.P.; Visualization, P.K.M.; Writing—original draft; P.K.M., A.M.K., F.M., R.E.P.,M.V, C.N.A.; Writing—review & editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Montso, P.K., Kropinski, A.M., Mokoena, F. et al. Comparative genomics and proteomics analysis of phages infecting multi-drug resistant Escherichia coli O177 isolated from cattle faeces. Sci Rep 13, 21426 (2023). https://doi.org/10.1038/s41598-023-48788-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-48788-w
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.