Abstract
Hepatocellular carcinoma (HCC) is an inflammatory problematic issue with higher mortality among different ethnic populations. The telomerase reverse transcriptase (TERT) gene has an imperative role in the proliferation of various cancerous illnesses, particularly HCC. Moreover, the TERT (rs2736098 and rs2739100) variants were correlated with the HCC susceptibility and telomere shortening, but with unconvincing outcomes. The main purpose of this outward work is to assess the correlation between these significant variants within the TERT gene and the elevated risk of HCC with the aid of various computational bioinformatics tools. This study included 233 participants [125 cancer-free controls and 108 HCC patients] from the same locality. In addition, 81.5% of HCC patients were positive for HCV autoantibodies, while 73.1% of HCC patients were positive for cirrhotic liver. Genomic DNA of the TERT (rs2736098 and rs2736100) variants were characterized utilizing the PCR–RFLP method. Interestingly, the frequencies of TERT (rs2736098*A allele) and TERT (rs2736100*T allele) conferred a significant correlation with increased risk of HCC compared to healthy controls (p-value = 0.002, and 0.016, respectively). The TERT (rs2736098*A/A) genotype indicated a definite association with positive smoking and splenomegaly (p-value < 0.05), while the TERT (rs2736100*T/T) genotype observed a significant difference with higher levels of HCV autoantibodies (p-value = 0.009). In conclusion, this significant work confirmed the contribution of the TERT (rs2736098*A and rs2736100*T) alleles with elevated risk of HCC progression and telomere shortening among Egyptian subjects.
Similar content being viewed by others
Introduction
Hepatic cancer is the seventh incidence cancer distributed worldwide occupying about 8.7% of cancer-related deaths with higher mortality recorded within developing countries1,2. Universally, hepatocellular carcinoma (HCC) is a global issue that requires a particular strategy to decline the mortality percentages associated with elevated incidence of this type of carcinoma3,4. The awareness of the molecular basis of HCC development is essential to recognize the pathogenesis mechanisms that correlate with the adverse effects of this type of hepatic-associated cancer, (Fig. 1)5,6. In Egypt, HCC is the second cancer distributed locally compared to other carcinomas, with incidence rates of 20.7% of the total cancerous patients identified among Egyptian subjects2,7. In addition, numerous reports with technological improvements indicated the potential impact of genetic background on the pathogenesis of hepatic carcinogenesis8,9,10.
The possible pathogenesis mechanisms for the HCC development with telomere length. The TERT gene encoded a ribonuclease telomerase enzyme that maintained the chromosomal ends from attrition by the addition of repetitive nucleotide units (TTAGGG) at the 3′ prime ends of the genome. The immortal cancerous tissues in the hepatocytes prevent cellular apoptosis by increasing the telomerase expression at early-onset HCC and decreasing the telomerase expression at late-onset HCC and thus enhancing the telomere shortening. The TERT (rs2736098; A allele) and TERT (rs2736100; T allele) were associated with telomere shortening and increased risk of hepatocellular carcinoma. Abbreviations: HCC, Hepatocellular carcinoma; TERT, telomerase reverse transcriptase.
The human telomerase reverse transcriptase (TERT) gene has a crucial function in the proliferation of different numbers of cancerous illnesses, particularly hepatic carcinoma6,11,12. The TERT gene (OMIM#: 187270, other synonyms include TRT, TP2, hEST2, and EST2) is located along the reverse strand on chromosome 5p15.33 and comprised of seven splice variants with the main transcript (TERT-201; ENST00000310581.10) that included 16 exons and 15 introns encoding a powerful ribonuclease enzyme which is referred as telomerase with 1132 amino acids13,14,15. This ribonuclease telomerase enzyme is a multicomplex protein with two main parts: telomerase reverse transcriptase (TERT) along with its core template, telomerase RNA component16,17. Furthermore, the telomerase has a significant function in the maintenance of the integrity of small structural elements at the terminal of the chromosomes that are called telomeres through the addition of repetitive nucleotide units (TTAGGG) at the 3′ prime ends of the genome18,19. These terminal sequences of telomeres have starring roles in the capping of the chromosomal terminus to prevent fusion and recombination of the chromosomes8,20.
Moreover, the attrition of these repeated units through cell cycle, genome replication, and aging could increase the levels of genomic instability, cell arresting, and apoptosis21,22. Nevertheless, the immortal cancerous tissues could overwhelm this inevitable fate by stimulating the expression rate of telomerase to lengthen the telomere structures at the chromosomal terminus16,23. Two potential genetic variants within the TERT gene involving TERT*(rs2736098; c.915G > A) and TERT*(rs2736100; c.1574-3777G > T) have a critical role in the elevated risk of several cancerous disorders including rectal cancer24, renal cell carcinoma19, prostate cancer25, and hepatocellular carcinoma26,27,28, but with inconclusive conclusions.
Intriguingly, hepatic cancer is considered a hyperinflammatory problem with adverse complications that could lead to cellular apoptosis and genomic variations 5,8. During the late-onset HCC, the immortal tumor tissues could decrease the telomerase activity within the hepatic tissues leading to telomere shortening and liver cirrhosis 29,30. Moreover, limited reports clarified the contributions of the synonymous TERT*(rs2736098*A allele) and the intron TERT*(rs2736100*T allele) variants with telomere shortening and increased risk of HCC progression among different ethnic populations 6,26,31. Due to the limited data concerning the role of the TERT gene in the progression of HCC, our team was motivated to design this research to estimate the association of these significant variants within the TERT gene and the HCC susceptibility among Egyptian subjects with the practice of various computational bioinformatics tools.
Materials and methods
Ethical statement approval
This work was authorized and achieved an approval announcement from the ethical committee board [IRB #: R.22.10.1899] at the Faculty of Medicine, Mansoura University, Egypt. In addition, the study protocol commenced according to the declaration guidelines of Helsinki recommendations. All enrolled subjects were asked to allocate an informed written consent upon participation in this work.
Study participants
This retrospective case–control study included a sum of 233 participants: comprising 108 HCC patients [81.5% males, and 18.5% females], together with 125 unrelated cancer-free controls [73.6% males, and 26.4% females] that matched with age, gender, and geographical district. All HCC patients were collected from the Oncology Clinics at Gastrointestinal Surgery Center, Mansoura University, Egypt, as formerly described5. The diagnostic criteria for the selection of HCC patients were executed based on imaging techniques involving computerized tomography (CT) and/or magnetic resonance imaging (MRI)32. Furthermore, HCC patients who experienced previous history of malignancies, autoimmune disorders, hepatic problems, renal diseases, diabetic mellitus, alcohol abuse, metabolic diseases, and/or other endocrine issues were omitted from this research. The clinical and demographic characteristics were extracted from medical archives.
Sample collection and genomic DNA extraction
Under complete aseptic conditions, a total of 10 ml of venous blood was gathered and obtained from all participants in this study. The blood samples were sectioned into two aliquots; the first portion was collected in a vacutainer tube containing EDTA anticoagulant for hematological and genetic purposes, while the subsequent portion was processed within serum separator tubes and subjected to centrifugation to separate serum aliquot for biochemical and serological measurements. Furthermore, the identification of biochemical assessments was performed using Roche Cobas c501 biochemical analyzer (Roche Diagnostics, Manheim, Germany), while the evaluation of serological examinations was carried out utilizing ADVIA Centaur® CP Immunoassay system (Siemens Healthineers, Germany) including alpha-fetoprotein (AFP), and hepatitis C virus autoantibodies (Anti-HCV). Additionally, the estimation of hematological parameters was substantiated using Abbott CELL-DYN 3700 SL hematology analyzer (Abbott Diagnostics, USA)33.
Genomic DNA extraction and purification
The manipulation of genomic DNA from peripheral EDTA blood was extracted utilizing Mini kit GeneJET Whole Blood Genomic DNA (Thermo Fischer Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. The degree of purity of extracted genomic DNA was evaluated utilizing the NanoDrop™ ND-1000 Spectrophotometer technology34.
Amplification of the TERT*(rs2736098; c.915G > A) variant
The genotyping of the TERT*(rs2736098; c.915G > A) and TERT*(rs2736100; c.1574-3777G > T) variants were executed using polymerase chain reaction with restriction fragment length polymorphism (PCR–RFLP) through pertaining enzymatic cleavage technique35,36. For the TERT*(rs2736098; c.915G > A) variant, the two primers used were [forward primer (F1): 5′- AGG ACG CGT GGA CCG AGT GAC-3′, and reverse primer (R1): 5′-GGA ACC CAG AAA GAT GGT CTC-3′, respectively]36. The PCR protocol was processed within a thermal cycler in total volume of 28 µl comprising of 4 µl of template genomic DNA, 4 µl of forward primer, 4 µl of reverse primer, 12.5 µl of 2X PCR Master Mix, 3.5 µl nuclease-free sterile water (ddH2O). The reaction conditions were computed with an initial denaturation step at 95 °C for 5 min, followed by 30 repetitive cycles of 95 °C for 30 s, 67 °C for 30 s, and 72 °C for 28 s, and a last extension step at 72 °C for 10 min. The amplified product of the TERT*(rs2736098; c.915G > A) variant generated a 324-bp fragment that was subjected to endonuclease cleavage by a restriction enzyme Bsp120I/PspOMI (New England Biolabs, Ipswich, MA, USA) at 37 °C for 60 min and could identify a cutting site (G/GGCCC). The G allele was digested and produced two fragments at 137-bp and 187-bp, while the undigested A allele generated a single band at 324-bp, (Figure S1)36.
Amplification of the TERT*(rs2736100; c.1574-3777G > T) variant
Alternatively, the two primers used in the amplification of TERT*(rs2736100; c.1574-3777G > T) variant were [forward primer (F2): 5′-GCT GTT TTC CCT GCT GAC TT-3′, and reverse primer (R2): 5′-AGA ACC ACG CAA AGG ACA AG-3′, respectively] 35. Moreover, the reaction protocol was handled in a total volume of 30 µl comprising 4 µl of template genomic DNA, 4 µl of forward primer, 4 µl of reverse primer, 12.5 µl of 2X PCR Master Mix, 5.5 µl nuclease-free sterile water (ddH2O). The PCR conditions were adjusted with an initial denaturation step at 94 °C for 5 min, followed by 35 repetitive cycles of 94 °C for 30 s, 59.5 °C for 30 s, and 72 °C for 30 s, and a last extension step at 72 °C for 7 min. The amplified fragment for the TERT*(rs2736100; c.1574-3777G > T) variant produced a 194-bp band that is exposed to endonuclease cleavage by a restriction enzyme SfcI (New England Biolabs, Ipswich, MA, USA) at 37 °C for 60 min and could distinguish a cutting site (C/TRYAG). The digested fragments could generate two fragments for the wildtype G allele at 101-bp and 93-bp, while the rare T allele remained undigested with a single fragment at 194-bp, (Figure S2)35. The digested fragments were electrophoresed utilizing 2.5% agarose gel including 0.5 μg/ml ethidium bromide to facilitate the photographing process with UV illumination. Additionally, about one-tenth of the samples were independently subjected to repeat genotyping to achieve a degree of quality assurance under strict blind conditions and the corresponding results were acceptable with a percentage of 100%.
Statistical analysis
In the beginning, the manipulation and tabulation of data extracted from this survey were accomplished using Stata Statistical Software, Release #17 (StataCorp. 2021, College Station, TX: LLC). The calculation of sample size of this presented work was computed utilizing the G*power software version 3.1.9.7, suggesting that the observed power (1–β error probability) attained a value of 82% with an effect size of 0.2433. The Shapiro–Wilk test was managed to assess the normality of the quantitative variables that were presented as median (IQR) for the nonparametric data. The allelic/genotypic distributions of the TERT*(rs2736098 and rs2736100) variants were computed utilizing Fisher's exact and Chi-squared test, as formerly described37. The web-established tool SNP analysis was conducted for the calculation of the Odds ratios (OR) and 95% confidence interval (CI) for the genetic models and haplotypic frequencies of the TERT*(rs2736098 and rs2736100) variants (www.snpstats.net/start.htm; accessed on 27 July 2022)38. According to the distribution of data types, the recessive model of the TERT (rs2736098*A/A and rs2736100*T/T) variants was analyzed with different clinical and laboratory parameters using Chi-square and two-sample Wilcoxon rank-sum tests. Moreover, the multivariate analysis including principal component analysis and correlation matrix was accomplished with the aid of R programming language software version 4.2.0 with R studio version 2022.07.1 Build 55439, while the meta-analysis design was established using Stata Statistical Software, Release #17. The level of statistical analysis was considered significantly beneficial with p-value < 0.05.
Ethical approval
All procedures performed in studies involving human participants were subjected to following the ethical standards of the institutional research committee and with the 1964 Helsinki Declaration and its later amendments.
Results
The fundamental characteristics of the study population
This study included 233 participants that clustered into two subgroups comprising 108 HCC patients with a median age (IQR) was 53.0 (45.0–61.5) years, together with 125 cancer-free controls from the same locality with a median age (IQR) was 54.0 (49.0–58.0) years. There was no remarkable correlation among HCC patients compared to control subjects regarding age, gender, consanguinity, family history, cirrhotic liver, hypertension, ascites status, and splenomegaly (p-value > 0.05). Interestingly, HCC patients with positive smoking and high weights revealed a statistically significant compared with control subjects (p-value < 0.05). Based on viral infection, 88 HCC patients (81.5%) were positive for HCV autoantibodies, while 79 HCC patients (73.1%) were positive due to cirrhotic issues. Furthermore, higher levels of ALT, AST, total bilirubin, INR, AFP, and HCV autoantibodies were observed among HCC patients compared with control subjects (p-value < 0.001). Absolutely not, lower concentrations of albumin, RBCs, WBCs, hemoglobin, hematocrit, and platelets count were noticed among HCC patients compared with control subjects (p-value < 0.001) (Table 1).
The genotypic and allelic frequencies of the TERT variants
The expected and observed frequencies of the TERT*(rs2736100; c.1574-3777G > T) variant for cancer-free controls and HCC patients were in alignment with the Hardy–Weinberg equilibrium (HWE) [p-value > 0.05], while those of the TERT*(rs2736098; c.915G > A) variant were mismatched with HWE among HCC patients [p-value = 0.02], and this could be imputed to the higher frequency of the TERT (rs2736098*G/A) genotype. Interestingly, the mutant genotypes of the TERT (rs2736098*A/A and rs2736100*T/T) were considerably elevated among HCC patients compared with control subjects [TERT (rs2736098*A/A): 16.7% vs. 8.0%, OR = 3.97, 95% CI = 1.60–9.89, p-value = 0.003; and TERT (rs2736100*T/T): 35.2% vs. 22.4%, OR = 2.30, 95% CI = 1.13–4.68, p-value = 0.023, respectively], (Table 2). Furthermore, the minor alleles of the TERT (rs2736098*A allele and rs2736100*T allele) were substantially associated with elevated risk of HCC compared with cancer-free controls [TERT (rs2736098*A allele): 47.2% vs. 32.8%, OR = 1.83, 95% CI = 1.26–2.67, p-value = 0.002; and TERT (rs2736100*T allele): 56.9% vs. 45.6%, OR = 1.58, 95% CI = 1.09–2.28, p-value = 0.016, respectively], (Fig. 2A & 2B). Besides that, the combined genotypes of TERT (rs2736098*G/A + rs2736100*T/T) and TERT (rs2736098*A/A + rs2736100*G/G) were statistically significant with an elevated risk of HCC compared to cancer-free controls [p-value = 0.004 and 0.047, respectively], (Fig. 2C). Upon establishing haplotype analysis of TERT*(rs2736098 and rs2736100) variants, our team identified a perceptible association of TERT (rs2736098*A and rs2736100*T) haplotypes among HCC patients compared with cancer-free controls [24.5% vs. 14.8%, OR = 1.87, 95% CI = 1.17–2.98, p-value = 0.009], (Table 2). Surprisingly, the minor allele frequency (MAF) among cancer-free controls for the TERT (rs2736098*A allele) was 0.33, while the minor allele frequency for the TERT (rs2736100*T allele) was 0.46. Based on the final phase of the 1000 genome project, the minor allele frequency of TERT (rs2736098*A allele) was matched with similar observations among East Asian (0.37) subjects, while those of TERT (rs2736100*T allele) was correlated with European (0.50) subjects, (Fig. 2D).
Allelic and genotypic distributions of the study population. (A): The allelic and genotypic frequencies of the TERT (rs2736098; c.915G > A) variant among HCC patients compared with cancer-free controls. (B): The allelic and genotypic frequencies of the TERT (rs2736100; c.1574-3777G > T) variant among HCC patients compared with cancer-free controls. (C): The combined genotypic frequencies of The TERT (rs2736098 and rs2736100) variants among HCC patients compared with cancer-free controls. (D) The 1000 Genome Project Phase 3 of the allelic frequencies for the present study compared with various ethnic populations (https://www.internationalgenome.org/). Abbreviations: AFR, Africa; AMR, America; EUR, Europe; EAS, East Asia; SAS, South Asia; HCC, Hepatocellular carcinoma.
Association of TERT variants with the susceptibility for HCC
Essentially, the distributions of the TERT*(rs2736098; c.915G > A) variant were significantly associated with elevated risk of HCC compared with control subjects under dominant [OR = 2.54, 95% CI = 1.43–4.53, P-value = 0.001], recessive [OR = 2.31, 95% CI = 1.01–5.27, P-value = 0.042], and homozygote [OR = 3.95, 95% CI = 1.58–9.85, P-value = 0.002] models. Moreover, the frequencies of the TERT*(rs2736100; c.1574-3777G > T) variant revealed a statistical difference with increased risk of HCC compared to control subjects under recessive [OR = 1.84, 95% CI = 1.03–3.29, P-value = 0.039] model, (Table 3).
Association of TERT variants with the clinical and laboratory variables
Remarkably, the TERT*(rs2736098; c.915G > A) variant was significantly correlated with higher levels of ALT, AST, and creatinine, (Fig. 3). Additionally, the TERT (rs2736098*A/A) genotype revealed a noticeable association with positive smoking and splenomegaly (p-value < 0.05). Alternatively, the TERT (rs2736100*T/T) genotype identified a significant association with elevated levels of HCV autoantibodies (p-value = 0.009), (Table S1).
Multivariate analysis of TERT variants
Firstly, our team performed the principal component analysis (PCA) that clustered the study participants into two categories involving cancer-free controls and HCC patients, (Fig. 4A). The PCA technique revealed an obvious distinction between the two groups with elongated arrows exhibiting a significant impact on the clinical, serological, laboratory, and hematological variables. By observation, the TERT*(rs2736098; c.915G > A) and TERT*(rs2736100; c.1574-3777G > T) variants exhibited a significant difference with an elevated risk of hepatocellular carcinoma. On the other hand, the correlation matrix was carried out utilizing Pearson’s correlation test to assess the conventional possibilities among HCC patients for the clinical characteristics, and laboratory investigations, (Fig. 4B).
Multivariate analysis of the TERT (rs2736098 and rs2736100) variants. (A): Principal component analysis (PCA) of the study participants revealed a definite distinction between HCC patients and cancer-free controls. Elongated arrows represented elevated impact of clinical, serological, and laboratory investigations among the study participants. Visually, the TERT (rs2736098; c.915G > A) and TERT (rs2736100; c.1574-3777G > T) variants exhibited a significant association with elevated risk of hepatocellular carcinoma. (B): Correlation matrix using Pearson’s correlation test of the clinical, serological measurements, and laboratory variables among HCC patients.
In silico data analysis using computational bioinformatics tools
The chromosomal localization of the TERT gene (ENSG00000164362) is positioned on 5p15.33 and spanned about 41,922 bases [Chr.5: 1253147–1295068] that is orientated along the minus strand. The genomic localization of the TERT gene comprised of seven splice variants with the main transcript (TERT-201) including 16 exons and 15 introns, with 1132 amino acids. Besides that, the TERT*(rs2736098; c.915G > A) variant is located on the second exon (Chr.5: 1293971), while the TERT*(rs2736100; c.1574-3777G > T) variant is situated on the second intron (Chr.5: 1286401) (Fig. 5). The somatic mutation frequency of the TERT gene that was extracted from several databases was 7.9%, indicating the presence of the TERT*(rs2736098; p.Ala305 =) variant at the second exon of the gene. The crystalline structure and amino acid residues of the TERT protein (O14746-1) and its domains representing the location of the TERT (rs2736098; p.Ala305 =) [Data source: Protter database, UniProt database, and Protein Data Bank]. Moreover, the Kaplan–Meier plotter database of hepatic cancer indicates elevated TERT expression with a worse prognosis (p-value = 0.032). The gene–gene associations of the TERT gene revealed the contribution of this gene in telomere maintenance, telomere lengthening, cell aging, and protein localization to chromosomes. The protein–protein frameworks identified the potential role of the TERT protein in telomere maintenance, protein localization to telomeric region, telomerase activity, transcription factor binding, and telomere extension by telomerase. The subcellular localizations of the TERT protein suggest high levels of profusion in the nucleus, cytosol, mitochondrion, and cytoskeleton. Additionally, mutual exclusive analysis of the TERT expression within different cancerous diseases implicated the higher rate of liver hepatocellular carcinoma. Lastly, our team identified the crucial role of the TERT gene in the development of cancer hallmarks.
The computational bioinformatic tools of the TERT gene. (A): The chromosomal localization and genomic structure of the TERT gene. It is positioned on the chromosome number 5p15.33 and spanned about 41,922 bases [Chr.5:1253147–1295068] that is orientated along the minus strand. The genomic structure of TERT gene identified that is comprised of seven splice transcripts with the main one (TERT-201) including 16 exons and 15 introns. (B): Lollipop diagram of the TERT gene mutations derived from several databases for studies indicating the presence of different somatic mutation with a frequency of 7.9% and observing the TERT (rs2736098; p.Ala305 =) variant at the second exon of the gene. (C): The crystalline structure of human TERT protein using Protein Data Bank. (D): Amino acid residues of the TERT protein (O14746-1) and its domains showing the locations of the TERT (rs2736098; c.915G > A; p.Ala305 =). (E): Survival analysis during high and low TERT expression. (F): Gene–gene interactions of the TERT gene using GeneMANIA database. (G): Protein–protein interactions of the TERT protein using the STRING database. (H): Subcellular localization of the TERT protein, with darker colors suggesting further abundance. (I): Mutual exclusive analysis of the TERT expression in different cancerous diseases. (J): TERT is involved in the development of cancer hallmarks. [Data source: Ensembl.org, NCBI database, UniProt database, Compartment database, Protter database, Kaplan‐Meier plotter database, GeneMania, STRING version 11.0, cBioPortal cancer Genomics database, Cancer Hallmarks Analytics tool, and Protein Data Bank].
Discussion
Recently, three crucial factors were thought to control the telomere lengthening involving telomerase activity, high mitotic rate, and telomerase-dependent variables6,40. However, the ability of the TERT gene to encode a regulatable enzyme that controls the telomerase expression and telomere lengthening through inserting repetitive nucleotide units (5′-TTAGGG-3′) at the chromosomal terminus could determine the proliferation level of carcinoma 18,24. Upon thorough literature searching for the reports identifying the association of the TERT*(rs2736098 and rs2736100) variants with the progression of hepatocellular carcinoma, our team confirmed that this work is the innovative one for testing the correlation of the TERT*(rs2736100; c.1574-3777G > T) variant with elevated risk of HCC among Egyptian subjects.
Remarkably, our findings revealed a significant difference for the TERT*(rs2736098; c.915G > A) variant and elevated risk of HCC under allelic, recessive, and dominant models (OR = 1.83, 2.31, and 2.54, respectively). Obviously, HCC patients carrying TERT (rs2736098*AA) genotype exposed a statistically significant for positive smoking, and levels of AST/ALT compared to control subjects (p-value < 0.05). On the other hand, the TERT*(rs2736100; c.1574-3777G > T) variant was identified significantly associated with elevated risk of HCC under allelic (rs2736100*T), and recessive (rs2736100*TT) models (OR = 1.58, and 1.84, respectively). Additionally, HCC patients carrying the TERT (rs2736100*TT) genotype conferred a significant difference with higher levels of autoantibodies for anti-HCV compared to control subjects (p-value = 0.009). Interestingly, the higher levels of ALT, AST, and HCV autoantibodies with HCC development were confirmed and studied in previous reports41,42,43. Several meta-analysis reports investigated the association of the TERT gene variants with the susceptibility for hepatocellular carcinoma, but with challenging interpretations6,18,44. A recent meta-analysis study indicated the contribution of the TERT*(rs2736098; c.915G > A) variant with the susceptibility for HCC among overall subjects under the dominant model (GA + AA vs. GG; OR = 1.38)44. On the contrary, a meta-analysis report revealed a strong association between the TERT*(rs2736098 and rs2736100) variants with increased risk of different types of carcinomas among overall subjects6. However, they failed to indicate a significant correlation between the TERT*(rs2736098; c.915G > A) and TERT*(rs2736100; c.1574-3777G > T) variants and the HCC progression among Asian and Caucasian subjects6. Another meta-analysis study confirmed no evidence for the association between TERT*(rs2736100; c.1574-3777G > T) variant and the susceptibility of different digestive cancers under various genetic models within Asian and Caucasian subjects18.
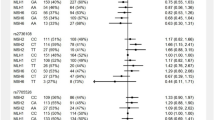
Owing the inconsistency of findings, our team was motivated to construct a systematic comparison for all the articles investigated the correlation of the TERT*(rs2736098 and rs2736100) variants with the HCC susceptibility among different ethnic groups (Table 4). Interestingly, two comparisons identified a significant association between the TERT*(rs2736098; c.915G > A) variant and elevated risk of HCC among Chinese subjects (OR = 1.88 and 1.59, respectively)45,46. On the other hand, three reports confirmed no significant difference for the TERT*(rs2736098; c.915G > A) variant and increased risk of HCC among Chinese subjects26,28, and Egyptian subjects27 under different genetic models. Limited data is available concerning the contribution of the TERT*(rs2736100; c.1574-3777G > T) variant with the progression of HCC among various ethnic groups. A single report done among Chinese subjects revealed a statistically significant for the TERT*(rs2736100; c.1574-3777G > T) variant with increased risk of HCC under dominant model (OR = 1.61, p-value = 0.047)26. Conversely, another study among Chinese subjects observed no indication of association for the TERT*(rs2736100; c.1574-3777G > T) variant with the HCC progression under dominant and recessive models28. As exemplified in the forest plots shown in (Figure S3) for the pooled data concerning the association between TERT*(rs2736098 and rs2736100) variants and HCC progression, our team revealed a strong association for the TERT*(rs2736098; c.915G > A) variant with elevated risk of HCC under dominant model (OR = 1.37, p-value = 0.041).
Upon precise searching for online electronic databases that could predict the potential impact of the intronic variant of the TERT*(rs2736100; c.1574-3777G > T) gene using RegulomeDB annotation for regulatory elements in non-coding regions, we could predict that this variant has a probability score equal to 0.1347. Another predicted tool for exploring annotations of the noncoding variants is HaploReg that extracted their information from linkage disequilibrium (LD) of the final phase of 1000 genome project 48. Interestingly, the TERT*(rs2736100; c.1574-3777G > T) variant revealed linkage disequilibrium with (r2 ≥ 0.8) and could induce the expression of transcriptional factor within the Foxa subfamily of Forkhead box protein A (Foxa) factors49. These Foxa factors also are called as hepatocyte nuclear factor 3 (HNF3) that have a potential function in hepatic specification through controlling in the hepatic organogenesis and repositioning chromatin and nucleosomes49,50. From the above-mentioned outcomes, we could consider that the TERT (rs2736098*A allele) and TERT (rs2736100*T allele) variants were correlated with elevated risk of HCC and telomere shortening in the late onset of the disease. Lastly, minor limitations were recognized during the establishment of this research including the relatively small sample size, formulating this work among Egyptian subjects, the case–control design, the lack of functional analysis, and the absence of accounting for potential confounding factors. Replication and validation of the findings in larger and more diverse cohorts, along with functional studies and control of confounding variables, would further strengthen the upcoming studies. Overall, this study contributes to the understanding of the potential role of TERT gene variants in HCC susceptibility among Egyptian subjects, highlighting the need for further research in this area.
Conclusion
This work indicated the potential role of the TERT (rs2736098*A and rs2736100*T) alleles with elevated risk of HCC among Egyptian patients. Interestingly, the TERT (rs2736098*A) and TERT (rs2736100*T) alleles were correlated with telomere shortening in the late onset of the disease. Thus, the TERT*(rs2736098; c.915G > A) and TERT*(rs2736100; c.1574-3777G > T) variants could be thought as independent risk factors with the progression of HCC.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
McGlynn, K. A., Petrick, J. L. & El-Serag, H. B. Epidemiology of Hepatocellular Carcinoma. Hepatology 73(Suppl 1), 4–13. https://doi.org/10.1002/hep.31288 (2021).
Sung, H. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 71, 209–249. https://doi.org/10.3322/caac.21660 (2021).
Zhou, S. L. et al. Whole-genome sequencing reveals the evolutionary trajectory of HBV-related hepatocellular carcinoma early recurrence. Signal Transduct. Target. Ther. 7, doi:https://doi.org/10.1038/s41392-021-00838-3 (2022).
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin. 68, 7–30. https://doi.org/10.3322/caac.21442 (2018).
Saad, A. M., Abdel-Megied, A. E. S., Elbaz, R. A., Hassab El-Nabi, S. E. & Elshazli, R. M. Genetic variants of APEX1 p.Asp148Glu and XRCC1 p.Gln399Arg with the susceptibility of hepatocellular carcinoma. J Med Virol 93, 6278–6291. https://doi.org/10.1002/jmv.27217 (2021).
Zhang, X., Chen, Y., Yan, D., Han, J. & Zhu, L. TERT Gene rs2736100 and rs2736098 polymorphisms are associated with increased cancer risk: A meta-analysis. Biochem. Genet. 60, 241–266. https://doi.org/10.1007/s10528-021-10097-0 (2022).
Ezzat, R., Eltabbakh, M. & El Kassas, M. Unique situation of hepatocellular carcinoma in Egypt: A review of epidemiology and control measures. World J. Gastrointest. Oncol. 13, 1919–1938. https://doi.org/10.4251/wjgo.v13.i12.1919 (2021).
Ningarhari, M. et al. Telomere length is key to hepatocellular carcinoma diversity and telomerase addiction is an actionable therapeutic target. J. Hepatol. 74, 1155–1166. https://doi.org/10.1016/j.jhep.2020.11.052 (2021).
Dratwa, M., Wysoczańska, B., Łacina, P., Kubik, T. & Bogunia-Kubik, K. TERT—Regulation and Roles in Cancer Formation. Front. Immunol. 11. https://doi.org/10.3389/fimmu.2020.589929 (2020).
Amer, T. et al. Genetic polymorphisms of IL-23R (rs7517847) and LEP (rs7799039) among Egyptian patients with hepatocellular carcinoma. Arch. Physiol. Biochem. 123, 279–285. https://doi.org/10.1080/13813455.2017.1320680 (2017).
Jang, J.-W. et al. Significance of TERT genetic alterations and telomere length in hepatocellular carcinoma. Cancers 13, 2160 (2021).
Robinson, N. J. & Schiemann, W. P. Telomerase in cancer: Function, regulation, and clinical translation. Cancers. https://doi.org/10.3390/cancers14030808 (2022).
Wu, S. et al. Telomerase RNA TERC and the PI3K-AKT pathway form a positive feedback loop to regulate cell proliferation independent of telomerase activity. Nucleic Acids Res. 50, 3764–3776. https://doi.org/10.1093/nar/gkac179 (2022).
Nguyen, E., Richerolle, A., Sánchez-Bellver, J., Varennes, J. & Ségal-Bendirdjian, E. hTERT DNA methylation analysis identifies a biomarker for retinoic acid-induced hTERT repression in breast cancer cell lines. Biomedicines https://doi.org/10.3390/biomedicines10030695 (2022).
Yu, J., Li, X., Zhou, B. & Yan, A. Polymorphisms of the TERT-CLPTM1L gene are associated with pharynx-larynx cancer. DNA Cell Biol. 38, 915–921. https://doi.org/10.1089/dna.2019.4744 (2019).
Yang, L. et al. Tumorigenic effect of TERT and its potential therapeutic target in NSCLC (Review). Oncol. Rep. https://doi.org/10.3892/OR.2021.8133 (2021).
Colebatch, A. J., Dobrovic, A. & Cooper, W. A. TERT gene: its function and dysregulation in cancer. J. Clin. Pathol. 72, 281–284. https://doi.org/10.1136/jclinpath-2018-205653 (2019).
Song, H. et al. Association between the rs2736100 polymorphisms of telomerase reverse transcriptase gene and digestive cancers: A meta-analysis. J. Cancer Res. Ther. 17, 1202–1208. https://doi.org/10.4103/jcrt.jcrt_1102_21 (2021).
Ma, R. et al. The TERT locus genotypes of rs2736100-CC/CA and rs2736098-AA predict shorter survival in renal cell carcinoma. Urol. Oncol. Semin. Orig. Invest. 37(301), e301-301.e310. https://doi.org/10.1016/j.urolonc.2019.01.014 (2019).
Yamaguchi, H. et al. Mutations in TERT, the gene for telomerase reverse transcriptase, in aplastic anemia. N. Engl. J. Med. 352, 1413–1424. https://doi.org/10.1056/NEJMoa042980 (2005).
Furuie, H., Arimura-Omori, M., Hamada, N., Yanagihara, T. & Kiyohara, C. The association of aging-related polymorphisms with susceptibility to lung cancer: A case-control study in a Japanese population. Asian Pac. J. Cancer Preven. 22, 1279–1285. https://doi.org/10.31557/APJCP.2021.22.4.1279 (2021).
Yuan, X., Larsson, C. & Xu, D. Mechanisms underlying the activation of TERT transcription and telomerase activity in human cancer: old actors and new players. Oncogene 38, 6172–6183. https://doi.org/10.1038/s41388-019-0872-9 (2019).
Kim, N. W. et al. Specific association of human telomerase activity with immortal cells and cancer. Science 266, 2011–2015. https://doi.org/10.1126/science.7605428 (1994).
Rampazzo, E. et al. Genetic variants of the TERT gene, telomere length, and circulating tert as prognostic markers in rectal cancer patients. Cancers 12, 1–15. https://doi.org/10.3390/cancers12113115 (2020).
Shadrina, A. S. et al. TERT polymorphisms rs2853669 and rs7726159 influence on prostate cancer risk in Russian population. Tumor Biol. 36, 841–847. https://doi.org/10.1007/s13277-014-2688-0 (2015).
Yuan, X. et al. The TERT promoter mutation incidence is modified by germline TERT rs2736098 and rs2736100 polymorphisms in hepatocellular carcinoma. Oncotarget 8, 23120–23129. https://doi.org/10.18632/oncotarget.15498 (2017).
Sharaf, A., Elhalafawy, I. A., Abdel Aziz, A. A. & Sakr, M. A. Association of p53 codon 72 polymorphism and hTERT polymorphism (rs2736098) with risk of hepatocellular carcinoma. A pilot study in Egyptian patients. Gene Rep. https://doi.org/10.1016/j.genrep.2021.101255 (2021).
Ding, C. Y., Hu, L. M., Hu, Z. B. & Shen, H. B. The relationship between gene polymorphism of telomerase reverse transcriptase and susceptibility to hepatocellular carcinoma. Zhonghua Yu Fang Yi Xue Za Zhi 45, 593–596 (2011).
In der Stroth, L., Tharehalli, U., Gunes, C. & Lechel, A. Telomeres and Telomerase in the Development of Liver Cancer. Cancers. https://doi.org/10.3390/cancers12082048 (2020).
Urabe, Y. et al. Telomere length in human liver diseases. Liver 16, 293–297. https://doi.org/10.1111/j.1600-0676.1996.tb00748.x (1996).
Rafnar, T. et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat. Genet. 41, 221–227. https://doi.org/10.1038/ng.296 (2009).
Marrero, J. A. et al. Diagnosis, staging, and management of hepatocellular carcinoma: 2018 Practice guidance by the American association for the study of liver diseases. Hepatology 68, 723–750. https://doi.org/10.1002/hep.29913 (2018).
Galal, A. A., Abd Elmajeed, A. A., Elbaz, R. A., Wafa, A. M. & Elshazli, R. M. Association of apolipoprotein E gene polymorphism with the risk of T2DM and obesity among Egyptian subjects. Gene 769, 145223. https://doi.org/10.1016/j.gene.2020.145223 (2021).
Yahia, S. et al. Genetic variant in the 5’ untranslated region of endothelin1 (EDN1) gene in children with primary nephrotic syndrome. J. Biochem. Mol. Toxicol. 36, e22963. https://doi.org/10.1002/jbt.22963 (2022).
Jannuzzi, A. T., Karaman, E., Oztas, E., Yanar, H. T. & Ozhan, G. Telomerase reverse transcriptase (TERT) gene variations and susceptibility of colorectal cancer. Genet. Test. Mol. Biomark. 19, 692–697. https://doi.org/10.1089/gtmb.2015.0150 (2015).
Hashemi, M. et al. Association between hTERT polymorphisms and the risk of breast cancer in a sample of Southeast Iranian population. BMC Res. Notes 7, 895. https://doi.org/10.1186/1756-0500-7-895 (2014).
Elsaid, A., Elshazli, R., El-Tarapely, F., Darwish, H. & Abdel-Malak, C. Association of monoallelic MUTYH mutation among Egyptian patients with colorectal cancer. Fam. Cancer 16, 83–90. https://doi.org/10.1007/s10689-016-9927-z (2017).
Elsalahaty, M. I. et al. Unleash multifunctional role of miRNA biogenesis gene variants (XPO5*rs34324334 and RAN*rs14035) with susceptibility to hepatocellular carcinoma. J. Pers. Med. https://doi.org/10.3390/jpm13060959 (2023).
Elsaid, A. M. et al. The potential impact of CYP2D6 (*2/*4/*10) gene variants among Egyptian epileptic children: a preliminary study. Gene 832, 146585. https://doi.org/10.1016/j.gene.2022.146585 (2022).
Choi, B. J. et al. Influence of the hTERT rs2736100 polymorphism on telomere length in gastric cancer. World J. Gastroenterol 21, 9328–9336. https://doi.org/10.3748/wjg.v21.i31.9328 (2015).
Jearth, V. et al. Correlation of clinicopathological profile, prognostic factors, and survival outcomes with baseline alfa-fetoprotein levels in patients with hepatocellular carcinoma: A biomarker that is bruised but not broken. J. Clin. Exp. Hepatol. 12, 841–852. https://doi.org/10.1016/j.jceh.2021.11.006 (2022).
Kobeisy, M. A. et al. Clinical significance of elevated alpha-foetoprotein (AFP) in patients with chronic hepatitis C without hepatocellular carcinoma in upper EGYPT. Arab J. Gastroenterol. 13, 49–53. https://doi.org/10.1016/j.ajg.2012.06.004 (2012).
Wu, H. C. et al. Incidence of hepatocellular carcinoma in a community-based Taiwanese population without chronic HBV/HCV infection. JHEP Rep 4, 100410. https://doi.org/10.1016/j.jhepr.2021.100410 (2022).
Li, T., Xian, Y., Tian, T., Zhuang, X. & Chu, M. New evidence of TERT rs2736098 polymorphism and cancer risk: An updated meta-analysis. J. Buon. 21, 491–497 (2016).
Su, L. Y. et al. Polymorphisms of TERT and CLPTM1L and the Risk of hepatocellular carcinoma in Chinese males. Asian Pac. J. Cancer Preven. 15, 8197–8201. https://doi.org/10.7314/APJCP.2014.15.19.8197 (2014).
Zhang, C. et al. hTERT rs2736098 genetic variants and susceptibility of hepatocellular carcinoma in the Chinese population: A case-control study. Hepatobiliary Pancreat. Dis. Int. 12, 74–79. https://doi.org/10.1016/s1499-3872(13)60009-0 (2013).
Dong, S. & Boyle, A. P. Predicting functional variants in enhancer and promoter elements using RegulomeDB. Hum. Mutat. 40, 1292–1298. https://doi.org/10.1002/humu.23791 (2019).
Ward, L. D. & Kellis, M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 44, D877–D881. https://doi.org/10.1093/nar/gkv1340 (2015).
Zhao, Y. & Li, Z. Interplay of estrogen receptors and FOXA factors in the liver cancer. Mol. Cell Endocrinol. 418(Pt 3), 334–339. https://doi.org/10.1016/j.mce.2015.01.043 (2015).
McPherson, C. E., Shim, E. Y., Friedman, D. S. & Zaret, K. S. An active tissue-specific enhancer and bound transcription factors existing in a precisely positioned nucleosomal array. Cell 75, 387–398. https://doi.org/10.1016/0092-8674(93)80079-t (1993).
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB). This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
W.R.S.E.: Conceptualization, Methodology, Investigation, Writing original draft. E.A.S.: Project administration, Conceptualization, Data curation, Formal analysis, Validation, Methodology, Writing—review & editing. A.M.: Formal analysis, Writing original draft, Data curation. R.M.E.: Project administration, Conceptualization, Methodology, Software, Formal analysis, Data curation, Validation, Investigation, Writing—review & editing. E.A.S. and R.M.E. supervised the work. All authors approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Seif Eldin, W.R., Saad, E.A., Monier, A. et al. Association of TERT (rs2736098 and rs2736100) genetic variants with elevated risk of hepatocellular carcinoma: a retrospective case–control study. Sci Rep 13, 18382 (2023). https://doi.org/10.1038/s41598-023-45716-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-45716-w
This article is cited by
-
Genetic Variants of AGO1*rs595961 and AGO2*rs4961280 with Susceptibility to Bladder Carcinoma
Indian Journal of Clinical Biochemistry (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.