Abstract
Any reliable biomarker has to be specific, generalizable, and reproducible across individuals and contexts. The exact values of such a biomarker must represent similar health states in different individuals and at different times within the same individual to result in the minimum possible false-positive and false-negative rates. The application of standard cut-off points and risk scores across populations hinges upon the assumption of such generalizability. Such generalizability, in turn, hinges upon this condition that the phenomenon investigated by current statistical methods is ergodic, i.e., its statistical measures converge over individuals and time within the finite limit of observations. However, emerging evidence indicates that biological processes abound with nonergodicity, threatening this generalizability. Here, we present a solution for how to make generalizable inferences by deriving ergodic descriptions of nonergodic phenomena. For this aim, we proposed capturing the origin of ergodicity-breaking in many biological processes: cascade dynamics. To assess our hypotheses, we embraced the challenge of identifying reliable biomarkers for heart disease and stroke, which, despite being the leading cause of death worldwide and decades of research, lacks reliable biomarkers and risk stratification tools. We showed that raw R-R interval data and its common descriptors based on mean and variance are nonergodic and non-specific. On the other hand, the cascade-dynamical descriptors, the Hurst exponent encoding linear temporal correlations, and multifractal nonlinearity encoding nonlinear interactions across scales described the nonergodic heart rate variability more ergodically and were specific. This study inaugurates applying the critical concept of ergodicity in discovering and applying digital biomarkers of health and disease.
Similar content being viewed by others
Introduction
Heart disease and stroke are the leading causes of disease and disability globally and in the United States, claiming 655,000 American lives every year—one in four deaths1,2. This staggering toll of cardiovascular diseases does not end here, as it costs the nation over \(\$200\) billion annually in direct medical expenses and lost productivity. This colossal burden highlights the importance of early diagnosis and intervention of heart disease and stroke. One of the primary requisites for effective diagnosis is the availability of specific and reliable biomarkers. Although numerous biomarkers, risk stratification models, and risk scores for various cardiovascular diseases have been proposed over the past decades, effective diagnostic and prognostic digital biomarkers are still missing3,4,5. The urgency of addressing this need is amplified by the rise and ever-growing expansion of diverse digital health and telehealth solutions in recent years, specifically in the cardiovascular field6,7,8. Such solutions, like mobile applications (mhealth), smart watches, wearable devices, implantable electronic devices, and implantable hemodynamic monitors, enable the gathering of vast amounts of data for everyone; however, the lack of diagnostic and prognostic biomarkers lays waste to this ability as such valuable amounts of data cannot be appropriately used. Lack of evidence of effect has been cited as one of the reasons why digital health technologies have not been widely employed in clinical settings9. The lack of reliable digital biomarkers can be considered one of the main contributors to this lack of evidence.
Heart rate variability (HRV) has been one of the key noninvasive biomarkers of cardiovascular health10. It measures the fluctuations and variations in time intervals between successive heartbeats or R-R intervals (RRi). HRV is an emergent phenomenon that emerges out of the complex and nonlinear interactions between the cardiovascular and nervous systems11,12,13 and represents the peripheral output of the central autonomic network (CAN) and the capacity for behavioral adaption to environmental stresses14,15,16,17,18,19,20,21,22. Because it emerges from such complex and integral interactions, HRV can be a representative marker of cardiovascular health. Healthy human HRV indicates desirable balance and interaction between the functions of the sympathetic and parasympathetic nervous systems23,24,25. Group-level findings have shown that HRV might be superior to many other biomarkers in representing the overall state of health and well-being26,27.
Although the emergence of HRV out of complex and intricate interactions confers HRV such an ability to represent the state of the body, it also makes its appropriate application as a digital biomarker replete with nuances. Analyses of heartbeat dynamics and HRV reveal significant nonlinearity, non-Gaussianity, and chaotic behaviors in the RRi series28,29,30,31,32,33,34,35,36,37,38,39,40,41,42. These statistical signatures of nonlinearity, non-Gaussianity, and chaotic behaviors in RRi can be interpreted as manifestations of the emergence of HRV from interdependent and bidirectional interactions across multiple timescales. Such processes which lead to multiplicative fluctuations and dynamics have been termed multifractal cascades43,44,45,46,47. The cascade dynamical nature of HRV, like many other behavioral and physiological functions48,49,50,51,52,53,54, inclines many of its measurements and descriptors toward a characteristic that has been, unfortunately, grossly overlooked in the biomedical literature: ergodicity. We believe the overlooking of ergodicity has hindered the broad application of HRV probably much more than the other challenges that have been discussed regarding HRV, like analytical challenges associated with data variability, missing data and artifacts, and lack of theory for data interpretation55,56,57,58,59,60,61,62,63,64,65,66.
Ergodicity is an essential requirement of a digital biomarker to be applied reliably in current medical practice. Similar values of a digital biomarker across different individuals must represent similar bodily states. In other words, standard cut-off points of such a biomarker must reliably separate the states of health and disease in each different individual67,68. Based on these practices, most medical research, similar to most biological, psychological, and social research, has aggregated the data gathered from randomly selected groups of individuals and used group-based statistical methods to reach conclusions. Such conclusions are then deemed generalizable to the behaviors of different individuals across different contexts. However, ergodicity is a requisite of this generalizability from group-level data to an individual’s behaviors. In nonergodic measurements, the behaviors of an individual at a specific time diverge from the average of that measurement across a group of individuals and also the average of that individual’s behaviors over an extended period69,70,71,72. Ergodicity refers to the convergence of these two averages: the finite-ensemble average and the finite-time average (Fig. 1). The finite-ensemble average, which is also recognized as the “sample average,” is
where \(x_{i}(t)\) is the ith of N individual cases of x(t) included in the finite-ensemble average. The finite-time average, which biomedical discourse recognizes as the “average performance/trajectory of the individual,” is
for continuous change. The finite-time average when the measured behavior x changes at \(T=\Delta t/\delta t\) discrete times \(t+\delta t,t+2\delta t,\dots\) is
So, ergodicity is an equivalence between these two averages,
Nonergodicity refers to the lack of equivalence between finite-ensemble and finite-time averages. The finite-ensemble average, which biomedical discourse recognizes as the “sample average,” is \(\langle x_{i}(t)\rangle _{N}=\frac{1}{N}\sum _{i=1}^{N}x_{i}(t)\), where \(x_{i}(t)\) is the ith of N individual cases of x(t) included in the finite-ensemble average. The finite-time average when the measured behavior x changes at \(T=\Delta t/\delta t\) discrete times \(t+\delta t,t+2\delta t,\dots\) is \(\overline{x_{\Delta t}}=\frac{1}{T\delta t}\sum _{\tau =1}^{T}x(t+\tau \delta t)\).
Another phrasing of the concept of ergodicity is that ergodic systems visit all of their possible states—in a sense, ergodic systems do not have a deep sense of “history.” The criterion of “mixing” emphasizes this addition to the traditional interpretation of ergodicity. Mixing denotes independence of the states of a system across time in a way that all values of a stochastic process across all times would have equal probabilities73. This concept clarifies why emerging experimental data suggests that the processes related to organisms teem with, and probably are even dominated by, nonergodicity74,75,76, although the inferences of the majority of biological, psychological, and social studies in the past century have been based on this implicit presupposition that the processes they study and their measurements are ergodic. Biological processes teem with properties like interactions across space and time scales43,47, historical contingency77,78, and context dependency to break ergodicity71. Consider the exemplary biological process we have chosen in this study: HRV. As we mentioned earlier, data strongly suggests that heartbeat dynamics and HRV have a cascade dynamical nature and emerge from interdependent and bidirectional interactions across scales28,29,30,31,32,33,34,35,36,37,38,39,40,41,42. Also, HRV and many of its descriptors highly depend on various individual, contextual, and measurement factors such as sex and age65. Such historical contingency and context-dependency of HRV and other biological processes generally lead to nonergodicity and lack of generalizability from group-level findings to individuals71,72.
The concern for ergodicity is evident in the application of HRV. An appropriate diagnosis and risk stratification based on HRV depends on two conditions: First, the limited data gathered during the visits, consultations, or laboratory assays should sufficiently represent the states of the individual’s body over time. Second, the standard and established principles and cut-off points used to make decisions should be generalizable to that individual. These conditions have been taken for granted until now. However, as we discussed, evidence suggests that nonergodicity probably violates these conditions. Neglecting this violation can be detrimental; for instance, if screening is conducted on the entire general population, a minor increase in false positive rate can hugely raise subsequent medical tests and expenses58,79,80. An increased false-negative rate also implies delayed anticoagulant medication and increased risk of stroke in symptomatic or high-risk patients.
Our concern for ergodicity is not restricted to the application of HRV. We72,73,81,82, alongside a few others69,70,71,74, believe that ergodicity is an integral concept that undermines how scientific research across diverse fields has tried to identify cause-effect relationships. The breaking of ergodicity is abundant in biological processes and invalidates many conclusions of group-based research designs and statistical methods. Indeed, neglecting this nonergodicity and lack of generalizability could be the leading cause of the reproducibility crisis72,74, which currently encompasses diverse fields from biomedical and psychological sciences to social sciences and economics83,84. Specifically, in applying HRV as a biomarker of health and disease, some studies have suggested that the irreproducibility of results could be a critical problem85,86,87,88,89,90,91,92,93,94.
This study is an attempt in continuation of our previous works to obtain a solution to the problem of making generalizable inferences about nonergodic processes. In this series of works, we first tried identifying sources of nonergodicity in biological processes. Having recognized the abundance of multifractal and cascade dynamics in biological processes48,49,50,51,52,53,54, we hypothesized that a potential source of nonergodicity could be the emergence of many biological processes out of interdependent and bidirectional interactions across spatial and temporal scales, as in cascades. We observed phenomena that corroborated this hypothesis73,81,82,95. Afterward, interestingly, we observed that descriptors that could capture the cascade-dynamical sources of ergodicity breaking in a process might provide ergodic descriptions of that process73,81,82,95.
Here, prompted by the huge amount of evidence that had suggested the multifractal and cascade-dynamical nature of HRV28,29,30,31,32,33,34,35,36,37,38,39,40,41,42, We hypothesized that this nature of HRV leads to the nonergodicity of this phenomenon. Consequently, We predicted that the linear commonly used descriptors of HRV and raw RRi series, like sample means and variances, would be nonergodic and lack generalizability and reproducibility. Afterward, we hypothesized that descriptors that would capture the source of the nonergodicity of HRV might provide ergodic descriptions of this nonergodic phenomenon. Descriptors of the nonlinear, non-Gaussian, multifractal, and cascade-dynamical behaviors of HRV, some of which we had developed in our previous works, seemed worthy candidates29,33,34,96,96,97,98,99,100,101. For this study, we chose descriptors of long-range correlations, \(H_{fGn}\), and multifractal nonlinearity, \(t_{MF}\). We found strong support for our hypotheses.
Results
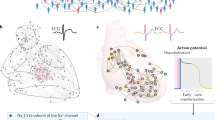
We analyzed the long-term ambulatory HRV in 108 chronic heart failure (CHF) patients—69 survivors (age (mean±SD) = \(64\pm 15\) years; 27 women) and 39 nonsurvivors (\(70\pm 14\) years; 20 women)—who died due to any cause within the follow-up period of \(33\pm 17\) months, and 115 age-matched healthy older adults (\(47.7\pm 18.2\) years; 25 women). The endpoint was all-cause mortality. The majority of deaths (34/39) were cardiac-related, including death from progressive heart failure (\(n=23\)), sudden death (\(n=10\)), and acute myocardial infarction (\(n=1\)). The remaining five patients died of sepsis (\(n=1\)), pneumonia (\(n=3\)), and stroke (\(n=1\)). We reanalyzed HRV data from one of our previous published studies99. Table 1 summarizes the demographic and baseline clinical characteristics of the CHF patients.
HRV breaks ergodicity
To examine the ergodic properties of the RRi series (exemplified in Fig. 2a–c), we submitted the original RRi series and the corresponding shuffled versions to the Thirumalai-Mountain analysis102,103, which yields a dimensionless metric called the ergodicity breaking factor, \(E_{B}\),
where \(\overline{\delta ^{2}(x(t))}=\int _{0}^{t-\Delta }[x(t^{\prime }+\Delta )-x(t^{\prime })]^{2}dt^{\prime }\bigl /(t-\Delta )\) is the time average mean-squared displacement of the stochastic series x(t) for lag time \(\Delta\). Rapid decay of \(E_{B}\) to a finite asymptotic value for progressively larger samples, i.e., \(E_{B}\rightarrow 0\) as \(t \rightarrow \infty\) implies ergodicity. Slower decay indicates less ergodic systems in which trajectories are less reproducible. No decay or convergence to a finite asymptotic value indicates strong ergodicity breaking104,105. \(E_{B}(x(t))\) thus allows testing whether a given series breaks ergodicity. \(E_{B}\) for the original RRi series did not decay at all with t in the finite range of 1000 secs, essentially remaining unchanged over a progressively longer time for healthy controls as well as the two patient groups (\(E_{B}(x(t))=-0.0183\frac{\Delta }{t},0.0194\frac{\Delta }{t}\), and \(-0.0306\frac{\Delta }{t}\) for healthy controls, CHF nonsurvivors, and CHF survivors, respectively; colored lines in Fig. 2d–f). These values of \(E_{B}(x(t))\) indicate strong ergodicity breaking in the original RRi series. In contrast, \(E_{B}\) for the shuffled RRi series rapidly decayed to a finite asymptotic value in the finite range of 1000 secs, indicating ergodicity (\(E_{B}(x(t))=-1.0274\frac{\Delta }{t}, -1.0475\frac{\Delta }{t}\), and \(-1.0029\frac{\Delta }{t}\) for healthy controls, CHF nonsurvivors, and CHF survivors, respectively; grey lines in Fig. 2d–f). As by shuffling the original RRi series, the temporal structure and information of the RRi series are removed, these values of \(E_{B}(x(t))\) suggest that the very temporal structure of HRV is the source of nonergodicity in HRV.
The raw R–R interval (RRi) series are nonergodic. (a–c) Representative examples of the original and shuffled RRi series (colored lines and grey lines, respectively). (a) The RRi series for a healthy control (a 54-year-old woman). (b) The RRi series for a 74-year-old man with congestive heart failure (CHF) who died 101 days after the measurement. (c) The RRi series for an 82-year-old woman who survived CHF. The original RRi series, for healthy controls (d) as well as the two patient groups (e, f), show no change in the ergodicity breaking parameter, \(E_{B}\), over progressively longer periods, reflecting that HRV breaks ergodicity (colored lines). Shuffling the original RRi series produces an RRi series that is ergodic, as indicated by the rapid decay in \(E_{B}\) over progressively longer periods (grey lines). Thin lines and thick lines in (d–f) represent ergodicity breaking for individuals and mean ergodicity breaking for the three groups, respectively.
Linear descriptors based on mean and variance are nonergodic
Now that we have witnessed ergodicity breaking in the raw RRi series, let us investigate the ergodic properties of some of the linear descriptors widely used in cardiovascular digital medicine: HRV parameters based on mean and variance65. Here, we chose the mean and root mean square of successive RR intervals, hereinafter noted as M and RMS, respectively. M and RMS were computed by dividing the RRi series into nonoverlapping epochs comprising 500 beats. So, for example, a raw RRi series of 100,000 beats yielded 2000 nonoverlapping epochs, each comprising 500 beats, ultimately yielding M and RMS series of 2000 samples each. Similar to the behavior of \(E_{B}\) for the raw RRi series, \(E_{B}\) for M and RMS did not decay at all over epochs in the narrow range of 20 epochs (\(E_{B}(M(\textrm{epoch}))=-0.0848\frac{\Delta }{\textrm{epoch}}, -0.1461\frac{\Delta }{\textrm{epoch}}\;\text {and}\;-0.0935\frac{\Delta }{\textrm{epoch}}\);\(E_{B}(RMS(\textrm{epoch}))=-0.0846\frac{\Delta }{\textrm{epoch}},-0.1480\frac{\Delta }{\textrm{epoch}}\;\text {and}\;-0.0937\frac{\Delta }{\textrm{epoch}}\) for healthy controls, CHF nonsurvivors, and CHF survivors, respectively). \(E_{B}\) remained unchanged over a progressively larger number of epochs for all three groups (colored lines in Fig. 3a,c). In contrast, \(E_{B}\) for M and RMS of the shuffled RRi series rapidly decayed to a finite asymptotic value in the narrow range of 20 epochs (\(E_{B}(M(\textrm{epoch}))=-1.2337\frac{\Delta }{\textrm{epoch}},-1.2890\frac{\Delta }{\textrm{epoch}}\;\text {and}\;-1.2275\frac{\Delta }{\textrm{epoch}}\);\(E_{B}(RMS(\textrm{epoch}))=-1.2400\frac{\Delta }{\textrm{epoch}},-1.2951\frac{\Delta }{\textrm{epoch}}\;\text {and}\;-1.2321\frac{\Delta }{\textrm{epoch}}\) for healthy controls, CHF nonsurvivors, and CHF survivors, respectively; grey lines in Fig. 3a,c). In other words, M and RMS-based HRV parameters failed to provide ergodic descriptions of HRV. Furthermore, the contrast between behaviors of \(E_{B}\) for the original and the shuffled RRi series indicates that the very temporal structure of HRV contributes to this failure.
Commonly used linear descriptors of HRV based on mean and root mean square are nonergodic. (a) The ergodicity breaking parameter, \(E_{B}\), did not change for the mean of successive RR intervals, M, in the original RRi series over a progressively larger number of epochs (colored lines). In contrast, \(E_{B}\) decayed rapidly for the M of the shuffled RRi series (grey lines). (b) Null hypothesis significance testing (NHST) for M across the three groups. One-way ANOVAs failed to detect reduced M of HRV due to the CHF, \(36.8\%\) and \(12.7\%\) times in nonsurvivors (red histogram) and survivors (green histogram), respectively, compared to healthy controls. (c) \(E_{B}\), did not change for the root mean square of successive RR interval differences, RMS, in the original RRi series over a progressively larger number of epochs (colored lines). In contrast, \(E_{B}\) decayed rapidly for the RMS of the shuffled RRi series (grey lines). (d) Null hypothesis significance testing (NHST) for RMS across the three groups. One-way ANOVAs failed to detect reduced M of HRV due to the CHF, \(34.6\%\) and \(11.3\%\) times in nonsurvivors (red histogram) and survivors (green histogram), respectively, compared to healthy controls. Thin lines and thick lines in (a,c) represent \(E_{B}\) for individuals and mean \(E_{B}\) for the three groups, respectively.
To test the specificity and reliability of these HRV parameters, we also performed Monte Carlo simulations by randomly sampling the 1000-sample RRi series from the 24-hour recordings for each individual and performing one-way ANOVA tests separately on these series’ M and RMS values. We repeated this process 1000 times. One-way ANOVAs failed to detect reduced M of HRV due to the CHF, \(36.8\%\) and \(12.7\%\) times in nonsurvivors (red histogram in Fig. 3b) and survivors (green histogram in Fig. 3b), respectively, compared to healthy controls. Likewise, one-way ANOVAs failed to detect reduced RMS of HRV due to the CHF, \(34.6\%\) and \(11.3\%\) times in nonsurvivors (red histogram in Fig. 3d) and survivors (green histogram in Fig. 3d), respectively, compared to healthy controls. In other words, we found a high likelihood of failing to identify statistically significant differences among the three groups’ M and RMS. These results confirm that linear descriptors M and RMS cannot be used as reliable HRV parameters for digital biomarkers of health and disease.
Linear descriptors \(\textrm{NN50}\) and \(\textrm{pNN50}\) are only weakly ergodic but not specific
The number of adjacent RR intervals that differ by more than 50 milliseconds and the percentage of such RR intervals are two other linear descriptors widely used as HRV parameters65. \(E_{B}\) for \(\textrm{NN50}\) and \(\textrm{pNN50}\) had similar behavior to that of the shuffled RRi series; however, \(E_{B}\) had a shallower initial decay for \(\textrm{NN50}\) and \(\textrm{pNN50}\) with a progressively larger number of epochs in the narrow range of 20 epochs (\(E_{B}(\textrm{NN50}(\textrm{epoch}))=-0.4297\frac{\Delta }{\textrm{epoch}},-0.4281\frac{\Delta }{\textrm{epoch}}\;\text {and}\;-0.7090\frac{\Delta }{\textrm{epoch}}\); \(E_{B}(\textrm{pNN50}(\textrm{epoch}))=-0.4295\frac{\Delta }{\textrm{epoch}},-0.4340\frac{\Delta }{\textrm{epoch}}\;\text {and}\;-0.7124\frac{\Delta }{\textrm{epoch}}\) for healthy controls, CHF nonsurvivors, and CHF survivors, respectively). Eventually, \(E_{B}\) reached an asymptotic finite but a relatively larger value over a progressively larger number of epochs (colored lines in Fig. 4a,c). These \(E_{B}\textrm{NN50}(\textrm{epoch}))\) and \(E_{B}(\mathrm {\textrm{pNN50}} (\textrm{epoch}))\) curves were only marginally shallower than those for the epoch series of \(\textrm{NN50}\) and \(\textrm{pNN50}\) for the shuffled RRi series in the narrow range of 20 epochs (\(E_{B}(\textrm{NN50}(\textrm{epoch}))=-1.2289\frac{\Delta }{\textrm{epoch}},-1.3169\frac{\Delta }{\textrm{epoch}}\;\text {and}\;-1.2557\frac{\Delta }{\textrm{epoch}}\);\(E_{B}(\textrm{pNN50}(\textrm{epoch}))=-1.2286\frac{\Delta }{\textrm{epoch}},-1.3166\frac{\Delta }{\textrm{epoch}}\;\text {and}\;-1.2556\frac{\Delta }{\textrm{epoch}}\) for healthy controls, CHF nonsurvivors, and CHF survivors, respectively; grey lines in Fig. 4a,c). Thus, \(\textrm{NN50}\) and \(\textrm{pNN50}\) break ergodicity only weakly, providing more ergodic descriptions of the nonergodic HRV than the previous two linear descriptors, M and RMS. Again, the contrast between the original and shuffled RRi series indicates that the very temporal structure of HRV contributes to this weak ergodicity breaking by \(\textrm{NN50}\) and \(\textrm{pNN50}\).
NN50 and pNN50 are specific but only weakly ergodic. (a) The epoch series of \(\textrm{NN50}\) describing the original RRi series show an initial decay in the ergodicity breaking parameter, \(E_{B}\), with epochs (colored lines), albeit shallower compared to the epoch series of \(\textrm{NN50}\) describing the shuffled RRi series (grey lines). (b) Null hypothesis significance testing (NHST) for \(\textrm{NN50}\) across the three groups. One-way ANOVAs revealed that NN50 of HRV did not differ between either patient populations and healthy controls: CHF nonsurvivors and survivors (red histogram and green histogram, respectively). (c) The epoch series of \(\textrm{pNN50}\) describing the original RRi series show an initial decay in \(E_{B}\) over epochs (colored lines), albeit shallower compared to the epoch series of \(\textrm{pNN50}\) describing the shuffled RRi series (grey lines). (d) NHST for \(\textrm{pNN50}\) across the three groups. One-way ANOVAs revealed that pNN50 of HRV did not differ between either patient populations and healthy controls: CHF nonsurvivors and survivors (red histogram and green histogram, respectively). Thin lines and thick lines in (a,c) represent \(E_{B}\) for individuals and mean \(E_{B}\) for the three groups, respectively.
To test the specificity and reliability of these parameters, we performed Monte Carlo simulations by randomly sampling 1000-sample RRi series from 24-hour recordings for each individual and performing one-way ANOVA tests separately on these series’ \(\textrm{NN50}\) and \(\textrm{pNN50}\) values. We repeated this process 1000 times. One-way ANOVAs revealed that NN50 of HRV did not differ between either patient populations and healthy controls: CHF nonsurvivors and survivors (red histogram and green histogram, respectively, in Fig. 4b). Likewise, one-way ANOVAs revealed that pNN50 of HRV did not differ between either patient populations and healthy controls: CHF nonsurvivors and survivors (red histogram and green histogram, respectively, in Fig. 4d). Hence, \(\textrm{NN50}\) and \(\textrm{pNN50}\) might only weakly break ergodicity but also not diagnose CHF.
Cascade-dynamical descriptors \(H_{fGn}\) and \(t_{MF}\) are both ergodic and specific
We hypothesized that cascade-dynamical descriptors might provide ergodic descriptions of the nonergodic HRV by capturing the source of ergodicity breaking. The most compelling descriptions of cascading dynamics come from multifractal geometry43,47,106. Simulations of cascade processes show two critical features: long-range linear temporal correlations and nonlinear correlations involving interactions across timescales. The former feature appears most frequently as a fractional Gaussian noise (fGn) in which the standard deviation increases as a power function of the timescale. The fractional power in this function is known as the Hurst exponent, \(H_{fGn}\). \(H_{fGn}\) and has already been shown to be sensitive to differences in HRV due to congestive heart failure38. The latter feature of nonlinear correlations concerns the effects spreading across the hierarchical organization of biological structures, producing a variation in \(H_{fGn}\), i.e., multifractality. We can estimate this nonlinear variation by estimating the variation in power functions over time and then comparing this multifractal variation to what a linear model of the underlying RRi series can produce. That is, by comparing the multifractality (i.e., the number of power functions) estimable for the original RRi series to the same multifractal property for a sample of synthetic RRi series. The one-sample t-test comparing the multifractality of the original to the synthetic RRi series provides a t-statistic, multifractal nonlinearity, \(t_{MF}\), which quantifies nonlinear correlations due to cascade dynamics107. \(H_{fGn}\) and \(t_{MF}\) have been shown to provide ergodic descriptions of the nonergodic series of both simulated and empirical biological measurements73,81,82. We aim to determine whether \(H_{fGn}\) and \(t_{MF}\) can adequately describe the nonergodic HRV. Furthermore, we test whether \(H_{fGn}\) and \(t_{MF}\) provide specificity.
\(H_{fGn}\) and \(t_{MF}\) were computed by first interpolating the RRi series to 2 Hz and then dividing it into nonoverlapping epochs comprising 1000 samples (i.e., spanning 500 s). So, for example, a raw RRi series of 100, 000 samples yielded 1000 nonoverlapping epochs, each comprising 1000 samples, ultimately yielding \(H_{fGn}\) and \(t_{MF}\) series of 2000 samples each. The behavior of \(E_{B}\) for the epoch series of \(H_{fGn}\) and \(t_{MF}\) bears a strong resemblance to the behavior of \(E_{B}\) for the shuffled raw RRi series. For \(H_{fGn}\), \(E_{B}\) had an initial rapid decay in the narrow range of 20 epochs that became shallower over a progressively larger number of epochs (\(E_{B}(H_{fGn}(\textrm{epoch}))=-0.4640\frac{\Delta }{\textrm{epoch}},-0.1648\frac{\Delta }{\textrm{epoch}}\;\text {and}\;-0.2121\frac{\Delta }{\textrm{epoch}}\); colored lines in Fig. 5a). For \(t_{MF}\), \(E_{B}\) rapidly decayed initially over a progressively larger number of epochs in the narrow range of 20 epochs (\(E_{B}(t_{MF}(\textrm{epoch}))=-0.8372\frac{\Delta }{\textrm{epoch}},-1.0238\frac{\Delta }{\textrm{epoch}}\;\text {and}\;-1.1759\frac{\Delta }{\textrm{epoch}}\); colored lines in Fig. 5c). These \(E_{B}(H_{fGn}(\textrm{epoch}))\) curves show a faster decay for the shuffled RRi series in the narrow range of 20 epochs (\(E_{B}(H_{fGn}(\textrm{epoch}))=-1.2423\frac{\Delta }{\textrm{epoch}},-1.2434\frac{\Delta }{\textrm{epoch}}\;\text {and}\;-1.2002\frac{\Delta }{\textrm{epoch}}\); grey lines in Fig. 5a). However, the decay rate of \(E_{B}(t_{MF}(\textrm{epoch}))\) curves for the shuffled RRi series was comparable to that of the original RRi series in the narrow range of 20 epochs (\(E_{B}(t_{MF}(\textrm{epoch}))=-1.2091\frac{\Delta }{\textrm{epoch}},-1.1555\frac{\Delta }{\textrm{epoch}}\;\text {and}\;-0.9862\frac{\Delta }{\textrm{epoch}}\) for healthy controls, CHF nonsurvivors, and CHF survivors, respectively; grey lines in Fig. 5c). As epoch size increased, CHF survivors and CHF nonsurvivors exhibited an \(E_{B}\) decay fully comparable with shuffled versions of the series for short to medium epoch sizes and all epoch sizes, respectively. Healthy patients showed much shallower decay for the original RRi series than for the shuffled RRi series. These results show that the cascade-dynamical descriptors, \(H_{fGn}\) and \(t_{MF}\), provide more ergodic descriptions of the nonergodic HRV but only over a narrow range of short epoch sizes for healthy cases and only for longer epochs in CHF cases. The cascade-dynamical nature of HRV that contributed to the nonergodicity of linear descriptors like M and RMS was thus captured by cascade-dynamical descriptors \(H_{fGn}\) and \(t_{MF}\) marginally more than traditional linear descriptors. But the truth is that these \(E_{B}\)-vs.-epoch curves show a heterogeneity across epochs that we had not previously observed in theoretical simulations. Hence, the success of ergodic characterization by cascade-dynamical descriptors is mixed at best and encouraging only in contrast to the much poorer performance of the linear descriptors. In the Discussion, we reflect on what these mixed results could mean for methodological and theoretical concerns moving forward.
Cascade-dynamical descriptors of long-range correlations, \(H_{fGn}\), and multifractal nonlinearity, \(t_{MF}\), are ergodic and specific. (a) The ergodicity breaking parameter, \(E_{B}\), decayed initially for the \(H_{fGn}\) of the original RRi series over short epochs (colored lines). However, this decay was shallower compared to that of the \(H_{fGn}\) of the shuffled RRi series(grey lines) overall longer epochs. (b) NHST of \(H_{fGn}\) across the three groups. One-way ANOVAs failed to detect reduced \(H_{fGn}\) of HRV due to the CHF, only \(0.2\%\) and \(1.8\%\) times in nonsurvivors (red histogram) and survivors (green histogram), respectively, compared to healthy controls. (c) \(E_{B}\) decayed rapidly over epochs for the \(t_{MF}\) of both the original RRi series (colored lines) and the shuffled RRi series (grey lines) but only for the CHF nonsurvivors. Rapid decays of \(E_{B}\) for the original RRi series comparable to the shuffled RRi series only held for extremely short epochs in the healthy case and for small to medium epoch sizes in the CHF survivors. (d) One-way ANOVAs failed to detect reduced \(t_{MF}\) of HRV due to the CHF, \(13.2\%\) and \(2.4\%\) times in nonsurvivors (red histogram) and survivors (green histogram), respectively, compared to healthy controls. Thin lines and thick lines in (a,c) represent \(E_{B}\) for individuals and mean \(E_{B}\) for the three groups, respectively.
To test the specificity of these cascade-dynamical descriptors, we performed Monte Carlo simulations by randomly sampling 1000-sample RRi series from 24-hour recordings for each individual and performing one-way ANOVA tests separately on these series’ \(H_{fGn}\) and \(t_{MF}\) values. We repeated this process 1000 times. One-way ANOVAs failed to detect reduced \(H_{fGn}\) of HRV due to the CHF, \(0.2\%\) and \(1.8\%\) times in nonsurvivors (red histogram in Fig. 5b) and survivors (green histogram in Fig. 5b), respectively, compared to healthy controls. Likewise, one-way ANOVAs failed to detect reduced M of HRV due to the CHF, \(13.2\%\) and \(2.4\%\) times in nonsurvivors (red histogram in Fig. 5d) and survivors (green histogram in Fig. 5d), respectively, compared to healthy controls. Thus, \(H_{fGn}\) and \(t_{MF}\) provide ergodic descriptions of the nonergodic HRV and can specifically differentiate clinical groups with high reliability. These results support our proposal of capitalizing cascade-dynamical descriptors as generalizable and reproducible HRV parameters for digital biomarkers of health and disease.
To assess the prognostic potential of the cascade-dynamical descriptors \(H_{fGn}\) and \(t_{MF}\) in predicting mortality among CHF patients108,109, survival analysis was conducted. Kaplan-Meier cumulative survival curves utilizing \(H_{fGn}\) and \(t_{MF}\) as predictive variables are depicted in Fig. 6. The dichotomization of these descriptors was determined by their respective medians (Mdn). However, the analysis revealed that \(H_{fGn}\) and \(t_{MF}\) failed to predict mortality effectively. Mantel-Haenszel log-rank statistics were calculated, yielding hazard ratios of \(0.814\;[0.619,1.010]\) with a p-value of 0.295 for \(H_{fGn}\) and \(0.846\;[0.652,1.040]\) with a p-value of 0.391 for \(t_{MF}\). These results suggest that neither of these descriptors exhibited significant prognostic capacities in this cohort of CHF patients, highlighting their limited utility. Therefore, incorporating cascade-dynamical descriptors like \(H_{fGn}\) and \(t_{MF}\) alongside conventional ones could be advantageous for optimizing predictivity, specificity, generalizability, and reproducibility in post-CHF prognosis until further advancements enhance the capabilities of these descriptors.
Kaplan-Meier cumulative survival curves of patients with congestive heart failure—both nonsurvivors and survivors. (a) Stratified to patients with long-range correlations in HRV, \(H_{fGn}\le Mdn\) and \(H_{fGn}>Mdn\), with log-rank statistics. (b) Stratified to patients with multifractal nonlinearity in HRV, \(t_{MF}\le Mdn\) and \(t_{MF}>Mdn\), with log-rank statistics. The cutoff points for dichotomization were determined by the respective descriptor’s median (Mdn). Shaded areas indicate \(95\%\) confidence intervals. n denotes the number of patients in a subgroup, with the number of deaths during the observation period in parentheses. The obtained hazard ratios \(0.814 [0.619,1.010], p=0.295\) for \(H_{fGn}\) and \(0.846 [0.652,1.040],p=0.391\) for \(t_{MF}\) indicate that these descriptors are not sufficient for reliable prognostics.
Discussion
Here, we surveyed various descriptors that could be used by traditional and digital medicine to inform the diagnosis and prognosis of cardiovascular conditions such as CHF to identify those descriptors that could provide predictive, specific, generalizable, and reproducible assessments. The primary risk we have highlighted is that the raw RRi series breaks ergodicity. This nonergodicity of HRV is a liability to clinical care because the raw RRi series fails to converge toward an average. Without this convergence, any sequence of RRi cannot be deemed sufficiently representative—whether of the patient’s long-term HRV or groups with a definitive clinical diagnosis or prognosis. We found that many of the most conventional linear descriptors are all as nonergodic as the raw RRi series they summarize. We also identified that the primary source of these nonergodicities is the very temporal structure of HRV and its cascade-dynamical nature. Afterward, we hypothesized that this very origin of nonergodicities might hold the key to the ergodic descriptions of HRV. The cascade-dynamical descriptors \(H_{fGn}\) and \(t_{MF}\) confirmed this hypothesis, provided ergodic descriptions of the nonergodic HRV, and could also specifically differentiate clinical groups. However, our survival analysis indicated that even \(H_{fGn}\) and \(t_{MF}\) cannot sufficiently predict post-CHF prognosis. Previous works exploring the usefulness of cascade-dynamical descriptors have often relied on a suite of descriptors, including those sensitive to the non-Gaussian statistics of HRV33,34,96,97,98,99,100,101. This suggests that it might be best to employ cascade-dynamical descriptors, like \(H_{fGn}\) and \(t_{MF}\), alongside more traditional descriptors to achieve maximum predictivity, specificity, generalizability, and reproducibility.
The widespread adoption of smartwatches and other wearable biosensors with heart-rate monitoring capabilities has sparked hope for the early detection of cardiovascular diseases. However, the belief that more data is sufficient to improve predictions is overly optimistic and misguided. Machine learning/artificial intelligence (ML/AI) models are being developed to achieve highly accurate, sensitive, and specific measures of cardiovascular health; however, to aptly capitalize on these powerful tools, the need for a theoretical understanding of heart rate variability must be addressed. Despite the availability of vast amounts of data, the advancement in understanding the nature of heart rate variability has been modest despite years of work and thousands of scientific publications. This limitation prevents us from making meaningful inferences using ML/AI models. The current reliance on manual or automatic feature extraction110,111,112,113,114,115 is problematic since these features may not suitably reflect the primary causal mechanisms and be too much dependent on contextual variables116,117,118,119. This study emphasizes that the current optimism surrounding the use of wearables and ML/AI models to detect cardiovascular diseases must be accompanied by a deeper understanding of the ergodicity-breaking behavior of HRV.
Our conclusions merit urgent attention as they show the unreliability of prevalent linear descriptors of HRV-like mean-based parameters. We have shown that some of the most intuitive conventional descriptors of HRV, like mean-based descriptors, are nonergodic. In contrast, cascade-dynamical descriptors, such as \(t_{MF}\), can improve the assessment of cardiovascular health when used along with traditional linear descriptors. These results align with those of previous studies that had reported that nonlinear descriptors could provide additional prognostic information compared to conventional linear descriptors39,42,97,99,101, e.g., short-term scaling exponent is a better predictor of mortality or other primary endpoints in cardiovascular patients120,121. Moreover, such nonlinear descriptors have even been found to be reproducible across different populations122,123,124 and contexts, e.g., receiving or not receiving beta-blockers125, and different times and methods of measurement39,123.
Our results also show that a descriptor’s ergodicity is necessary but insufficient for its prognostic capability. Although \(H_{fGn}\) and \(t_{MF}\) provided ergodic descriptions of HRV, they failed to predict mortality in CHF patients. Thus, although ergodicity is necessary for generalizable and reproducible inferences, to reach the utmost specific, generalizable, and reproducible assessment, combining descriptors that provide ergodic descriptions, like the cascade-dynamical descriptors investigated here, with other descriptors, like the conventional ones. These cascade dynamics descriptors encapsulate a pivotal facet of physiological functionality and remain firmly rooted in theoretical validity. Although the precise governing mechanism underlying the intricate heart rate dynamics remains elusive, a study by Lin and Hughson37 has drawn attention to a captivating analogy between heart rate dynamics and turbulence. This analogy is unveiled through the revelation of structural parallels within the realm of multifractal formalism126—specifically, Lin and Hughson established a correlation between heart rate increments and spatial velocity differences within a stochastic cascade process, which serves as a model for hydrodynamic turbulence. In our present investigation, we have harnessed this concept together with long-range correlations characterized by the fractal Hurst exponent to delineate the heart’s operation at a critical juncture127,128. Fluctuations occurring within systems operating near critical points are inherently entwined with scale invariance and universal behavior, as encapsulated by the scaling function129,130. Consequently, we could access the ergodic manifestation of these scale-invariant structures quantified by our cascade-dynamical descriptors73,81,82.
To also compare the nonlinear descriptors investigated here, it must be noted that \(H_{fGn}\) is primarily a monofractal descriptor and is best suited to describe series generated based on one fractal-scaling exponent. However, the modeling of cascade dynamics due to nonlinear interactions across scales inherent to HRV is beyond the scope of \(H_{fGn}\) and requires multifractal formalism47,106,131. Monofractal fluctuations such as fGn are ideally defined exhaustively by single fractional exponents \(H_{fGn}\) and fall cleanly within the linear model through an autocorrelation function indicative of fractional integration132,133. The nonlinearity of interactions across scales requires not only one but many fractional scaling exponents in addition to strictly linear long-range correlations. Hence, multifractal modeling is necessary to analyze the putative cascade-dynamical route to nonergodicity thoroughly73,81,82, i.e., the inherently multifractal descriptor, \(t_{MF}\), is superior to \(H_{fGn}\) in encoding nonlinear interactions across scales, which is characteristic of HRV.
It is important to recognize that ergodicity functions on various levels, including those of the individual and the group. When referring to an individual, ergodicity refers to the capacity to generalize through time. On the other hand, it entails extrapolating from a group level to an individual level when observed in a group setting. The former is ideal since it is compatible with personalized medicine; however, the latter situation is less preferable unless we support non-personalized medicine and more inclusive species-wide strategies. We mainly relied on the ergodicity breaking parameter \(E_{B}\)102,103,134, which reflects a strictly intraindividual analysis. Specifically, the \(E_{B}\) metric refers to the time average across various epochs within the same measurement series, i.e., intra-series variation of the mean, not inter-series variability. When we compare \(E_{B}\) to shuffled versions of the same measurement series, this comparison again uses the original measurement to construct the standard. In other words, \(E_{B}\) is never calculated by comparing one participant to another, let alone to any population parameters. Nonetheless, our stance is justified because ergodicity-breaking implies the absence of either type of ergodicity at the individual and group levels.
At first glance, it can seem absurd to seek ergodicity in a physiological measurement because biological and psychological processes routinely break ergodicity69,70,72,74,135,136,137,138,139,140. However, biological and physiological sciences explicitly identify those scales and spans where ergodicity holds. They do so because ergodicity supports the clearest information-theoretic and predictive modeling that our statistics could marshal for understanding biology and physiology141. This carving out of ranges where ergodicity holds might be why we still have many valuable biomarkers used in clinical practice and established through extensive group data. Indeed, at clinically relevant timescales, the best biomarkers have been those with little temporal change, allowing the expectation of consistent repetition throughout time for each individual and accurately reflecting their biological status. We usually prefer such intraindividual ergodicity rather than the less likely ergodicity at the scale of a whole population of organisms. Whereas ergodicity is likely to fail at the scale of a species population, the best hope for biomarkers is with ergodicity within the intraindividual variation, such as might support a more context-sensitive, personalized clinical approach.
Some other points also warrant further attention. For more comprehensive employment of nonlinear descriptors, such as those proposed here, especially in traditional medical settings, it might be necessary to provide more intuitive interpretations for clinicians and educate clinicians so that the biological basis of these mathematical parameters is clear28. Also, cascade dynamics is only one of the mechanisms that can lead to ergodicity-breaking physiological variabilities. It is still being determined whether all such mechanisms could be modeled as cascade processes (e.g.,95). Despite the central role of cascade processes in biomedical explanation142, we hope that future investigations examine a broader class of anomalous diffusion regimes143,144,145,146,147,148,149,150,151,152 that can also lead to ergodicity-breaking physiological variabilities. Further work is needed to determine whether cascade-dynamical descriptors enable reproducible health assessment when the sources of ergodicity breaking are more nuanced. The statistical modeling framework presented in the present study will be fundamental in guiding these investigations.
This study presents several salient limitations warranting scrutiny: First and foremost, excluding patients with cardiac pacemakers represents a notable constraint. However, we must recognize that patients possessing dual-chamber devices, thoughtfully programmed to forestall atrial pacing, could have been judiciously integrated into the analytical framework. In such instances, the sinus rate and rhythm regulation would have mirrored those observed in non-paced patients. This consideration bears particular relevance in light of the escalating prevalence of CHF patients undergoing cardiac resynchronization therapy. Secondly, it is conceivable that the prognostic potential of the cascade-dynamical descriptor may have been obscured by the relatively modest sample size, comprising a mere cohort of 108 CHF patients. Consequently, it becomes imperative to embark on further inquiries to unravel the prognostic import of the cascade-dynamical descriptors within the realm of CHF prognosis. In summation, notwithstanding the commendable contributions engendered by this study, it is paramount to acknowledge and systematically address these limitations. Such endeavors are quintessential for fostering a more encompassing comprehension of the repercussions and applicability of the findings, particularly within the context of biomarker research.
Eventually, the challenges faced in this study and our proposed solutions should be unrestricted to the case of HRV and cardiovascular health. nonergodicity and cascade dynamics abound in biological processes and are regularities—not exceptions. Much more attention must be paid to the ergodicity of investigated biological phenomena. Moreover, in cases of ergodicity breaking, we have shown here and in previous studies73,81,82 that cascade dynamics should be considered one of the primary candidates for its origin and that capturing this origin through nonlinear, multifractal, and cascade-dynamical descriptors may be the key to ergodically describing nonergodic phenomena. The importance of these insights cannot be exaggerated as they are crucial for reliable and reproducible diagnosis and prognosis across all fields. nonergodicity may be a signature of life, but seeking ergodicity in our generalizations and causal reasoning is pivotal for arriving at generalizable and reproducible digital biomarkers of health and disease.
In pragmatic terms, our investigation illuminates the inherent constraints of conventional biomarker descriptors predicated on the mean and variance calculations derived from raw R-R interval data. These erstwhile markers exhibit a pronounced nonergodic and non-specific character. In contrast, cascade-dynamical descriptors, exemplified by the Hurst exponent and multifractal nonlinearity, furnish a conspicuously more ergodic and precise portrayal of HRV. The discerned outcome carries profound implications, as it intimates that our cascade dynamics-centered methodology can unearth biomarkers of superior reliability germane to the domains of heart disease and stroke. This addresses a critical lacuna endemic to contemporary clinical practice, charting the course for integrating ergodicity paradigms into digital biomarker exploration. This paradigm shift holds the promise of catalyzing advancements in risk stratification and diagnostic precision, thereby auguring tangible enhancements in the quality of patient care within the intricate realm of cardiovascular health. Future inquiries should delve further into the comparative evaluation of our cascade-dynamical HRV descriptors vis-á-vis established clinical biomarkers, scrutinizing their synergistic potential to yield an amalgamated diagnostic and prognostic arsenal of heightened potency.
Theoretical and practical implications of ergodicity in mining physiological data for biomarkers: the curious choice of RRi series
A significant challenge in using the RRi series to yield a biomarker is the requirement for ergodicity. The standard linear descriptors fail to offer an ergodic description of nonergodic HRV. Theoretical work had previously found that time series like HRV with temporal correlations or non-Gaussian histograms thwart ergodic characterization by linear descriptors73,81,82. The same theoretical work had also found that descriptors derived for cascade-like dynamics, \(H_{fGN}\) and \(t_{MF}\), could do better. We see in the present results that the ergodic description these cascade-dynamical parameters provide is, at once, better than that from linear descriptors but also only suitable for short time windows. The \(E_{B}\) for \(H_{fGn}\) and \(t_{MF}\) for the original RRi series had decays comparable to the shuffled RRi series only for the smallest epochs. Beyond short epochs on the order of 5 epochs, \(H_{fGn}\) series describing the RRi series over longer timescales have comparable ergodicity breaking as the prior linear descriptors. We could punt this limitation back to the fact that the linear model has room to encompass the linear autocorrelation encoded by \(H_{fGn}\). By such logic, \(t_{MF}\) should be better at ergodically describing the nonergodic HRV because it encodes the nonlinear interactivity capable of breaking ergodicity81. However, the ergodicity breaking of the RRi series is so rampant that \(t_{MF}\) now only provides a fleeting improvement over \(H_{fGn}\)—\(t_{MF}\) provides more ergodic description across all epochs than all prior descriptors but only for the surviving CHF case. This group showed \(E_{B}\) for the original RRi series that decayed comparably to the shuffled RRi series across all epochs only in the CHF nonsurviving case. In healthy participants, \(t_{MF}\) was scarcely better than \(H_{fGn}\), yielding \(E_{B}\)-vs.-epoch cures resembling results for \(H_{fGn}\) with the decay of \(E_{B}\) resembling those for the shuffled RRi series for slightly longer epochs. In CHF nonsurviving case, \(t_{MF}\) exhibited ergodicity breaking similar to \(H_{fGn}\) across all epochs, with the \(E_{B}\)-vs.-epoch cures for the original RRi series exhibiting shallow decay than the shuffled RRi series over short to medium epochs and even more shallow decay over the longer epochs.
These results reflect a convergence of multiple constraints all at once. First, finite-size limitations are a perennial constraint on empirical work, preventing a clean resemblance to the theoretical work. For instance, even theoretical work using simulated time series with lengths customary to most empirical work rarely shows \(E_{B}\) converging to zero, even for the shuffled time series73,81,82. Second, previous theoretical work has already shown that the non-Gaussianity and temporal correlations implicit in cascade dynamics can together make the ergodic characterization by \(H_{fGn}\) mediocre, i.e., showing neither the non-decay of \(E_{B}\) for characteristically nonergodic process nor the same rapid decay of \(E_{B}\) for \(t_{MF}\)81. Third, the non-Gaussianity of the HRV could be so excessive as to introduce asymmetric multifractal spectra (e.g.,153), and such asymmetry could be such as to cloud the \(t_{MF}\) with surplus meaning. For instance, the \(t_{MF}\) offers a way to test for multifractality arising from nonlinearity as the marginal difference in spectrum width between original and surrogate spectra (e.g., ref47). Nevertheless, this proposed difference comes traditionally without clarifying how each side of the multifractal spectrum contributes to that marginal difference between original and surrogates. It is meanwhile well known that the left side of the spectrum is often more stable than the right for finite-length empirical series154,155,156,157,158,159. Hence, for time series with such multifaceted sources of ergodicity breaking as the RRi series, it is likely that \(t_{MF}\) is a crude simplification of a multifractal spectrum with more asymmetric nuance than a simple t-test can convey. Non-Gaussianity and finite-size limitations on the RRi series may increase \(t_{MF}\) for reasons that do not reflect nonlinearity. This point warrants reexamination of how we use t-tests for multifractal tests of nonlinearity, let alone how we use such tests as biomarkers.
These results point to two broader concerns about how we even begin approaching the theoretical and methodological work implicit in mining physiological data for biomarkers. First, it may be worth considering the rationale for using the RRi series in the first place—given current wisdom about the RRi series as containing nonlinear correlations across multiple timescales, treating each RR interval as an isolated event is a curious choice. The reduction of the ECG time series to an interval series through first-differences (i.e., subtractions of previous R-event time from current R-event time) is a methodological choice that could have yet-unknown implications for the reliability of any signal-processing outcomes that follow (e.g., ref160). Although this choice of how to reduce a raw series for subsequent analysis may be explicitly theoretical or may reflect convenience or habit, the fact that it can have implications for the reproducibility of a result may give us pause. Indeed, it is alarming that, at this late date, it remains an ongoing research question how to classify and detect the peaks in the QRS complex of an ECG record161,162.
Second, and more deeply theoretically, the heterogeneity of \(E_{B}\)-vs.-epoch curves for empirical applications instead of theoretical demonstrations raises old and persistent questions about how to envision ergodicity. In effect, what sort of variable is ergodicity? Is it a dichotomy in which systems are or are not ergodic, with no gradation or grey areas between (e.g., ref163)? Such a position feels formally clean, but it may raise questions that reflect a pretheoretical choice instead of a conclusion informed by empirical tests. This choice presents some steep challenges for future scientific enterprise164. Then again, is ergodicity more of a continuum? There has been a decades-long tradition of dabbling in considering ergodicity as a continuous property that can become “more” or “less”—or even “quasi”165,166,167,168,169,170,171,172,173,174,175,176,177,178. This latter position sometimes deals in quotation marks around these terms as though to dodge the critique of being mush-mouthed or nonspecific—as though defending against or reacting to the claims to the clarity of theories enlisting ergodicity-as-dichotomy. The time may come when the practical need for biomarkers brings enough empirical scientists to the theoretical concern of ergodicity. Measurements like the RRi series and \(E_{B}\) curves they yield may be an essential catalyst for the following dialogue. We suspect that the theoretical clarity from dichotomy may need to give way to some compromise with ergodicity-as-continuum if we are to confront the practicalities of diagnosis. Theoretical clarity and empirical/applied practicality reflect different motivations. Still, ergodicity-as-continuum may offer no less and more theoretical clarity—new theoretical clarity that could advise the interpretation of such heterogeneous \(E_{B}\) curves.
Methods
Each patient gave informed written consent with full knowledge of the details. The ethics committee of Fujita Health University approved the research, which followed the guidelines stated in the Declaration of Helsinki. All data were fully anonymized before we accessed them.
Subjects
Based on the data of one of our previous studies99, we retrospectively enrolled the patients referred to the hospital of the Fujita Health University from January 2000 to December 2001 for assessment or treatment of CHF. 24-hour monitoring of Holter ECG was conducted before their hospital discharge. To be eligible for this study, the patients had to be in normal sinus rhythm and had Holter ECG recordings whose periods taken up by artifacts or noise were less than \(5\%\). No intravenous positive-inotropic agents or vasodilators were administered during the Holter ECG recordings. We excluded patients with chronic or paroxysmal atrial fibrillation, permanent or temporary cardiac pacemakers, active thyroid disease, or malignancy.
Follow-up and endpoint
We recorded the baseline data upon hospital discharge and the time-to-event information for each subject in a database. We then periodically sent questionnaires to patients or their families during the follow-up period and conducted telephone interviews to gather mortality information. Death from progressive heart failure was defined as death resulting from multi-organ failure caused by the progression of pump failure, and sudden death was defined as either witnessed cardiac arrest or death within one hour of onset of acute symptoms or the unexpected death of a patient known to have been well within the previous 24 hours.
Analysis of holter ECG
Using proprietary software, we digitized ECG signals at 125 Hz and 12 bits (Cardy Analyzer II, Suzuken Co., Ltd., Nagoya, Japan). We included only recordings with at least 22 hours of data in the analysis and \(>95\%\) of quantified sinus beats. Although the Cardy Analyzer II software had detected and labeled all QRS complexes in each recording, we manually corrected any errors in R-wave detection and QRS labeling. We then exported the individual files containing the duration of individual RRi intervals and morphology classifications of individual QRS complexes (normal, supraventricular, and ventricular premature complexes, supraventricular, and ventricular escape beats). We analyzed the 24-hour sequence of intervals between two successive R waves of sinus rhythm (i.e., heart rate variability or HRV). To avoid the adverse effects of any remaining errors in detecting the R wave, we reviewed large (\(>20\%\)) consecutive RRi interval differences until all errors were corrected. In addition, when we encountered atrial or ventricular premature complexes, we interpolated the corresponding RRi intervals by the median of the two successive beat-to-beat intervals. We also confirmed that no sustained tachyarrhythmias were present in the HRV recordings. We then interpolated the observed RRi series with a cubic spline function and resampled at an interval (\(\Delta t\)) of 500 ms (2 Hz), yielding interpolated RRi series.
A previous study employing the same dataset, as reported by Kiyono et al.99, did not reveal any noticeable distinctions related to sex, disease severity based on the New York Heart Association classification, prevalence of ischemic heart disease, or ventricular premature beat frequency when comparing survivors and nonsurvivors. Furthermore, no significant differences were observed in key heart rate parameters, including mean RRi, time- and frequency-domain heart rate variability (HRV) measures, or the fractal exponents \(\alpha _{1}\) and \(\alpha _{2}\), between the two groups.
Estimating descriptors of HRV for epoch series
We computed the following descriptors of HRV—linear descriptors over nonoverlapping 500-beat epochs extracted from the RRi series and fractal and multifractal descriptors over nonoverlapping 1000-sample epochs extracted from the interpolated RRi series. Hence, we computed fractal and multifractal descriptors in the time domain, as both are time-domain analytical methods. We computed these descriptors for the original (i.e., unshuffled) and a shuffled counterpart (i.e., a version with the temporal information destroyed) of each RRi series.
Conventional linear descriptors
We computed four linear descriptors of HRV. (i) Mean of successive RR intervals (M). (ii) Root mean square of successive RR intervals (RMS) mathematically defined as
(iii) Number of pairs of successive RRi intervals that differ by more than 50 ms (\(\textrm{NN50}\)). (iv) The percentage of successive RRi intervals that differ from each other by more than 50 ms (\(\textrm{pNN50}\)).
Fractal-scaling descriptor of long-range correlations using monofractal detrended fluctuation analysis
Detrended fluctuation analysis (DFA) computes the Hurst exponent, \(H_{fGn}\), quantifying the strength of long-range correlations in series179,180 using the first-order integration of T-length series x(t):
DFA computes root mean square (RMS; i.e., averaging the residuals) for each linear trend \(y_{n}(t)\) fit to \(N_n\) nonoverlapping n-length bins to build a fluctuation function:
f(n) is a power law,
where \(H_{fGn}\) is the scaling exponent estimable using logarithmic transformation:
Higher \(H_{fGn}\) corresponds to stronger long-range correlations.
Multifractal spectrum width based on the direct estimation of singularity spectrum
Chhabra and Jensen’s181 direct method estimates multifractal spectrum width \(\Delta \alpha\) by sampling a series x(t) at progressively larger scales using the proportion of signal \(P_{i}(n)\) falling within the vth bin of scale n as
As n increases, \(P_{v}(n)\) represents a progressively larger proportion of x(t),
suggesting a growth of the proportion according to one “singularity” strength \(\alpha\)133. P(n) exhibits multifractal dynamics when it grows heterogeneously across time scales n according to multiple singularity strengths, such that
whereby each vth bin may show a distinct relationship of P(n) with n. The width of this singularity spectrum, \(\Delta \alpha =(\alpha _{max}-\alpha _{min})\), indicates the heterogeneity of these relationships182,183.
Chhabra and Jensen’s181 method estimates P(n) for \(N_{n}\) nonoverlapping bins of n-sizes and transforms them into a “mass” \(\mu (q)\) using a q parameter emphasizing higher or lower P(n) for \(q>1\) and \(q<1\), respectively, in the form
Then, \(\alpha (q)\) is the singularity for mass \(\mu\)-weighted P(n) estimated as
Each estimated value of \(\alpha (q)\) belongs to the multifractal spectrum only when the Shannon entropy of \(\mu (q,n)\) scales with n according to the Hausdorff dimension f(q)181, where
For values of q yielding a strong relationship between Eqs. (15) and (16)—in this study, correlation coefficient \(r > 0.9975\), the parametric curve \((\alpha (q),f(q))\) or \((\alpha ,f(\alpha ))\) constitutes the multifractal spectrum and \(\Delta \alpha\) (i.e., \(\alpha _{max}-\alpha _{min}\)) constitutes the multifractal spectrum width. r determines that only scaling relationships of comparable strength can support the estimation of the multifractal spectrum, whether generated as cascades or surrogates. Using a correlation benchmark aims to operationalize previously raised concerns about mis-specifications of the multifractal spectrum184.
Surrogate testing using Iterated Amplitude Adjusted Fourier Transformation (IAAFT) generated t-statistic, \(t_{MF}\)
While multifractality is necessary for cascade-like interactivity, multifractality is not conclusive evidence of cascade-like interactivity, as it can follow from other sources, e.g., linear autocorrelation and outliers in the histogram185. To identify whether non-zero multifractal spectrum width (i.e., \(\Delta \alpha >0\)) reflected multifractality due to nonlinear interactions across scales, we compared \(\Delta \alpha\) for the original and shuffled RRi series to \(\Delta \alpha\) for 32 iterated amplitude adjusted Fourier transform (IAAFT) surrogates186,187. IAAFT randomizes original values time-symmetrically around the autoregressive structure, generating surrogates with randomized phase ordering of the series’ spectral amplitudes while preserving linear temporal correlations. We refer interesting readers to Kelty-Stephen et al.107 for a step-by-step guide to generating the IAAFT surrogates for any series. The resulting surrogate series should thus have the same values as the original series and thus the same mean and variance. It should also have the same amplitude spectrum and autocorrelation function as the original series. The one-sample t-statistic, \(t_{MF}\) takes the subtractive difference between \(\Delta \alpha\) for the original series and that for 32 surrogates, dividing by the standard error of \(\Delta \alpha\) for the surrogates.
Estimating ergodicity breaking parameter, \(E_{B}\)
Ergodicity can be quantified using a dimensionless statistic of ergodicity breaking parameter, \(E_{B}\), also known as the Thirumalai-Mountain metric102,103 and already mentioned by Rytov et al.134, computed as
where \(\overline{\delta ^{2}(x(t))}=\int _{0}^{t-\Delta }[x(t^{\prime }+\Delta )-x(t^{\prime })]^{2}dt^{\prime }\bigl /(t-\Delta )\) is the time average mean-squared displacement of the stochastic series x(t) for lag time \(\Delta\). This relationship is effectively the variance of sample variance divided by the total-sample squared variance. Rapid decay of \(E_{B}\) to a finite asymptotic value for progressively larger samples, i.e., \(E_{B}\rightarrow 0\) as \(t\rightarrow \infty\) implies ergodicity. Thus, for Brownian motion \(E_{B}(x(t))=\frac{4}{3}(\frac{\Delta }{t})\)148,188. Slower decay indicates less ergodic systems in which trajectories are less reproducible, and no decay or convergence to a finite asymptotic value indicates strong ergodicity breaking104,105. \(E_{B}(x(t))\) thus allows testing whether a given series fulfills ergodic assumptions or breaks ergodicity. For instance, it has been shown that for fractional Brownian motion (FBM)104,105,
The present work is less focused on firmly meeting the criterion of \(E_{B}\) converging to zero within our finite samples. Instead, we compared the original and shuffled RRi series to assess ergodicity breaking instead of strict convergence of \(E_{B}\) to zero. We computed \(E_{B}\) for each original and shuffled RRi series (range \(=T/50\); lag \(\Delta =10\)) and for each epoch series of M, RMS, \(\textrm{NN50}\), \(\textrm{PNN50}\), \(H_{fGn}\), and \(t_{MF}\) for the original and shuffled RRi series (range \(=N_{\textrm{epochs}}/2\); lag \(\Delta =1\)).
Monte Carlo simulations
We performed Monte Carlo simulations to test our hypothesis that ergodicity breaking by various linear and cascade-dynamical HRV descriptors could compromise these descriptors’ reliability as diagnostic biomarkers. We randomly sampled 1000-sample RRi series from 24-hour recordings for each individual and performed linear mixed-effects models separately on M, RMS, \(\textrm{NN50}\), \(\textrm{pNN50}\), \(H_{fGn}\), \(t_{MF}\), values calculated from these series. We used linear mixed-effects models with each descriptor as the dependent variable and the participant group as the independent variable. The t-statistic and the resultant p value were saved across the 1000 iterations. We performed all mixed-effects modeling in MATLAB 2022b (Mathworks, Inc., Natick, MA) using the function fitlme().
Survival analysis
We examined whether \(H_{fGn}\) and \(t_{MF}\) were predictive of death using univariate Cox proportional hazards regression analysis108,109. We used the Mantel-Haenszel log-rank test to compare Kaplan-Meier cumulative survival curves to examine the impact of identified risk factors on survival. We performed all survival analysis in R189 using the function coxph() from the package “survival”190. The sex ratio among the survivors, as presented in Table 1, exhibits a notable skew towards males (42M/27F) when contrasted with the non-survivors (19M/20F). Given that sex constitutes a pivotal physiological determinant in cardiac health, it merits consideration in interpreting results. Nonetheless, it is worth noting that a prior study utilizing the same dataset failed to uncover any discernible disparities concerning sex between survivors and nonsurvivors99. Consequently, we lacked a compelling rationale to anticipate that sex would influence any facet of the current analysis.
Data availability
The data that support the results reported herein can be obtained upon request from the corresponding author.
References
Tsao, C. W. et al. Heart disease and stroke statistics–2023 update: A report from the American heart association. Circulation 147, e93–e621. https://doi.org/10.1161/CIR.0000000000001123 (2023).
Roth, G. A. et al. Global burden of cardiovascular diseases and risk factors, 1990–2019: Update from the gbd 2019 study. J. Am. Coll. Cardiol. 76, 2982–3021. https://doi.org/10.1016/j.jacc.2020.11.010 (2020).
Omland, T. & White, H. D. State of the art: Blood biomarkers for risk stratification in patients with stable ischemic heart disease. Clin. Chem. 63, 165–176. https://doi.org/10.1373/clinchem.2016.255190 (2017).
Mănescu, I.-B., Pál, K., Lupu, S. & Dobreanu, M. Conventional biomarkers for predicting clinical outcomes in patients with heart disease. Life 12, 2112. https://doi.org/10.3390/life12122112 (2022).
Pál, K., Mănescu, I.-B., Lupu, S. & Dobreanu, M. Emerging biomarkers for predicting clinical outcomes in patients with heart disease. Life 13, 230 (2023).
Zwack, C. C. et al. The evolution of digital health technologies in cardiovascular disease research. NPJ Digit. Med. 6, 1. https://doi.org/10.1038/s41746-022-00734-2 (2023).
Yeung, A. W. K. et al. Research on digital technology use in cardiology: Bibliometric analysis. J. Med. Internet Res. 24, e36086. https://doi.org/10.2196/36086 (2022).
Hughes, A., Shandhi, M. M. H., Master, H., Dunn, J. & Brittain, E. Wearable devices in cardiovascular medicine. Circ. Res. 132, 652–670. https://doi.org/10.1161/CIRCRESAHA.122.322389 (2023).
Nes, L. S. Digital health in cardiology: Time for action. Cardiology 145, 106–109. https://doi.org/10.1159/000504797 (2020).
Karemaker, J. M. An introduction into autonomic nervous function. Physiol. Meas. 38, R89. https://doi.org/10.1088/1361-6579/aa6782 (2017).
Agliari, E. et al. Detecting cardiac pathologies via machine learning on heart-rate variability time series and related markers. Sci. Rep. 10, 8845. https://doi.org/10.1038/s41598-020-64083-4 (2020).
Freeman, J. V., Dewey, F. E., Hadley, D. M., Myers, J. & Froelicher, V. F. Autonomic nervous system interaction with the cardiovascular system during exercise. Prog. Cardiovasc. Dis. 48, 342–362. https://doi.org/10.1016/j.pcad.2005.11.003 (2006).
Zhong, Y. et al. Autonomic nervous nonlinear interactions lead to frequency modulation between low-and high-frequency bands of the heart rate variability spectrum. Am. J. Physiol.-Regul. Integrat. Comp. Physiol. 293, R1961–R1968. https://doi.org/10.1152/ajpregu.00362.2007 (2007).
Beissner, F., Meissner, K., Bär, K.-J. & Napadow, V. The autonomic brain: An activation likelihood estimation meta-analysis for central processing of autonomic function. J. Neurosci. 33, 10503–10511. https://doi.org/10.1523/JNEUROSCI.1103-13.2013 (2013).
Brosschot, J. F., Verkuil, B. & Thayer, J. F. Exposed to events that never happen: Generalized unsafety, the default stress response, and prolonged autonomic activity. Neurosci. Biobehav. Rev. 74, 287–296. https://doi.org/10.1016/j.neubiorev.2016.07.019 (2017).
Cechetto, D. F. & Shoemaker, J. K. Functional neuroanatomy of autonomic regulation. Neuroimage 47, 795–803. https://doi.org/10.1016/j.neuroimage.2009.05.024 (2009).
Guyenet, P. G. The sympathetic control of blood pressure. Nat. Rev. Neurosci. 7, 335–346. https://doi.org/10.1038/nrn1902 (2006).
Ruiz Vargas, E., Sörös, P., Shoemaker, J. K. & Hachinski, V. Human cerebral circuitry related to cardiac control: A neuroimaging meta-analysis. Ann. Neurol. 79, 709–716. https://doi.org/10.1002/ana.24642 (2016).
Thayer, J. F. & Lane, R. D. Claude Bernard and the heart-brain connection: Further elaboration of a model of neurovisceral integration. Neurosci. Biobehav. Rev. 33, 81–88. https://doi.org/10.1016/j.neubiorev.2008.08.004 (2009).
Thayer, J. F., Åhs, F., Fredrikson, M., Sollers, J. J. III. & Wager, T. D. A meta-analysis of heart rate variability and neuroimaging studies: Implications for heart rate variability as a marker of stress and health. Neurosci. Biobehav. Rev. 36, 747–756. https://doi.org/10.1016/j.neubiorev.2011.11.009 (2012).
Thayer, J. F., Mather, M. & Koenig, J. Stress and aging: A neurovisceral integration perspective. Psychophysiology 58, e13804. https://doi.org/10.1111/psyp.13804 (2021).
Wulsin, L., Herman, J. & Thayer, J. F. Stress, autonomic imbalance, and the prediction of metabolic risk: A model and a proposal for research. Neurosci. Biobehav. Rev. 86, 12–20. https://doi.org/10.1016/j.neubiorev.2017.12.010 (2018).
Bootsma, M. et al. Heart rate and heart rate variability as indexes of sympathovagal balance. Am. J. Physiol.-Heart Circ. Physiol. 266, H1565–H1571. https://doi.org/10.1152/ajpheart.1994.266.4.H1565 (1994).
Khan, A. A., Lip, G. Y. & Shantsila, A. Heart rate variability in atrial fibrillation: The balance between sympathetic and parasympathetic nervous system. Eur. J. Clin. Invest. 49, e13174. https://doi.org/10.1111/eci.13174 (2019).
van Ravenswaaij-Arts, C. M., Kollee, L. A., Hopman, J. C., Stoelinga, G. B. & van Geijn, H. P. Heart rate variability. Ann. Int. Med. 118, 436–447. https://doi.org/10.7326/0003-4819-118-6-199303150-00008 (1993).
Jarczok, M. N., Li, J., Mauss, D., Fischer, J. E. & Thayer, J. F. Heart rate variability is associated with glycemic status after controlling for components of the metabolic syndrome. Int. J. Cardiol. 167, 855–861. https://doi.org/10.1016/j.ijcard.2012.02.002 (2013).
Jarczok, M. N. et al. Investigating the associations of self-rated health: Heart rate variability is more strongly associated than inflammatory and other frequently used biomarkers in a cross sectional occupational sample. PLoS ONE 10, e0117196. https://doi.org/10.1371/journal.pone.0117196 (2015).
Captur, G., Karperien, A. L., Hughes, A. D., Francis, D. P. & Moon, J. C. The fractal heart-Embracing mathematics in the cardiology clinic. Nat. Rev. Cardiol. 14, 56–64. https://doi.org/10.1038/nrcardio.2016.161 (2017).
Gierałtowski, J., Żebrowski, J. & Baranowski, R. Multiscale multifractal analysis of heart rate variability recordings with a large number of occurrences of arrhythmia. Phys. Rev. E 85, 021915. https://doi.org/10.1103/PhysRevE.85.021915 (2012).
Goldberger, A. L. et al. Fractal dynamics in physiology: Alterations with disease and aging. Proc. Natl. Acad. Sci. 99, 2466–2472. https://doi.org/10.1073/pnas.012579499 (2002).
Ivanov, P. C. et al. Scaling behaviour of heartbeat intervals obtained by wavelet-based time-series analysis. Nature 383, 323–327. https://doi.org/10.1038/383323a0 (1996).
Ivanov, P. C. et al. From 1/f noise to multifractal cascades in heartbeat dynamics. Chaos 11, 641–652. https://doi.org/10.1063/1.1395631 (2001).
Kiyono, K. et al. Critical scale invariance in a healthy human heart rate. Phys. Rev. Lett. 93, 178103. https://doi.org/10.1103/PhysRevLett.93.178103 (2004).
Kiyono, K., Struzik, Z. R., Aoyagi, N., Togo, F. & Yamamoto, Y. Phase transition in a healthy human heart rate. Phys. Rev. Lett. 95, 058101. https://doi.org/10.1103/PhysRevLett.95.058101 (2005).
Kurths, J. et al. Quantitative analysis of heart rate variability. Chaos 5, 88–94. https://doi.org/10.1063/1.166090 (1995).
Lefebvre, J., Goodings, D., Kamath, M. & Fallen, E. Predictability of normal heart rhythms and deterministic chaos. Chaos 3, 267–276. https://doi.org/10.1063/1.165990 (1993).
Lin, D. & Hughson, R. Modeling heart rate variability in healthy humans: A turbulence analogy. Phys. Rev. Lett. 86, 1650. https://doi.org/10.1103/PhysRevLett.86.1650 (2001).
Peng, C.-K. et al. Fractal mechanisms and heart rate dynamics: Long-range correlations and their breakdown with disease. J. Electrocardiol. 28, 59–65. https://doi.org/10.1016/S0022-0736(95)80017-4 (1995).
Perkiömäki, J. S. Heart rate variability and non-linear dynamics in risk stratification. Front. Physiol. 2, 81. https://doi.org/10.3389/fphys.2011.00081 (2011).
Sugihara, G., Allan, W., Sobel, D. & Allan, K. D. Nonlinear control of heart rate variability in human infants. Proc. Natl. Acad. Sci. 93, 2608–2613. https://doi.org/10.1073/pnas.93.6.2608 (1996).
Tan, C. O., Cohen, M. A., Eckberg, D. L. & Taylor, J. A. Fractal properties of human heart period variability: Physiological and methodological implications. J. Physiol. 587, 3929–3941. https://doi.org/10.1113/jphysiol.2009.169219 (2009).
Voss, A., Schulz, S., Schroeder, R., Baumert, M. & Caminal, P. Methods derived from nonlinear dynamics for analysing heart rate variability. Philos. Trans. R. Soc. A: Math. Phys. Eng. Sci. 367, 277–296. https://doi.org/10.1098/rsta.2008.0232 (2009).
Kelty-Stephen, D. G., Palatinus, K., Saltzman, E. & Dixon, J. A. A tutorial on multifractality, cascades, and interactivity for empirical time series in ecological science. Ecol. Psychol. 25, 1–62. https://doi.org/10.1080/10407413.2013.753804 (2013).
Turcotte, D. L., Malamud, B. D., Guzzetti, F. & Reichenbach, P. Self-organization, the cascade model, and natural hazards. Proc. Natl. Acad. Sci. 99, 2530–2537 (2002).
Lovejoy, S. & Schertzer, D. Multifractals, cloud radiances and rain. J. Hydrol. 322, 59–88. https://doi.org/10.1016/j.jhydrol.2005.02.042 (2006).
Olsson, J., Persson, M. & Jinno, K. Analysis and modeling of solute transport dynamics by breakdown coefficients and random cascades. Water Resour. Res. 43, W03417. https://doi.org/10.1029/2005WR004631 (2007).
Ihlen, E. A. & Vereijken, B. Interaction-dominant dynamics in human cognition: Beyond \(1/f^{\alpha }\) fluctuation. J. Experim. Psychol.: Gen. 139, 436–463. https://doi.org/10.1037/a0019098 (2010).
Bloomfield, L., Lane, E., Mangalam, M. & Kelty-Stephen, D. G. Perceiving and remembering speech depend on multifractal nonlinearity in movements producing and exploring speech. J. R. Soc. Interface 18, 20210272. https://doi.org/10.1098/rsif.2021.0272 (2021).
Kelty-Stephen, D. G., Lee, I. C., Carver, N. S., Newell, K. M. & Mangalam, M. Multifractal roots of suprapostural dexterity. Hum. Mov. Sci. 76, 102771. https://doi.org/10.1016/j.humov.2021.102771 (2021).
Kelty-Stephen, D. G. & Mangalam, M. Turing’s cascade instability supports the coordination of the mind, brain, and behavior. Neurosci. Biobehav. Rev. 141, 104810. https://doi.org/10.1016/j.neubiorev.2022.104810 (2022).
Kelty-Stephen, D. G., Lee, J., Cole, K. R., Shields, R. K. & Mangalam, M. Multifractal nonlinearity moderates feedforward and feedback responses to suprapostural perturbations. Percept. Mot. Skills 130, 622–657. https://doi.org/10.1177/00315125221149147 (2023).
Mangalam, M., Carver, N. S. & Kelty-Stephen, D. G. Global broadcasting of local fractal fluctuations in a bodywide distributed system supports perception via effortful touch. Chaos Solitons Fract. 135, 109740. https://doi.org/10.1016/j.chaos.2020.109740 (2020).
Mangalam, M., Carver, N. S. & Kelty-Stephen, D. G. Multifractal signatures of perceptual processing on anatomical sleeves of the human body. J. R. Soc. Interface 17, 20200328. https://doi.org/10.1098/rsif.2020.0328 (2020).
Mangalam, M. & Kelty-Stephen, D. G. Multiplicative-cascade dynamics supports whole-body coordination for perception via effortful touch. Hum. Mov. Sci. 70, 102595. https://doi.org/10.1016/j.humov.2020.102595 (2020).
Biswas, D., Simões-Capela, N., Van Hoof, C. & Van Helleputte, N. Heart rate estimation from wrist-worn photoplethysmography: A review. IEEE Sens. J. 19, 6560–6570. https://doi.org/10.1109/JSEN.2019.2914166 (2019).
Cajal, D. et al. Effects of missing data on heart rate variability metrics. Sensors 22, 5774. https://doi.org/10.3390/s22155774 (2022).
Georgiou, K. et al. Can wearable devices accurately measure heart rate variability? A systematic review. Folia Medica 60, 7–20. https://doi.org/10.2478/folmed-2018-0012 (2018).
Isakadze, N. & Martin, S. S. How useful is the smartwatch ECG?. Trends Cardiovasc. Med. 30, 442–448. https://doi.org/10.1016/j.tcm.2019.10.010 (2020).
Karemaker, J. M. Interpretation of heart rate variability: The art of looking through a keyhole. Front. Neurosci. 14, 609570. https://doi.org/10.3389/fnins.2020.609570 (2020).
Karemaker, J. M. The multibranched nerve: Vagal function beyond heart rate variability. Biol. Psychol. 172, 108378. https://doi.org/10.1016/j.biopsycho.2022.108378 (2022).
Nelson, B. W. & Allen, N. B. Accuracy of consumer wearable heart rate measurement during an ecologically valid 24-hour period: Intraindividual validation study. JMIR Mhealth Uhealth 7, e10828 (2019).
Raja, J. M. et al. Apple watch, wearables, and heart rhythm: Where do we stand?. Ann. Transl. Med. 7, 417 (2019).
Shaffer, F., McCraty, R. & Zerr, C. L. A healthy heart is not a metronome: An integrative review of the heart’s anatomy and heart rate variability. Front. Psychol. 5, 1040. https://doi.org/10.3389/fpsyg.2014.01040 (2014).
Hayano, J. & Yuda, E. Pitfalls of assessment of autonomic function by heart rate variability. J. Physiol. Anthropol. 38, 3. https://doi.org/10.1186/s40101-019-0193-2 (2019).
Shaffer, F. & Ginsberg, J. P. An overview of heart rate variability metrics and norms. Front. Public Health 5, 258. https://doi.org/10.3389/fpubh.2017.00258 (2017).
Thomas, B. L., Claassen, N., Becker, P. & Viljoen, M. Validity of commonly used heart rate variability markers of autonomic nervous system function. Neuropsychobiology 78, 14–26. https://doi.org/10.1159/000495519 (2019).
Galton, F. Inquiries into Human Faculty and Its Development (Macmillan, New York, NY, 1883).
Aldrich, J. The origins of modern statistics: The english statistical school. In Hájek, A. & Hitchcock, C. (eds.) The Oxford Handbook of Probability and Philosophy, 112–129 (Oxford University Press, Oxford, UK, 2016).
Molenaar, P. C. A manifesto on psychology as idiographic science: Bringing the person back into scientific psychology, this time forever. Measurement 2, 201–218. https://doi.org/10.1207/s15366359mea0204_1 (2004).
Molenaar, P. C. & Campbell, C. G. The new person-specific paradigm in psychology. Curr. Dir. Psychol. Sci. 18, 112–117. https://doi.org/10.1111/j.1467-8721.2009.01619.x (2009).
Voelkle, M. C., Brose, A., Schmiedek, F. & Lindenberger, U. Toward a unified framework for the study of between-person and within-person structures: Building a bridge between two research paradigms. Multivar. Behav. Res. 49, 193–213. https://doi.org/10.1080/00273171.2014.889593 (2014).
Mangalam, M. & Kelty-Stephen, D. G. Point estimates, Simpson’s paradox, and nonergodicity in biological sciences. Neurosci. Biobehav. Rev. 125, 98–107. https://doi.org/10.1016/j.neubiorev.2021.02.017 (2021).
Kelty-Stephen, D. G. & Mangalam, M. Fractal and multifractal descriptors restore ergodicity broken by non-Gaussianity in time series. Chaos Solitons Fract. 163, 112568. https://doi.org/10.1016/j.chaos.2022.112568 (2022).
Fisher, A. J., Medaglia, J. D. & Jeronimus, B. F. Lack of group-to-individual generalizability is a threat to human subjects research. Proc. Natl. Acad. Sci. 115, E6106–E6115. https://doi.org/10.1073/pnas.1711978115 (2018).
Li, J. et al. Non-ergodicity of a globular protein extending beyond its functional timescale. Chem. Sci. 13, 9668–9677. https://doi.org/10.1039/D2SC03069A (2022).
Weigel, A. V., Simon, B., Tamkun, M. M. & Krapf, D. Ergodic and nonergodic processes coexist in the plasma membrane as observed by single-molecule tracking. Proc. Natl. Acad. Sci. 108, 6438–6443. https://doi.org/10.1073/pnas.1016325108 (2011).
Card, K. J., LaBar, T., Gomez, J. B. & Lenski, R. E. Historical contingency in the evolution of antibiotic resistance after decades of relaxed selection. PLoS Biol. 17, e3000397. https://doi.org/10.1371/journal.pbio.3000397 (2019).
Xie, V. C., Pu, J., Metzger, B. P., Thornton, J. W. & Dickinson, B. C. Contingency and chance erase necessity in the experimental evolution of ancestral proteins. eLife10, e67336 (2021). https://doi.org/10.7554/eLife.67336.
Bayoumy, K. et al. Smart wearable devices in cardiovascular care: Where we are and how to move forward. Nat. Rev. Cardiol. 18, 581–599. https://doi.org/10.1038/s41569-021-00522-7 (2021).
Marcus, G. M. The Apple Watch can detect atrial fibrillation: So what now?. Nat. Rev. Cardiol. 17, 135–136. https://doi.org/10.1038/s41569-019-0330-y (2020).
Kelty-Stephen, D. G. & Mangalam, M. Multifractal descriptors ergodically characterize non-ergodic multiplicative cascade processes. Physica A 616, 128651. https://doi.org/10.1016/j.physa.2023.128651 (2023).
Mangalam, M. & Kelty-Stephen, D. G. Ergodic descriptors of non-ergodic stochastic processes. J. R. Soc. Interface 19, 20220095. https://doi.org/10.1098/rsif.2022.0095 (2022).
Hanin, L. Why statistical inference from clinical trials is likely to generate false and irreproducible results. BMC Med. Res. Methodol. 17, 127. https://doi.org/10.1186/s12874-017-0399-0 (2017).
Ioannidis, J. P. Why most published research findings are false. PLoS Med. 2, e124. https://doi.org/10.1371/journal.pmed.0020124 (2005).
Bassi, D. et al. Inter and intra-rater reliability of short-term measurement of heart rate variability on rest in diabetic type 2 patients. J. Med. Syst. 42, 1–7. https://doi.org/10.1007/s10916-018-1101-8 (2018).
Farah, B. Q. et al. Intra-individuals and inter-and intra-observer reliability of short-term heart rate variability in adolescents. Clin. Physiol. Funct. Imaging 36, 33–39. https://doi.org/10.1111/cpf.12190 (2016).
da Cruz, C. J. G. et al. Impact of heart rate on reproducibility of heart rate variability analysis in the supine and standing positions in healthy men. Clinics 74, e806. https://doi.org/10.6061/clinics/2019/e806 (2019).
Højgaard, M. V., Holstein-Rathlou, N.-H., Agner, E. & Kanters, J. K. Reproducibility of heart rate variability, blood pressure variability and baroreceptor sensitivity during rest and head-up tilt. Blood Press. Monit. 10, 19–24. https://doi.org/10.1097/00126097-200502000-00005 (2005).
Leicht, A. & Allen, G. Moderate-term reproducibility of heart rate variability during rest and light to moderate exercise in children. Braz. J. Med. Biol. Res. 41, 627–633. https://doi.org/10.1590/S0100-879X2008000700013 (2008).
Lord, S. et al. Low-frequency heart rate variability: Reproducibility in cardiac transplant recipients and normal subjects. Clin. Sci. 100, 43–46. https://doi.org/10.1042/cs1000043 (2001).
Pitzalis, M. V. et al. Short-and long-term reproducibility of time and frequency domain heart rate variability measurements in normal subjects. Cardiovasc. Res. 32, 226–233. https://doi.org/10.1016/0008-6363(96)00086-7 (1996).
Plaza-Florido, A. et al. Inter-and intra-researcher reproducibility of heart rate variability parameters in three human cohorts. Sci. Rep. 10, 11399. https://doi.org/10.1038/s41598-020-68197-7 (2020).
Sacha, J., Sobon, J., Sacha, K. & Barabach, S. Heart rate impact on the reproducibility of heart rate variability analysis. Int. J. Cardiol. 168, 4257–4259. https://doi.org/10.1016/j.ijcard.2013.04.160 (2013).
Sandercock, G. R., Bromley, P. D. & Brodie, D. A. The reliability of short-term measurements of heart rate variability. Int. J. Cardiol. 103, 238–247. https://doi.org/10.1016/j.ijcard.2004.09.013 (2005).
Mangalam, M., Metzler, R. & Kelty-Stephen, D. Ergodic characterization of non-ergodic anomalous diffusion. Phys. Rev. Res. 5(2), 023144. https://doi.org/10.1098/rsif.2022.0095 (2023).
Kiyono, K., Struzik, Z. R., Aoyagi, N. & Yamamoto, Y. Multiscale probability density function analysis: Non-Gaussian and scale-invariant fluctuations of healthy human heart rate. IEEE Trans. Biomed. Eng. 53, 95–102. https://doi.org/10.1109/TBME.2005.859804 (2005).
Hayano, J. et al. Increased non-Gaussianity of heart rate variability predicts cardiac mortality after an acute myocardial infarction. Front. Physiol. 2, 65. https://doi.org/10.3389/fphys.2011.00065 (2011).
Kiyono, K., Struzik, Z. R. & Yamamoto, Y. Estimator of a non-Gaussian parameter in multiplicative log-normal models. Phys. Rev. E 76, 041113. https://doi.org/10.1103/PhysRevE.76.041113 (2007).
Kiyono, K., Hayano, J., Watanabe, E., Struzik, Z. R. & Yamamoto, Y. Non-Gaussian heart rate as an independent predictor of mortality in patients with chronic heart failure. Heart Rhythm 5, 261–268. https://doi.org/10.1016/j.hrthm.2007.10.030 (2008).
Kiyono, K. Log-amplitude statistics of intermittent and non-Gaussian time series. Phys. Rev. E 79, 031129. https://doi.org/10.1103/PhysRevE.79.031129 (2009).
Kiyono, K., Hayano, J., Kwak, S., Watanabe, E. & Yamamoto, Y. Non-Gaussianity of low frequency heart rate variability and sympathetic activation: Lack of increases in multiple system atrophy and Parkinson disease. Front. Physiol. 3, 34. https://doi.org/10.3389/fphys.2012.00034 (2012).
He, Y., Burov, S., Metzler, R. & Barkai, E. Random time-scale invariant diffusion and transport coefficients. Phys. Rev. Lett. 101, 058101. https://doi.org/10.1103/PhysRevLett.101.058101 (2008).
Thirumalai, D., Mountain, R. D. & Kirkpatrick, T. Ergodic behavior in supercooled liquids and in glasses. Phys. Rev. A 39, 3563. https://doi.org/10.1103/PhysRevA.39.3563 (1989).
Deng, W. & Barkai, E. Ergodic properties of fractional Brownian–Langevin motion. Phys. Rev. E 79, 011112. https://doi.org/10.1103/PhysRevE.79.011112 (2009).
Wang, W. et al. Fractional Brownian motion with random diffusivity: Emerging residual nonergodicity below the correlation time. J. Phys. A: Math. Theor. 53, 474001. https://doi.org/10.1088/1751-8121/aba467 (2020).
Lovejoy, S. & Schertzer, D. The Weather and Climate: Emergent Laws and Multifractal Cascades (Cambridge University Press, Cambridge, UK, 2018).
Kelty-Stephen, D. G., Lane, E., Bloomfield, L. & Mangalam, M. Multifractal test for nonlinearity of interactions across scales in time series. Behav. Res. Methods 55, 2249–2282 (2023). https://doi.org/10.3758/s13428-022-01866-9.
Efron, B. Logistic regression, survival analysis, and the Kaplan-Meier curve. J. Am. Stat. Assoc. 83, 414–425 (1988).
Rich, J. T. et al. A practical guide to understanding Kaplan–Meier curves. Otolaryngol.-Head Neck Surg. 143, 331–336. https://doi.org/10.1016/j.otohns.2010.05.007 (2010).
Coutts, L. V., Plans, D., Brown, A. W. & Collomosse, J. Deep learning with wearable based heart rate variability for prediction of mental and general health. J. Biomed. Inform. 112, 103610. https://doi.org/10.1016/j.jbi.2020.103610 (2020).
Hannun, A. Y. et al. Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat. Med. 25, 65–69. https://doi.org/10.1038/s41591-018-0268-3 (2019).
Hong, S., Zhou, Y., Shang, J., Xiao, C. & Sun, J. Opportunities and challenges of deep learning methods for electrocardiogram data: A systematic review. Comput. Biol. Med. 122, 103801. https://doi.org/10.1016/j.compbiomed.2020.103801 (2020).
Kim, H., Ishag, M. I. M., Piao, M., Kwon, T. & Ryu, K. H. A data mining approach for cardiovascular disease diagnosis using heart rate variability and images of carotid arteries. Symmetry 8, 47. https://doi.org/10.3390/sym8060047 (2016).
Torres-Soto, J. & Ashley, E. A. Multi-task deep learning for cardiac rhythm detection in wearable devices. NPJ Digit. Med.3, 116 (2020). https://doi.org/10.1038/s41746-020-00320-4.
Wang, L. & Zhou, X. Detection of congestive heart failure based on LSTM-based deep network via short-term RR intervals. Sensors 19, 1502. https://doi.org/10.3390/s19071502 (2019).
Baek, H. J. & Shin, J. Effect of missing inter-beat interval data on heart rate variability analysis using wrist-worn wearables. J. Med. Syst. 41, 147. https://doi.org/10.1007/s10916-017-0796-2 (2017).
Faust, L., Feldman, K., Mattingly, S. M., Hachen, D. & V. Chawla, N. Deviations from normal bedtimes are associated with short-term increases in resting heart rate. NJP Digit. Med.3, 39 (2020). https://doi.org/10.1038/s41746-020-0250-6.
Kim, K. K., Kim, J. S., Lim, Y. G. & Park, K. S. The effect of missing RR-interval data on heart rate variability analysis in the frequency domain. Physiol. Meas. 30, 1039. https://doi.org/10.1111/j.1540-8159.2010.02841.x (2009).
Kim, K. K., Baek, H. J., Lim, Y. G. & Park, K. S. Effect of missing RR-interval data on nonlinear heart rate variability analysis. Comput. Methods Programs Biomed. 106, 210–218. https://doi.org/10.1016/j.cmpb.2010.11.011 (2012).
Mäkikallio, T. H. et al. Fractal analysis of heart rate dynamics as a predictor of mortality in patients with depressed left ventricular function after acute myocardial infarction. Am. J. Cardiol. 83, 836–839. https://doi.org/10.1016/s0002-9149(98)01076-5 (1999).
Huikuri, H. V. et al. Fractal correlation properties of rr interval dynamics and mortality in patients with depressed left ventricular function after an acute myocardial infarction. Circulation 101, 47–53. https://doi.org/10.1161/01.cir.101.1.47 (2000).
Maestri, R. et al. Assessing nonlinear properties of heart rate variability from short-term recordings: Are these measurements reliable?. Physiol. Meas. 28, 1067. https://doi.org/10.1088/0967-3334/28/9/008 (2007).
Perkiömäki, J. S. et al. Fractal and complexity measures of heart rate dynamics after acute myocardial infarction. Am. J. Cardiol. 88, 777–781. https://doi.org/10.1016/s0002-9149(01)01851-3 (2001).
Pikkujamsa, S. M., Makikallio, T. H., Airaksinen, K. J. & Huikuri, H. V. Determinants and interindividual variation of rr interval dynamics in healthy middle-aged subjects. Am. J. Physiol.-Heart Circ. Physiol. 280, H1400–H1406. https://doi.org/10.1152/ajpheart.2001.280.3.H1400 (2001).
Jokinen, V., Tapanainen, J. M., Seppänen, T. & Huikuri, H. V. Temporal changes and prognostic significance of measures of heart rate dynamics after acute myocardial infarction in the beta-blocking era. Am. J. Cardiol. 92, 907–912. https://doi.org/10.1016/S0002-9149(03)00968-8 (2003).
Muzy, J.-F., Bacry, E. & Arneodo, A. Multifractal formalism for fractal signals: The structure-function approach versus the wavelet-transform modulus-maxima method. Phys. Rev. E 47, 875. https://doi.org/10.1103/PhysRevE.47.875 (1993).
Hinrichsen, H., Stenull, O. & Janssen, H.-K. Multifractal current distribution in random-diode networks. Phys. Rev. E 65, 045104. https://doi.org/10.1103/PhysRevE.65.045104 (2002).
Kadanoff, L. P., Nagel, S. R., Wu, L. & Zhou, S.-M. Scaling and universality in avalanches. Phys. Rev. A 39, 6524. https://doi.org/10.1103/PhysRevA.39.6524 (1989).
Sethna, J. P., Dahmen, K. A. & Myers, C. R. Crackling noise. Nature 410, 242–250. https://doi.org/10.1038/35065675 (2001).
Stanley, H. E. Scaling, universality, and renormalization: Three pillars of modern critical phenomena. Rev. Mod. Phys. 71, S358. https://doi.org/10.1103/RevModPhys.71.S358 (1999).
Mandelbrot, B. B. Intermittent turbulence in self-similar cascades: Divergence of high moments and dimension of the carrier. J. Fluid Mech. 62, 331–358. https://doi.org/10.1017/S0022112074000711 (1974).
Mandelbrot, B. B. & Van Ness, J. W. Fractional Brownian motions, fractional noises and applications. SIAM Rev. 10, 422–437. https://doi.org/10.1137/1010093 (1968).
Mandelbrot, B. B. & Mandelbrot, B. B. The Fractal Geometry of Nature (WH Freeman, New York, NY, 1982).
Rytov, S. M., Kravtsov, Y. A. & Tatarskii, V. I. Principles of Statistical Radiophysics: Wave Propagation Through Random Media (Springer, Berlin, Germany, 1989).
Castro-Schilo, L. & Ferrer, E. Comparison of nomothetic versus idiographic-oriented methods for making predictions about distal outcomes from time series data. Multivar. Behav. Res. 48, 175–207. https://doi.org/10.1080/00273171.2012.736042 (2013).
Hamaker, E. L., Dolan, C. V. & Molenaar, P. C. Statistical modeling of the individual: Rationale and application of multivariate stationary time series analysis. Multivar. Behav. Res. 40, 207–233. https://doi.org/10.1207/s15327906mbr4002_3 (2005).
Lowie, W. M. & Verspoor, M. H. Individual differences and the ergodicity problem. Lang. Learn. 69, 184–206. https://doi.org/10.1111/lang.12324 (2019).
Medaglia, J. D., Ramanathan, D. M., Venkatesan, U. M. & Hillary, F. G. The challenge of non-ergodicity in network neuroscience. Netw.: Comput. Neural Syst.22, 148–153 (2011). https://doi.org/10.3109/09638237.2011.639604.
Molenaar, P. C. On the implications of the classical ergodic theorems: Analysis of developmental processes has to focus on intra-individual variation. Dev. Psychobiol. 50, 60–69. https://doi.org/10.1002/dev.20262 (2008).
Molenaar, P., Sinclair, K. O., Rovine, M. J., Ram, N. & Corneal, S. E. Analyzing developmental processes on an individual level using nonstationary time series modeling. Dev. Psychol.45, 260 (2009). https://psycnet.apa.org/doi/10.1037/a0014170.
McLeish, T. C. Are there ergodic limits to evolution? Ergodic exploration of genome space and convergence. Interface Focus 5, 20150041. https://doi.org/10.1098/rsfs.2015.0041 (2015).
Partridge, T. & Greenberg, G. Contemporary ideas in physics and biology in Gottlieb’s psychology. In Hood, K. E., Halpern, C. T., Greenberg, G. & Lerner, R. M. (eds.) Handbook of Developmental Science, Behavior, and Genetics, 121–145 (Wiley-Blackwell Oxford, UK, 1976).
Barkai, E., Garini, Y. & Metzler, R. Strange kinetics of single molecules in living cells. Phys. Today 65, 29. https://doi.org/10.1063/PT.3.1677 (2012).
Fuliński, A. Anomalous diffusion and weak nonergodicity. Phys. Rev. E 83, 061140. https://doi.org/10.1103/PhysRevE.83.061140 (2011).
Krapf, D. & Metzler, R. Strange interfacial molecular dynamics. Phys. Today 72, 48–55. https://doi.org/10.1063/PT.3.4294 (2019).
Magdziarz, M. & Weron, A. Anomalous diffusion: Testing ergodicity breaking in experimental data. Phys. Rev. E 84, 051138. https://doi.org/10.1103/PhysRevE.84.051138 (2011).
Metzler, R. & Jeon, J.-H. The role of ergodicity in anomalous stochastic processes: Analysis of single-particle trajectories. Phys. Scr. 86, 058510. https://doi.org/10.1088/0031-8949/86/05/058510 (2012).
Metzler, R., Jeon, J.-H., Cherstvy, A. G. & Barkai, E. Anomalous diffusion models and their properties: Non-stationarity, non-ergodicity, and ageing at the centenary of single particle tracking. Phys. Chem. Chem. Phys. 16, 24128–24164. https://doi.org/10.1039/C4CP03465A (2014).
Muñoz-Gil, G. et al. Objective comparison of methods to decode anomalous diffusion. Nat. Commun. 12, 6253. https://doi.org/10.1038/s41467-021-26320-w (2021).
Thiel, F. & Sokolov, I. M. Weak ergodicity breaking in an anomalous diffusion process of mixed origins. Phys. Rev. E 89, 012136. https://doi.org/10.1103/PhysRevE.89.012136 (2014).
Vinod, D., Cherstvy, A. G., Wang, W., Metzler, R. & Sokolov, I. M. Nonergodicity of reset geometric Brownian motion. Phys. Rev. E 105, L012106. https://doi.org/10.1103/PhysRevE.105.L012106 (2022).
Wang, W., Cherstvy, A. G., Metzler, R. & Sokolov, I. M. Restoring ergodicity of stochastically reset anomalous-diffusion processes. Phys. Rev. Res. 4, 013161. https://doi.org/10.1103/PhysRevResearch.4.013161 (2022).
Zhang, J. et al. Higher-order turbulence statistics in the sub-alfvénic solar wind observed by parker solar probe. Astrophys. J. 937, 70. https://doi.org/10.3847/1538-4357/ac8c34 (2022).
Benouioua, D., Candusso, D., Harel, F. & Oukhellou, L. PEMFC stack voltage singularity measurement and fault classification. Int. J. Hydrog. Energy 39, 21631–21637. https://doi.org/10.1016/j.ijhydene.2014.09.117 (2014).
Conlon, P. et al. Multifractal properties of evolving active regions. Solar Image Anal. Vis. 297, 87–99. https://doi.org/10.1007/s11207-007-9074-7 (2009).
de la Calleja Mora, E., Carrillo, J., Mendoza, M. & Donado, F. Structural transformations in magnetorheological slurries induced by perturbations. Eur. Phys. J. B86, 1–9 (2013). https://doi.org/10.1140/epjb/e2013-31014-8.
Matia, K., Ashkenazy, Y. & Stanley, H. E. Multifractal properties of price fluctuations of stocks and commodities. Europhys. Lett. 61, 422. https://doi.org/10.1209/epl/i2003-00194-y (2003).
Miranda, J. G. V., Montero, E., Alves, M. C., González, A. P. & Vázquez, E. V. Multifractal characterization of saprolite particle-size distributions after topsoil removal. Geoderma 134, 373–385. https://doi.org/10.1016/j.geoderma.2006.03.014 (2006).
Pinzón, J., Puente, C., Parlange, M. & Eichinger, W. A multifractal analysis of lidar measured water vapour. Bound.-Layer Meteorol. 76, 323–347. https://doi.org/10.1007/BF00709237 (1995).
Mahmoodi, K., Kerick, S. E., Grigolini, P., Franaszczuk, P. J. & West, B. J. Temporal complexity measure of reaction time series: Operational versus event time. Brain Behav. e3069 (2023). https://doi.org/10.1002/brb3.3069.
Nasimi, F., Khayyambashi, M. R. & Movahhedinia, N. Redundancy cancellation of compressed measurements by QRS complex alignment. PLoS ONE 17, e0262219. https://doi.org/10.1371/journal.pone.0262219 (2022).
Rao, A., Gupta, P. & Ghosh, P. K. P-and T-wave delineation in ECG signals using parametric mixture Gaussian and dynamic programming. Biomed. Signal Process. Control 51, 328–337. https://doi.org/10.1016/j.bspc.2019.03.001 (2019).
Kauffman, S. A. A World Beyond Physics: The Emergence and Evolution of Life (Oxford University Press, New York, NY, 2019).
van der Merwe, R. Enablement, the adjacent possible and the becoming of the biosphere. Metascience 29, 279–282. https://doi.org/10.1007/s11016-020-00525-z (2020).
Adolf, J. K. & Fried, E. I. Ergodicity is sufficient but not necessary for group-to-individual generalizability. Proc. Natl. Acad. Sci. 116, 6540–6541. https://doi.org/10.1073/pnas.1818675116 (2019).
Bäck, A., Nordholm, S. & Nyman, G. Investigation of ergodic character of quantized vibrational motion. J. Phys. Chem. A 108, 8782–8794. https://doi.org/10.1021/jp049113l (2004).
Calvo, F. & Yurtsever, E. Lyapunov instability in rotating systems from ergodic molecular dynamics simulations. Phys. Lett. A 266, 387–393. https://doi.org/10.1016/S0375-9601(00)00046-3 (2000).
Chakraborty, D. & Chattaraj, P. K. A quantum-classical correspondence in the dynamics around higher order saddle points: A Bohmian perspective. Theoret. Chem. Acc. 142, 18. https://doi.org/10.1007/s00214-023-02957-2 (2023).
Clamp, M. E., Baker, P., Stirling, C. & Brass, A. Hybrid Monte Carlo: An efficient algorithm for condensed matter simulation. J. Comput. Chem. 15, 838–846. https://doi.org/10.1002/jcc.540150805 (1994).
Isichenko, M. & Petviashvili, N. Ergodic mixing for turbulent drift motion in an inhomogeneous magnetic field. Phys. Plasmas 2, 3650–3654. https://doi.org/10.1063/1.871064 (1995).
Lomholt, M. A., Zaid, I. M. & Metzler, R. Subdiffusion and weak ergodicity breaking in the presence of a reactive boundary. Phys. Rev. Lett. 98, 200603. https://doi.org/10.1103/PhysRevLett.98.200603 (2007).
Mauro, J. C., Gupta, P. K. & Loucks, R. J. Continuously broken ergodicity. J. Chem. Phys. 126, 184511. https://doi.org/10.1063/1.2731774 (2007).
Palmer, R. G. Broken ergodicity. Adv. Phys. 31, 669–735. https://doi.org/10.1080/00018738200101438 (1982).
Russomanno, A., Fava, M. & Fazio, R. Weak ergodicity breaking in Josephson-junction arrays. Phys. Rev. B 106, 035123. https://doi.org/10.1103/PhysRevB.106.035123 (2022).
Sun, H., Chan, Y. L. & Kwok, K. Electric field-responsive photoluminescence color switching and reversible properties via tb/eu co-doped ergodic relaxor ferroelectrics. Phys. Chem. Chem. Phys. 21, 7567–7575. https://doi.org/10.1039/C9CP00324J (2019).
Zeifman, A. Quasi-ergodicity for non-homogeneous continuous-time Markov chains. J. Appl. Probab. 26, 643–648. https://doi.org/10.2307/3214422 (1989).
Zheng, Z., Hu, G. & Zhang, J. Ergodic property of a Henon–Heiles model with reflecting walls. Phys. Rev. E 52, 3440. https://doi.org/10.1103/PhysRevE.52.3440 (1995).
Zhou, P., Du, J., Zhou, K. & Wei, S. 2D mixed pseudo-random coupling ps map lattice and its application in S-box generation. Nonlinear Dyn. 103, 1151–1166. https://doi.org/10.1007/s11071-020-06098-0 (2021).
Peng, C.-K. et al. Mosaic organization of DNA nucleotides. Phys. Rev. E 49, 1685. https://doi.org/10.1103/PhysRevE.49.1685 (1994).
Peng, C.-K., Havlin, S., Stanley, H. E. & Goldberger, A. L. Quantification of scaling exponents and crossover phenomena in nonstationary heartbeat time series. Chaos 5, 82–87. https://doi.org/10.1063/1.166141 (1995).
Chhabra, A. & Jensen, R. V. Direct determination of the f (\(\alpha\)) singularity spectrum. Phys. Rev. Lett. 62, 1327. https://doi.org/10.1103/PhysRevLett.62.1327 (1989).
Halsey, T. C., Jensen, M. H., Kadanoff, L. P., Procaccia, I. & Shraiman, B. I. Fractal measures and their singularities: The characterization of strange sets. Phys. Rev. A 33, 1141. https://doi.org/10.1103/PhysRevA.33.1141 (1986).
Mandelbrot, B. B. Fractals and Scaling in Finance: Discontinuity, Concentration, Risk (Springer, New York, NY, 2013).
Zamir, M. Critique of the test of multifractality as applied to biological data. J. Theor. Biol. 225, 407–412. https://doi.org/10.1016/S0022-5193(03)00261-3 (2003).
Veneziano, D., Moglen, G. E. & Bras, R. L. Multifractal analysis: Pitfalls of standard procedures and alternatives. Phys. Rev. E 52, 1387. https://doi.org/10.1103/PhysRevE.52.1387 (1995).
Ihlen, E. A. Introduction to multifractal detrended fluctuation analysis in Matlab. Front. Physiol. 3, 141. https://doi.org/10.3389/fphys.2012.00141 (2012).
Schreiber, T. & Schmitz, A. Improved surrogate data for nonlinearity tests. Phys. Rev. Lett. 77, 635. https://doi.org/10.1103/PhysRevLett.77.635 (1996).
Cherstvy, A. G., Chechkin, A. V. & Metzler, R. Anomalous diffusion and ergodicity breaking in heterogeneous diffusion processes. New J. Phys. 15, 083039. https://doi.org/10.1088/1367-2630/15/8/083039 (2013).
R Core Team. R: A language and environment for statistical computing (2013). https://www.R-project.org/.
Therneau, T. M. & Lumley, T. Package ’survival’. R Top Doc128, 28–33 (2015). https://cran.r-project.org/package=survival.
Acknowledgements
This work was supported by the Center for Research in Human Movement Variability at the University of Nebraska at Omaha, funded by the NIH award P20GM109090.
Author information
Authors and Affiliations
Contributions
M.M.: Conceptualization, Methodology, Software, Validation, Formal analysis, Writing – Original draft, Writing – Review & Editing, Visualization, Project administration; A.S. Writing – Original draft, Writing – Review & Editing; J.H.: Investigation, Data curation; E.W.: Investigation, Resources, Data curation; K.K.: Conceptualization, Methodology, Writing – Review & Editing; D.G.K.-S.: Conceptualization, Methodology, Writing – Original draft, Writing – Review & Editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mangalam, M., Sadri, A., Hayano, J. et al. Multifractal foundations of biomarker discovery for heart disease and stroke. Sci Rep 13, 18316 (2023). https://doi.org/10.1038/s41598-023-45184-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-45184-2
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.