Abstract
Circular RNAs (circRNAs) play key roles in colorectal cancer (CRC) progression, but little is known about the biological functions of hsa_circRNA_001676 in CRC. Therefore, we explored the potential role of hsa_circRNA_001676 in CRC development. RT-qPCR was performed to determine hsa_circRNA_001676, miR-556-3p and Ras-GTPase-activating SH3 domain-binding-proteins 2 (G3BP2) levels in CRC tissues. Meanwhile, to evaluate the roles of hsa_circRNA_001676, miR-556-3p and G3BP2 on CRC, functional analysis of cell proliferation, migration and stemness were then performed. Our results showed that compared to normal tissues, hsa_circRNA_001676 and G3BP2 level was elevated, but miR-556-3p level was reduced in CRC tissues. Additionally, luciferase reporter results showed that hsa_circRNA_001676 was shown to target miR-556-3p, and G3BP2 was targeted by miR-556-3p. Hsa_circRNA_001676 or G3BP2 overexpression promoted CRC cell proliferation and migration. Conversely, miR-556-3p overexpression suppressed CRC cell proliferation and migration. Moreover, deficiency of hsa_circRNA_001676 or G3BP2 repressed the CRC cell proliferation, migration and stemness. Meanwhile, hsa_circRNA_001676 deficiency obviously reduced tumor growth and stemness in a CRC mouse xenograft model. Furthermore, hsa_circRNA_001676 deficiency notably reduced G3BP2 level, but elevated miR-556-3p level in tumor tissues from tumor-bearing mice. Mechanistically, hsa_circRNA_001676 targeted miR-556-3p to increase G3BP2 level, contributing to the progression of CRC. Collectively, hsa_circRNA_001676 was able to accelerate proliferation, migration and stemness in CRC through regulating miR-556-3p/G3BP2 axis, suggesting that hsa_circRNA_001676 may become a potential therapeutic target in treating CRC.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) is a common malignancy in the digestive system, and is the second leading cause of cancer-related deaths worldwide1. Notwithstanding that adjuvant chemotherapy, chemoradiotherapy, immunotherapy and targeted therapy have been applied for treating CRC2,3,4, the prognosis remains worse in CRC patients5,6. Therefore, improving the diagnosis of CRC and developing new treatments for CRC patients are urgently needed.
Circular RNAs (circRNAs), a type of non-coding RNA molecules (ncRNAs), are formed by covalently closed loops with no 3′ poly(A) tails or 5′ end caps7,8. CircRNAs play crucial regulatory roles in different biological processes including cell growth and migration through sponging microRNA (miRNAs)9. MiRNAs are another class of ncRNAs with 17–22 nucleotides in length10. MiRNAs are able to repress gene expression through breaking translation or promoting mRNA degradation11. Mechanically, circRNAs can work as miRNA sponges, thereby repressing the ability of miRNAs to its target genes12.
CircRNAs have been shown to play key roles in cancer development, including CRC13,14. Some circRNAs are differentially expressed between normal and cancer tissues and may function as oncogenes or tumor suppressors15. For example, Circ3823 could accelerate CRC tumorigenesis, metastasis and angiogenesis via targeting miR-30c-5p/TCF7 axis16. CircEZH2 could aggravate CRC progression through stabilization of CREB117. Meanwhile, circIFNGR2 notably facilitated CRC cell growth and increased cetuximab resistance through targeting miR-30b18. Conversely, hsa_circ_0001666 was able to restrain CRC progression through sponging miR-576-5p19. CircTMEM59 was found to target miR-668-3p/ID4 axis, thereby repressing CRC cell growth and metastasis20. In the present research, we identified that compared to normal tissues, hsa_circRNA_001676 was aberrantly upregulated in CRC tissues. Nevertheless, the role of hsa_circRNA_001676 in cancer progression remains largely unknown.
Furthermore, miR-556-3p was found to exert important roles in human cancers21,22. Additionally, G3BP2 protein was also found to be participated in the processes of tumor initiation and development23,24. However, the relationship among hsa_circRNA_001676, miR-556-3p and G3BP2 in CRC remains largely unknown. In this study, our results showed that miR-556-3p was targeted by hsa_circRNA_001676, and G3BP2 was targeted by miR-556-3p. Significantly, hsa_circRNA_001676 could affect CRC progression via targeting miR-556-3p/G3BP2 axis, suggesting that hsa_circRNA_001676 may become a potential therapeutic target in treating CRC.
Materials and methods
Clinical samples
A total of 3 pairs of CRC tissues and corresponding noncancerous tissues were obtained from the Peking University Cancer Hospital (Inner Mongolia Campus)/Affiliated Cancer Hospital of Inner Mongolia Medical University, conformed to the Declaration of Helsinki. Informed consent was got from all participants. Meanwhile, the Ethics Committee of Peking University Cancer Hospital (Inner Mongolia Campus)/Affiliated Cancer Hospital of Inner Mongolia Medical University approved this study.
Bioinformatics analysis
The CRC-related circRNA expression profiles were downloaded from the Gene Expression Omnibus (GEO) database (GSE142837). The differentially expressed circRNAs (DEcircRNAs) between CRC tissues and corresponding noncancerous tissues were screened by using the R language and the GEO2R tool. Threshold criteria were: |log (Fold Change (FC)) |> 1 and -log10 (P-value) > 1.3.
Cell culture
The normal colonic mucosa cell line NCM460 (GuangZhou Jennio Biotech Co., Ltd), HT29 and HCT116 cells (Procell), SW620 , SW480, LOVO and 293 T cells (Shanghai Cell Bank) were maintained in Dulbecco’s modified Eagle medium (DMEM) supplied with 10% fetal bovine serum (FBS) with 5% CO2.
Cell transfection
The sequences of mimics or inhibitor negative control (NC), miR-556-3p mimics or inhibitor were synthesized by HANBIO. The pcDNA3.1-NC and pcDNA3.1-G3BP2 plasmids and siRNA (si) specifically targeting G3BP2 (si-G3BP2) and siRNA NC (si-NC) were constructed by HANBIO. HT29 cells were transfected with mimics NC, miR-556-3p mimics, si-NC or si-G3BP2; SW480 cells were transfected with inhibitor NC, miR-556-3p inhibitor, pcDNA3.1-NC or pcDNA3.1-G3BP2 using the lipofectamine 2000, respectively.
The hsa_circRNA_001676-overexpressing lentivirus (hsa_circRNA_001676-OE), short hairpin RNAs targeting hsa_circRNA_001676 (sh-hsa_circRNA_001676) were obtained from HANBIO. HT29 cells were infected with sh-hsa_circRNA_001676 and SW480 cells were infected with hsa_circRNA_001676-OE for 48 h respectively, and then stable-expressing CRC cells were chose by puromycin.
Reverse transcription-quantitative PCR (RT-qPCR) assay
Total RNA from cells or tumor tissues were extracted using the Redzol reagent (SBS Genetech Co.,Ltd.). Next, the cDNA was synthesized using the Surescript™ First-Strand cDNA Synthesis Kit (iGeneBio). After that, qPCR was conducted with the SYBR Green qPCR Master Mix (None ROX) kit (Servicebio). U6 was used as an internal control for miR-556-3p, and GAPDH was used as an internal control for hsa_circRNA_001676, G3BP2, OCT4 and Nanog. The 2−ΔΔCt method was used for determining relative gene levels.
Cell counting Kit-8 (CCK-8) assay
HT29 and SW480 cells were seeded onto 96-well plates overnight at 37 °C. After indicated treatment, each well was added with CCK-8 reagent (10 μl, Beyotime). Next, after another 4 h of incubation at 37 °C, the optical density (OD) was measured under a micro-plate reader (DNM-9602; PERLONG), the wavelength was set to 490 nm.
Colony formation assay
HT29 and SW480 cells were loaded onto 12-well plates overnight at 37 °C. After indicated treatment, cells were then incubated for 1 week at 37 °C. Thereafter, cells were stained with 0.1% crystal violet solution for 30 min. Finally, the colonies were captured under a light microscope.
Wound healing assay
HT29 and SW480 cells were plated into 12-well plates overnight at 37 °C. The wounds were then created by using a sterilized pipet tip. After 0, 24 or 48 h of incubation, images were taken using a light microscope.
Transwell assay
Cell migration ability was assessed using a 24-well transwell chamber (Corning) with an 8 µm-pore size filter membrane. Cells suspended in DMEM without serum (200 µl) were loaded onto the upper Transwell cell inserts, and DMEM medium (500 µl) with 10% FBS was seeded onto the bottom chambers. After 24 h of incubation at 37 °C, crystal violet solution (0.1%) was used for staining the cells on the lower surface of the filter. Finally, the migrated cells were observed under a light microscope and counted in three randomly selected regions.
Spheroid formation assay
Cancer stemness was assessed by the spheroid formation assay. Cells were suspended in serum-free DMEM containing 2% B27 + 20 ng/ml epidermal growth factor (EGF) + 10 ng/ml of basic fibroblast growth factor (bFGF)25, and then added into a 6-well ultralow attachment plate. After 2 weeks of incubation at 37 °C, the spheres were photographed and counted under a light microscope. A Spheroid containing > 50 cells were counted.
Dual-Luciferase Reporter Assay
The wild-type (wt) or mutant (mut) 3′-untranslated regions (UTRs) of hsa_circRNA_001676 and G3BP2 were cloned into the pmirGLO luciferase reporter plasmids. Next, these plasmids and miR-556-3p mimics or inhibitor were co-transfected into 293 T cells using the lipofectamine 2000, respectively. Subsequently, the luciferase activity was assessed by a Dual Luciferase Reporter Assay kit (Beyotime) at 48 h.
Western blot assay
Protein samples were separated with 10% SDS-PAGE gels and followed by transferring onto a PVDF membrane. Later on, the membrane was probed with primary antibodies at 4 °C overnight including anti-Oct-4 (No. ab109250), anti-Nanog (No. ab205481), anti-G3BP2 (No. ab190011), anti-CD133 (No. ab19898), anti-CD44 (No. ab189524) and anti-GAPDH (No. Ab9485, Abcam) antibodies, followed by incubation with a HRP-conjugated secondary antibody (No. Ab6721, Abcam) at room temperature. Finally, blots were visualized using a LightningTM Chemiluminescence Reagent (PerkinElmer).
TUNEL staining assay
Paraffin-embedded tumor tissues were cut into 4-μm thick slices. Next, the TUNEL Apoptosis Detection Kit (FITC) purchased from BOSTER was used for detecting cell apoptosis in tumor tissues according to the manufacturer's instructions. The nuclei was counterstained with DAPI (Solarbio). Images were then observed under a fluorescence microscope (OLYMPUS).
Immunohistochemical (IHC) and immunofluorescence (IF) staining assays
For IHC assay, sections were blocked in normal goal serum at room temperature for 20 min and then probed with anti-G3BP2 antibody (No. ab190011), anti-Oct-4 antibody (No. ab200834), anti-Nanog antibody (No. ab109250, Abcam) overnight at 4 °C. Next, sections were stained with HRP-labeled secondary antibody (No. Ab6721) for 20 min at room temperature. After visualizing with DAB solution, photographs were taken with a light microscope (OLYMPUS).
For IF assay, sections were probed with anti-CD133 antibody (No. ab19898, Abcam) and anti-CD44 antibody (No. ab238464, Abcam) overnight at 4 °C and then probed with the fluorescently-labeled secondary antibody (No. ab150077). Finally, images were taken with a fluorescence microscope (OLYMPUS). The Image Pro Plus software was used for semiquantitative analysis.
Animal study
All animal experiments were approved by the Animal Care and Use Committee of the Peking University Cancer Hospital (Inner Mongolia Campus)/Affiliated Cancer Hospital of Inner Mongolia Medical University, and conducted in accordance with the National Institutes of Health Guidelines for Care and Use of Laboratory Animals and carried out in accordance with the ARRIVE guidelines. A total of 20 BALB/c nude mice (18–20 g, Vital River) were grouped into three groups randomly: Control, NC and sh-hsa_circRNA_001676 groups. Each mouse was injected subcutaneously with untransfected (control group; 2 × 106 cells) or transfected HT29 cells (NC group and sh-hsa_circRNA_001676 group; 2 × 106 cells). Tumor growth was monitored at indicated times. The tumor volume (V) was calculated as: tumor length × tumor width2/2. After 4 weeks, the mice were sacrificed by cervical dislocation under anesthesia (1% isoflurane inhalation), and tumors were dissected out from the sacrificed mice.
Hematoxylin and eosin (H&E) staining assay
Paraffin-embedded tumor tissues were cut into 4-μm thick slices. After dewaxing and rehydration, sections were then subjected to H&E staining. Finally, images were observed under a light microscope.
Statistical analysis
Each experiment was independently repeated at least three times. GraphPad Prism 8 software was used for statistical analysis. The differences between two groups were evaluating using an unpaired Student t‐test. Meanwhile, one‐way analysis of variance (ANOVA) was used for multiple comparison. Data are shown as mean ± standard deviation. P < 0.05 indicates statistical significance.
Ethical approval and consent to participate
Informed consent was got from all participants and the Ethics Committee of Peking University Cancer Hospital (Inner Mongolia Campus)/Affiliated Cancer Hospital of Inner Mongolia Medical University approved this study.
All animal experiments were approved by the Animal Care and Use Committee of the Peking University Cancer Hospital (Inner Mongolia Campus)/Affiliated Cancer Hospital of Inner Mongolia Medical University, and conducted in accordance with the National Institutes of Health Guidelines for Care and Use of Laboratory Animals and carried out in accordance with the ARRIVE guidelines.
Results
Hsa_circRNA_001676 level was elevated in CRC tissues
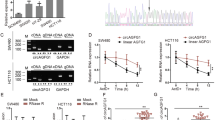
By comparing circRNA levels in five pairs of noncancerous tissues and CRC tissues from CRC patients, a circRNA expression profile was downloaded from the GSE142837 dataset. The volcano plot disclosed that compared to the matched normal tissues, hsa_circRNA_001676 level was obviously elevated in CRC samples (Fig. S1), which was verified by the RT-qPCR results in Fig. 1A. Moreover, compared to NCM460 cells, higher level of hsa_circRNA_001676 was detected in CRC cells (Fig. 1B). Meanwhile, among these five CRC cells, hsa_circRNA_001676 was expressed at the highest level in HT29 cells and expressed at the lowest level in SW480 cells (Fig. 1B). Thus, we reduced hsa_circRNA_001676 level in HT29 cells by using sh-Hsa_circRNA_001676, and overexpressed hsa_circRNA_001676 level in SW480 cells by using Hsa_circRNA_001676-OE (Fig. 1C and D). Significantly, hsa_circRNA_001676 expression was declined in HT29 cells transfected with sh-hsa_circRNA_001676 plasmids and increased in SW480 cells transfected with hsa_circRNA_001676-OE plasmids (Fig. 1C and D).
Hsa_circRNA_001676 enhanced CRC cell viability. (A) RT-qPCR analysis of hsa_circRNA_001676 level in CRC tissues (n = 3) and paired noncancerous tissues (n = 3). (B) RT-qPCR analysis of hsa_circRNA_001676 level in NCM460 cells and different CRC cells. (C) RT-qPCR analysis of hsa_circRNA_001676 level in HT29 cells transfected with sh-NC or sh-hsa_circRNA_001676. (D) RT-qPCR analysis of hsa_circRNA_001676 level in SW480 cells transfected with OE-NC or hsa_circRNA_001676-OE. (E, F) HT29 cells were transfected with sh-NC or sh-hsa_circRNA_001676 and SW480 cells were transfected with OE-NC or hsa_circRNA_001676-OE. (E) CCK-8 and (F) colony formation assays were applied for evaluating cell viability and proliferation, respectively. *P < 0.05; **P < 0.01; ***P < 0.001.
Hsa_circRNA_001676 enhanced CRC cell proliferation, migration and stemness
To explore the role of hsa_circRNA_001676 in CRC, CCK-8 and colony formation assays were conducted. Obviously, silenced hsa_circRNA_001676 strongly repressed HT29 cell viability and proliferation, whereas forced expression of hsa_circRNA_001676 enhanced SW480 cell viability and proliferation (Fig. 1E and F). Additionally, the results of transwell, wound healing and spheroid formation assays indicated that silenced hsa_circRNA_001676 weakened HT29 cell migration and sphere formation abilities, whereas hsa_circRNA_001676 overexpression displayed the opposite effects on SW480 cells (Fig. 2A–C). Collectively, hsa_circRNA_001676 could accelerate CRC cell growth, migration and stemness.
Hsa_circRNA_001676 enhanced CRC cell migration and stemness. HT29 cells were transfected with sh-NC or sh-hsa_circRNA_001676 and SW480 cells were transfected with OE-NC or hsa_circRNA_001676-OE. (A) Transwell and (B) wound healing assays were performed to assess cell migration (Scale bar = 100 μm). (C) Sphere formation efficiency was determined by using the sphere formation assay. **P < 0.05; ***P < 0.01.
Hsa_circRNA_001676 functioned as a sponge of miR-556-3p
Previous studies have shown that circRNAs could function as miRNA “sponges” to “adsorb” miRNAs26,27. This process greatly eliminated the inhibitory effects of miRNAs on target genes27. Therefore, to identify the downstream miRNAs of hsa_circRNA_001676, Circbank database (http://www.circbank.cn/) and CircInteractome database (https://circinteractome.irp.nia.nih.gov/) were used. The results of dual-luciferase reporter assay showed that the luciferase activity in wt-hsa_circRNA_001676-transfected 293 T cells was declined by miR-556-3p mimics and elevated by miR-556-3p inhibitor (Fig. 3A). Moreover, sh-hsa_circRNA_001676 greatly elevated miR-556-3p level in HT29 cells, whereas hsa_circRNA_001676-OE remarkably declined miR-556-3p level in SW480 cells (Fig. 3B). These results verified the interaction between miR-556-3p and hsa_circRNA_001676 in CRC.
Hsa_circRNA_001676 functioned as a sponge of miR-556-3p. (A) The luciferase activities were measured in 293 T cells co-transfected with hsa_circRNA_001676 3’UTR-wt or -mut and miR-556-3p mimics or inhibitor. (B) RT-qPCR analysis of miR-556-3p level in HT29 or SW480 cells transfected with sh-hsa_circRNA_001676 or hsa_circRNA_001676-OE, respectively. (C) RT-qPCR analysis of miR-556-3p level in CRC tissues (n = 3) and paired noncancerous tissues (n = 3). (D) RT-qPCR analysis of miR-556-3p level in NCM460 cells and different CRC cells. (E) RT-qPCR analysis of miR-556-3p level in HT29 and SW480 cells transfected with miR-556-3p mimics or inhibitor, respectively. ***P < 0.001.
Additionally, compared to normal noncancerous tissues and normal colonic mucosa cells, CRC tissues and CRC cells evidenced a lower miR-556-3p level respectively (Fig. 3C and D). MiR-556-3p mimics dramatically elevated miR-556-3p level in HT29 cells, whereas miR-556-3p inhibitor greatly reduced miR-556-3p level in SW480 cells (Fig. 3E). Moreover, miR-556-3p mimics markedly repressed HT29 cell viability, proliferation and migration; however, miR-556-3p inhibitor displayed the opposite effects on SW480 cells (Fig. 4A–D).
MiR-556-3p weakened the proliferation and migration of CRC cells. SW480 cells were transfected with miR-556-3p inhibitor and HT29 cells were transfected with miR-556-3p mimics. (A) CCK-8 and (B) colony formation assays were applied for evaluating cell viability and proliferation, respectively. (C) transwell and (D) wound healing assays were performed to assess cell migration (Scale bar = 100 μm). *P < 0.05; **P < 0.01; ***P < 0.001.
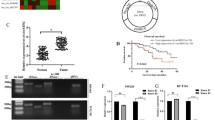
G3BP2 is a downstream binding target of miR-556-3p
Next, Starbase (https://starbase.sysu.edu.cn/) database predicted the downstream targets of miR-556-3p. Over 3000 target genes were listed in Starbase. Among these, G3BP2 could regulate cancer stemness via upregulating the expressions of stem cell markers Oct-4 and Nanog23. Our results found that compared to normal controls, cell apoptosis was attenuated in CRC tissues (Fig. S2A). Meanwhile, the expressions of Oct-4, Nanog, CD133, CD44 and G3BP2 were obviously elevated in CRC tissues (Figs. S2B–E), suggesting that these tumor tissues displayed high self-renewal capability and high metastatic potential28. Meanwhile, the effect of G3BP2 on stemness in CRC and the relationship between G3BP2 and hsa_circRNA_001676/miR-556-3p in CRC remain elusive. Thus, we focused on G3BP2 because of its role in cancer stemness in the study. The data in Starbase showed that 3’UTR of G3BP2 was complementary to miR-556-3p sequences (Fig. 5A). Additionally, miR-556-3p mimics notably repressed the luciferase activity in wt-G3BP2 group cells (Fig. 5B), suggesting G3BP2 could be a direct target of miR-556-3p.
G3BP2 was a downtream binding target of miR-556-3p. (A) The binding sites between G3BP2 and miR-556-3p was obtained from Starbase databases. (B) The luciferase activities were measured in 293 T cells co-transfected with G3BP2 3’UTR-wt or -mut and miR-556-3p mimics or inhibitor. (C) HT29 cells were transfected with si-G3BP2 and SW480 cells were transfected with pcDNA3.1-G3BP2. RT-qPCR was conducted to determine G3BP2 level in CRC cells. (D, E) CCK-8 and colony formation assays were applied for evaluating cell viability and proliferation, respectively. (F) The transwell assay were performed to assess cell migration (Scale bar = 100 μm). **P < 0.05; ***P < 0.01; ****P < 0.001.
Furthermore, silenced G3BP2 greatly declined G3BP2 level in HT29 cells, as well as repressed HT29 cell viability, proliferation and migration; conversely, G3BP2 overexpression strongly elevated G3BP2 level in SW480 cells, as well as facilitated SW480 cell viability, proliferation and migration (Figs. 5C-F and S3A-3B). Collectively, G3BP2 could act as an oncogene in CRC cells.
G3BP2 promoted CRC cell stemness
Next, we tested the effect of G3BP2 on CRC cell stemness in vitro. As revealed in Fig. 6A and B, Oct-4 and Nanog levels were declined in CRC cells by silencing of G3BP2, and increased by overexpression of G3BP2. Additionally, G3BP2 deficiency dramatically weakened the sphere formation ability of HT29 cells, whereas G3BP2 overexpression notably enhanced the sphere-forming capacity of SW480 cells (Fig. 6C). To sum up, G3BP2 could promote CRC cell stemness.
G3BP2 promoted CRC cell stemness. HT29 cells were transfected with si-G3BP2 and SW480 cells were transfected with pcDNA3.1-G3BP2. (A) RT-qPCR and (B) western blot analysis of Oct-4 and Nanog level in transfected cells. (C) Sphere formation efficiency was determined by using the sphere formation assay. *P < 0.05; **P < 0.01; ***P < 0.001.
Hsa_circRNA_001676 deficiency reduced tumor growth and stemness in CRC in vivo by regulating miR-556-3p/G3BP2 axis
A xenograft mouse model with CRC was constructed to further explore the role of hsa_circRNA_001676 in CRC. As shown in Fig. 7A and B, hsa_circRNA_001676 deficiency obviously reduced the tumor volume and triggered tumor cell apoptosis (Fig. 7A–C). Meanwhile, hsa_circRNA_001676 downregulation remarkably declined hsa_circRNA_001676 and G3BP2 levels, and elevated miR-556 level in tumor tissues (Fig. 7D–G). Furthermore, compared to the sh-NC group, downregulation of hsa_circRNA_001676 led to significant decreases in CD133, CD44, Oct-4 and Nanog levels in tumor tissues (Fig. 8A–D). Collectively, hsa_circRNA_001676 deficiency was able to hamper CRC progression in vivo by regulating miR-556-3p/G3BP2 axis.
Hsa_circRNA_001676 deficiency reduced tumor growth of CRC in vivo by regulating miR-556-3p/G3BP2 axis. (A) Photographs of xenograft tumors. (B) Curves of tumor volumes. (C) TUNEL staining analysis of cell apoptosis in tumor tissues (Scale bar = 100 μm). RT-qPCR analysis of (D) hsa_circRNA_001676, (E) miR-556-3p and (F) G3BP2 levels in tumor tissues. (G) H&E (upper panel) stained histological images of the tumor tissues. IHC (lower panel) analysis of G3BP2 protein expression in tumor tissues (Scale bar = 100 μm). *P < 0.05; ***P < 0.001.
Hsa_circRNA_001676 deficiency reduced the stemness of CRC in vivo by regulating miR-556-3p/G3BP2 axis. (A, B) IF analysis of CD133 and CD44 protein expressions in tumor tissues. (C) IHC and (D) RT-qPCR analysis of Oct-4 and Nanog levels in tumor tissues. Scale bar = 100 μm. **P < 0.01; ***P < 0.001.
Discussion
CircRNAs have been shown to exhibit a regulatory role in CRC progression29. However, the biological role of a large number of circRNAs in CRC progression remain not well clarified. In this study, we found that hsa_circRNA_001676 level was elevated in CRC tissues and CRC cell lines. Additionally, downregulation of hsa_circRNA_001676 could suppress tumor growth and stemness in CRC in vitro and in vivo. Conversely, hsa_circRNA_001676 overexpression strongly accelerated tumor growth and stemness in CRC in vitro. Our results showed that hsa_circRNA_001676 could function as a new oncogenic driver to facilitate tumor growth and stemness in CRC.
Cancer stemness has been recognized as the leading cause of cancer metastasis and relapse30. Evidence has shown that some cancer cells possess typical stemness properties, which are important for the initiation and metastasis of tumors31. Targeting cancer stemness is a promising approach to fight CRC32. CircRNAs play a key role in modulating cancer stemness33,34. For example, CircPTN enhanced glioma cell stemness and self-renewal via sponging miR-145-5p35. CircAGFG1 could facilitate the stemness and metastasis in CRC through targeting miR-4262 and miR-185-5p36. Circ_0030586 retarded bladder cancer cell growth and stemness through targeting miR-66537. Our data showed that deficiency of hsa_circRNA_001676 remarkably repressed the sphere-formation ability of CRC cells in vitro and reduced CD133, CD44, Oct-4 and Nanog levels in tumor tissues in vivo, suggesting that deficiency of hsa_circRNA_001676 could suppress CRC cell stemness. However, the mechanism by which hsa_circRNA_001676 affects the stemness in CRC remains unknown.
CircRNAs have been reported to exert their regulatory roles in cancer development via functioning as miRNA “sponges”38. In this study, Circbank and CircInteractome databases were applied to predict the targets of hsa_circRNA_001676. The data showed that hsa_circRNA_001676 could interact with miR-556-3p. Li et al. found that inhibition of miR-556-3p could enhance gastric cancer cell growth21. Conversely, diminished miR-556-3p repressed hemangioma cell proliferation through upregulating VEGFC22. These findings demonstrated that miR-556-3p may be expressed both as a tumor suppressor or an oncogene in different cancers. Our results showed that compared to normal colonic mucosa cells, miR-556-3p level was remarkably reduced in CRC cells. Forced miR-556-3p expression greatly repressed CRC cell proliferation and migration, suggesting that miR-556-3p might tumor suppressor gene in CRC.
Furthermore, in this study, miR-556-3p was shown to target G3BP2. It has been shown that G3BP2 often overexpressed in some cancers and acted as an oncogene39,40. Overexpression of G3BP2 could expedite ESCC cell migration and invasion39. Inhibition of G3BP2 could decline PD-L1 expression in cancer cells, thereby facilitating anticancer immunotherapy40. Consistent with previous studies, we verified that G3BP2 overexpression could enhance CRC cell proliferation and migration, indicating that G3BP2 could act as an oncogene in CRC. Additionally, G3BP2 could trigger tumor initiation in breast cancer via upregulation of Oct-4 and Nanog23. Meanwhile, Oct-4 and Nanog are important stemness-associated mediators in the maintenance of cancer stemness41,42. Reducing Oct-4 and Nanog levels in CRC cells could repress the cancer stemness properties43. Moreover, high Oct-4 and Nanog levels are related to worse prognosis in CRC44. These findings above suggested a relationship between G3BP2 and cancer stemness. For the first time, we found that G3BP2 overexpression could upregulate Oct-4 and Nanog levels in CRC cells, suggesting that G3BP2 could facilitate CRC cell stemness.
Furthermore, circHERC4 could elevate E-cadherin protein level in CRC through inactivating miR-556-5p, thereby facilitating tumor migration and metastasis45. Circ_0020378 could facilitate osteosarcoma cell migration and growth via sponging miR-556-5p46. Meanwhile, circ-ABCB10 could affect cisplatin sensibility in lung cancer through binding with miR-556-3p47. Moreover, circFNDC3B could repress oncogene G3BP2 via binding with miR-1178-3p, thus suppressing bladder cancer progression48. These findings showed the relationships between different circRNAs and miR-556 or G3BP2. In this research, forced hsa_circRNA_001676 expression notably reduced miR-556-3p level, and elevated G3BP2, Oct-4 and Nanog levels in tumor tissues. These results showed that hsa_circRNA_001676 could act as a miR-556-3p sponge to weaken the inhibitory effects of miR-556-3p on its target G3BP2, thereby upregulating Oct-4 and Nanog levels.
It has been shown that one miRNA can target multiple genes and one circRNA also can sponge various miRNAs49,50. In this study, we only illustrated that hsa_circRNA_001676 could affect CRC cell growth and stemness through miR-556-3p/G3BP2 axis. However, further study is needed to uncover more hsa_circRNA_001676-associated miRNAs or mRNAs, and this research might further illustrate the molecular mechanism of hsa_circRNA_001676 in CRC.
Conclusions
This study is the first to research hsa_circRNA_001676/miR-556-3p/G3BP2 axis in CRC. In summary, Hsa_circRNA_001676 was able to accelerate proliferation, migration and stemness in CRC through targeting miR-556-3p/G3BP2 axis, demonstrating that hsa_circRNA_001676 may serve as a potential target for treating CRC.
Data availability
The data supporting the conclusions of this study are included in this article.
References
Wang, H., Tian, T. & Zhang, J. Tumor-associated macrophages (TAMs) in colorectal cancer (CRC): From mechanism to therapy and prognosis. Int. J. Mol. Sci. 22(16), 8470 (2021).
Wan, J. et al. Incidence of chemotherapy- and chemoradiotherapy-induced amenorrhea in premenopausal women with stage II/III colorectal cancer. Clin. Colorectal Cancer 14(1), 31–34 (2015).
Yu, I., Dakwar, A. & Takabe, K. Immunotherapy: Recent advances and its future as a neoadjuvant, adjuvant, and primary treatment in colorectal cancer. Cells 12(2), 258 (2023).
Karan, C. et al. Human epidermal growth factor receptor 2-targeting approaches for colorectal cancer: Clinical implications of novel treatments and future therapeutic avenues. JCO Oncol. Pract. 18(8), 545–554 (2022).
Zekri, L. et al. An optimized IgG-based B7–H3xCD3 bispecific antibody for treatment of gastrointestinal cancers. Mol. Ther. 31(4), 1033–1045 (2023).
Chen, Y. et al. Anti-PD-1 combined with targeted therapy: Theory and practice in gastric and colorectal cancer. Biochim. Biophys. Acta Rev. Cancer 1877(5), 188775 (2022).
Kristensen, L. S. et al. The emerging roles of circRNAs in cancer and oncology. Nat. Rev. Clin. Oncol. 19(3), 188–206 (2022).
Zhu, Z. X. et al. Current evidence on circRNAs as potential theranostic markers for detecting chemoresistance in breast cancer: a systematic review and meta-analysis. Sci. Rep. 12(1), 22016 (2022).
Feng, H. et al. The comprehensive detection of miRNA and circRNA in the regulation of intramuscular and subcutaneous adipose tissue of Laiwu pig. Sci. Rep. 12(1), 16542 (2022).
Alkan, A. H. & Akgul, B. Endogenous miRNA sponges. Methods Mol. Biol. 2257, 91–104 (2022).
de Sousa, M. C. et al. Deciphering miRNAs’ Action through miRNA editing. Int. J. Mol. Sci. 20(24), 6249 (2019).
Du, W. W. et al. Identifying and characterizing circRNA-Protein Interaction. Theranostics 7(17), 4183–4191 (2017).
Cao, L. et al. Circular RNA circRNF20 promotes breast cancer tumorigenesis and Warburg effect through miR-487a/HIF-1alpha/HK2. Cell Death Dis. 11(2), 145 (2020).
Zheng, R. et al. Exosomal circLPAR1 functions in colorectal cancer diagnosis and tumorigenesis through suppressing BRD4 via METTL3-eIF3h interaction. Mol. Cancer 21(1), 49 (2022).
Long, F. et al. Comprehensive landscape and future perspectives of circular RNAs in colorectal cancer. Mol. Cancer 20(1), 26 (2021).
Guo, Y. et al. Circ3823 contributes to growth, metastasis and angiogenesis of colorectal cancer: Involvement of miR-30c-5p/TCF7 axis. Mol. Cancer 20(1), 93 (2021).
Yao, B. et al. CircEZH2/miR-133b/IGF2BP2 aggravates colorectal cancer progression via enhancing the stability of m(6)A-modified CREB1 mRNA. Mol. Cancer 21(1), 140 (2022).
Zhang, Q. et al. CircIFNGR2 enhances proliferation and migration of CRC and induces cetuximab resistance by indirectly targeting KRAS via sponging to MiR-30b. Cell Death Dis. 14(1), 24 (2023).
Zhou, J. et al. Hsa_circ_0001666 suppresses the progression of colorectal cancer through the miR-576-5p/PCDH10 axis. Clin. Transl. Med. 11(11), e565 (2021).
Feng, Y. et al. Upregulated circTMEM59 inhibits cell growth and metastasis by miR-668-3p/ID4 axis in colorectal cancer. Oxid. Med. Cell Longev. 2022, 7242124 (2022).
Li, B. et al. Tumor-derived extracellular vesicles shuttle c-Myc to promote gastric cancer growth and metastasis via the KCNQ1OT1/miR-556-3p/CLIC1 axis. Cell Death Dis. 13(3), 217 (2022).
Jin, W. et al. Down-regulation of miR-556-3p inhibits hemangioma cell proliferation and promotes apoptosis by targeting VEGFC. Cell Mol. Biol. 66(5), 204–207 (2020).
Gupta, N. et al. Stress granule-associated protein G3BP2 regulates breast tumor initiation. Proc. Natl. Acad. Sci. U S A 114(5), 1033–1038 (2017).
Chen, Y. et al. RIOK1 mediates p53 degradation and radioresistance in colorectal cancer through phosphorylation of G3BP2. Oncogene 41(25), 3433–3444 (2022).
Gheytanchi, E. et al. Morphological and molecular characteristics of spheroid formation in HT-29 and Caco-2 colorectal cancer cell lines. Cancer Cell Int. 21(1), 204 (2021).
Kan, Z. et al. Identification of circRNA-miRNA-mRNA regulatory network and crucial signaling pathway axis involved in tetralogy of fallot. Front. Genet. 13, 917454 (2022).
Bao, N. et al. Identification of circRNA-miRNA-mRNA networks to explore the molecular mechanism and immune regulation of postoperative neurocognitive disorder. Aging 14(20), 8374–8393 (2022).
Liou, G. Y. CD133 as a regulator of cancer metastasis through the cancer stem cells. Int. J. Biochem. Cell Biol. 106, 1–7 (2019).
Xu, H. et al. CircRNA_0000392 promotes colorectal cancer progression through the miR-193a-5p/PIK3R3/AKT axis. J. Exp. Clin. Cancer Res. 39(1), 283 (2020).
Chen, H. Y. & Cheng, A. J. Potential to eradicate cancer stemness by Targeting cell surface GRP78. Biomolecules 12(7), 941 (2022).
Zhao, B. et al. Inflammatory micro-environment contributes to stemness properties and metastatic potential of HCC via the NF-kappaB/miR-497/SALL4 axis. Mol. Ther. Oncolytics 15, 79–90 (2019).
Yin, H. et al. FUBP1 promotes colorectal cancer stemness and metastasis via DVL1-mediated activation of Wnt/beta-catenin signaling. Mol. Oncol. 15(12), 3490–3512 (2021).
Lagunas-Rangel, F. A. Circular RNAs and their participation in stemness of cancer. Med. Oncol. 37(5), 42 (2020).
Jia, L., Wang, Y. & Wang, C. Y. circFAT1 promotes cancer stemness and immune evasion by promoting STAT3 activation. Adv. Sci. (Weinh) 8(13), 2003376 (2021).
Chen, J. et al. circPTN sponges miR-145-5p/miR-330-5p to promote proliferation and stemness in glioma. J. Exp. Clin. Cancer Res. 38(1), 398 (2019).
Zhang, L. et al. CircAGFG1 drives metastasis and stemness in colorectal cancer by modulating YY1/CTNNB1. Cell Death Dis. 11(7), 542 (2020).
Fan, L. et al. Circ_0030586 inhibits cell proliferation and stemness in bladder cancer by inactivating the ERK signaling via miR-665/NR4A3 axis. Acta Histochem. 123(5), 151745 (2021).
Liu, Z. et al. Circular RNA hsa_circ_001783 regulates breast cancer progression via sponging miR-200c-3p. Cell Death Dis. 10(2), 55 (2019).
Zheng, Y. et al. G3BP2 regulated by the lncRNA LINC01554 facilitates esophageal squamous cell carcinoma metastasis through stabilizing HDGF transcript. Oncogene 41(4), 515–526 (2022).
Zhang, Y., et al. Repression of the stress granule protein G3BP2 inhibits immune checkpoint molecule PD-L1. Mol. Oncol. (2021).
Rad, S. M. et al. Transcription factor decoy against stem cells master regulators, Nanog and Oct-4: A possible approach for differentiation therapy. Tumour Biol. 36(4), 2621–2629 (2015).
Zhu, Y. D. et al. Celastrus orbiculatus extract reduces stemness of gastric cancer stem cells by targeting PDCD4 and EIF3H. Integr. Cancer Ther. 20, 15347354211058168 (2021).
Zhuang, Y. W. et al. Solasodine reverses stemness and epithelial-mesenchymal transition in human colorectal cancer. Biochem. Biophys. Res. Commun. 505(2), 485–491 (2018).
Yan, X. et al. Clinical significance of Fusobacterium nucleatum, epithelial-mesenchymal transition, and cancer stem cell markers in stage III/IV colorectal cancer patients. Oncol. Targets Ther. 10, 5031–5046 (2017).
He, J. et al. Circular RNA circHERC4 as a novel oncogenic driver to promote tumor metastasis via the miR-556-5p/CTBP2/E-cadherin axis in colorectal cancer. J. Hematol. Oncol. 14(1), 194 (2021).
Li, Z. et al. Hsa_circ_0020378 targets miR-556-5p/MAPK1 to regulate osteosarcoma cell proliferation and migration. Gene 856, 147135 (2023).
Wu, Z. et al. Knockdown of circ-ABCB10 promotes sensitivity of lung cancer cells to cisplatin via miR-556-3p/AK4 axis. BMC Pulm Med. 20(1), 10 (2020).
Liu, H. et al. Invasion-related circular RNA circFNDC3B inhibits bladder cancer progression through the miR-1178-3p/G3BP2/SRC/FAK axis. Mol. Cancer 17(1), 161 (2018).
Kabekkodu, S. P. et al. Clustered miRNAs and their role in biological functions and diseases. Biol. Rev. Camb. Philos. Soc. 93(4), 1955–1986 (2018).
Wang, J. et al. circRNA-002178 act as a ceRNA to promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis. 11(1), 32 (2020).
Acknowledgements
This study was supported by the Natural Science Foundation of Inner Mongolia (2021JQ09), the Inner Mongolia Autonomous Region Health Science and Technology Plan Project (202201256, 202201363), the Inner Mongolia Autonomous Region science and technology planning project (2022YFHH0095), the Scientific and Technological Innovative Research Team for Inner Mongolia Universities (NMGIRT2327), the Project of Inner Mongolia Medical University (YKD2021LH006, YKD2022ZD004), and the Zhi Yuan Talent Projects of Inner Mongolia Medical University (ZY0130014).
Funding
This study was supported by the Natural Science Foundation of Inner Mongolia (2021JQ09), the Inner Mongolia Autonomous Region Health Science and Technology Plan Project (202201256, 202201363, 202202222), the Inner Mongolia Autonomous Region science and technology planning project (201802152, 2022YFHH0095, 2023KJHZ0023), the Scientific and Technological Innovative Research Team for Inner Mongolia Universities (NMGIRT2327), the Project of Inner Mongolia Medical University (YKD2021LH006, YKD2022ZD004, YKD2022TD003, YKD2023TD010), and the Zhi Yuan Talent Projects of Inner Mongolia Medical University (ZY0130014).
Author information
Authors and Affiliations
Contributions
Q.H., M.Z. and Y.W. contributed to the study conception and design. Data collection and analysis were performed by Q.H., M.Z., Y.W., Y.G. and Y.Z.; Validation was conducted by Q.H., M.Z., Y.W. and W.L.; The first draft of the manuscript was written by Q.H., M.Z. and Y.W.; L.F. and Z.W. reviewed and edited the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hao, Q., Zhang, M., Wu, Y. et al. Hsa_circRNA_001676 accelerates the proliferation, migration and stemness in colorectal cancer through regulating miR-556-3p/G3BP2 axis. Sci Rep 13, 18353 (2023). https://doi.org/10.1038/s41598-023-45164-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-45164-6
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.