Abstract
Recently, prediction of gestational diabetes mellitus (GDM) using artificial intelligence (AI) from medical records has been reported. We aimed to evaluate GDM-predictive AI-based models using birth cohort data with a wide range of information and to explore factors contributing to GDM development. This investigation was conducted as a part of the Japan Environment and Children's Study. In total, 82,698 pregnant mothers who provided data on lifestyle, anthropometry, and socioeconomic status before pregnancy and the first trimester were included in the study. We employed machine learning methods as AI algorithms, such as random forest (RF), gradient boosting decision tree (GBDT), and support vector machine (SVM), along with logistic regression (LR) as a reference. GBDT displayed the highest accuracy, followed by LR, RF, and SVM. Exploratory analysis of the JECS data revealed that health-related quality of life in early pregnancy and maternal birthweight, which were rarely reported to be associated with GDM, were found along with variables that were reported to be associated with GDM. The results of decision tree-based algorithms, such as GBDT, have shown high accuracy, interpretability, and superiority for predicting GDM using birth cohort data.
Similar content being viewed by others
Introduction
Gestational diabetes mellitus (GDM) is the glucose intolerance that is first recognized during pregnancy. GDM is a common complication during pregnancy, affecting up to 15% of pregnant women worldwide, affecting both maternal and fetal status, and causing perinatal complications, including stillbirth, premature delivery, macrosomia, fetal hyperinsulinemia, and clinical neonatal hypoglycemia1. Fetuses exposed to a hyperglycemic environment have a higher risk of developing chronic diseases, such as obesity, diabetes, and cardiovascular disease2,3,4,5. Mothers are more likely to be diagnosed with GDM at 24–28 weeks of gestation. However, early interventions for GDM are effective in reducing its impact on mothers and fetuses6.
Diagnostic technology using artificial intelligence (AI) for disease evaluations has shown the equivalent performance to that of clinicians, depending on the intended use and combination of AI algorithms7. These results come from the application of several characteristic types of AI algorithms. For example, deep learning is effective for unstructured data (e.g., images or sound data)8. Actually, deep learning has a variety of training models in the fields of image, natural language, and audio data. However, Grinsztajn et al. reported that they are inferior to decision tree-based models for tabular data9. Because of problems with the interpretability of the model and results, this study considered other methods.
Other algorithms included support vector machine (SVM), random forest (RF), and the gradient boosting decision tree (GBDT), which are effective for database and other structured data10,11,12. These diagnostic technologies predict GDM with high accuracy13,14,15. However, the related studies used medical records as their data source16. Medical records provide accurate medical histories and conditions along with several blood test results obtained during pregnancy13,14. In contrast, birth cohort data, including information on lifestyle and living environment that is not typically included in medical records, are used for only a few AI studies examining the accuracy and efficacy of GDM prediction15,16. Birth cohort data include lifestyle and social factors, depending on the purpose of the survey17,18. A recent study reported that the modifiable risk factors for GDM during pregnancy include excessive weight gain, lifestyle behaviors, and poor mental health19. Analyzing a combination of medical records and lifestyle or living environment data can provide comprehensive evaluations of GDM prediction. Additionally, if lifestyle and living environment have a high impact on predictive accuracy, interventions targeting these factors or diagnostic algorithms considering these parameters may lead to early GDM prediction and prevention, thereby reducing GDM exacerbation and morbidity. Recently, a systematic review of the relationship between diet and physical activity and GDM has been reported20. In this report, the authors report that pre-pregnancy or early pregnancy intervention is effective in preventing GDM. This suggests that intervention can prevent GDM if it can be predicted at an early stage. For this purpose, it is important to investigate whether there are other factors in addition to the ones that are already reported that are predictive of GDM. The Japan Environment and Children’s Study (JECS) (2011–2014), a large birth cohort study, recruited participants from 15 Regional Centres in Japan. It enrolled pregnant women and collected a wide range of information (e.g., living environment and lifestyle factors) to reveal the environmental factors affecting children’s health and development17,18. Investigations using the JECS data have shown significant associations between GDM and various specific lifestyle factors (e.g. social capital)21. These were the projects that examined the relationship between limited variables and GDM. However, no study involved AI application to assess the relationship between GDM and comprehensive maternal data from a large cohort. Therefore, we aimed to evaluate the accuracy, predictive value, and utility of each AI algorithm in developing a GDM prediction model, and explore the factors contributing to GDM development using structured data, various parameters, and large data from the JECS.
Results
Of the 103,060 JECS cohort participants, excluding those not suitable for analysis, there were 82,698 participants. Of these, 624 were GDM in groups with a past history of GDM (GDM-PH(+)) and 82,074 were GDM in groups without a past history of GDM (GDM-PH(−)). Table 1 shows characteristics of the pregnant women. The incidence of GDM according to the study data was 2.7%. The incidence of GDM-PH(+) and GDM-PH(−) was 52.3% and 2.7%, respectively. The number of weeks at the time of completion of the Maternal questionnaire at study enrollment for GDM-PH(+) and GDM-PH(−) was 14.88 weeks and 13.99 weeks, respectively.
The results of comparing the predictive accuracy of each algorithm are shown in Table 2. In the SVM model, overfitting occurred in both datasets; these overfitted models classified all input data as non-GDM. The results of the RF and LR models for the GDM-PH(+) group showed an area under the receiver operating characteristic curve (AUC) of 0.52 (95% CI 0.43–0.61) and 0.56 (95% CI 0.46–0.67), respectively. In the GBDT model for the GDM-PH(+) group, the performance were as follows: AUC = 0.67 (95% CI 0.57–0.75) and True Positive Rate (TPR) = 0.52 (95% CI 0.15–0.68). In contrast, the TPR of the RF and LR models for GDM-PH(−) mothers were zero due to overfitting. In the GBDT model for the GDM-PH(−) group, the performance were as follows: AUC = 0.74 (95% CI 0.71–0.77) and TPR = 0.01(95% CI 0.00–0.02).
For the GDM-PH(−) group, wherein overfitting occurred frequently, the results of changing the sampling methods are shown in Table 3. The results obtained using the SVM model did not change, even after altering the sampling methods. However, the results of undersampling in the RF model improved; the TPR increased to 0.18 (95% CI 0.14–0.22). The TPR also improved in both undersampling and oversampling in the GBDT and LR models as follows: undersampling in GBDT, 0.35 (95% CI 0.34–0.38); oversampling in GBDT, 0.21 (95% CI 0.16–0.27); undersampling in LR, 0.24 (0.17–0.30); and oversampling in LR, 0.23 (0.17–0.28). Following changes in sampling methods, undersampling showed higher accuracy than oversampling in the GBDT, LR, and RF models (except the SVM models).
Using GBDT modeling for GDM-PH(−) group, the relationship between TPR, false-positive rate (FPR), and change in AUC on altering the risk threshold is shown in Fig. 1. When the risk threshold was reduced, the TPR increased faster than the FPR. The AUC yielded a unimodal graph with a maximum value of 0.66 when the risk threshold was 0.025. In other models, the probability of GDM occurrence was zero in most input data due to overfitting; thus, altering the risk threshold was ineffective.
Variables with high variable importance (VIP) identified in the analysis of the GBDT model without changing the sampling methods are shown in Table 4. Variables with high VIP in the GDM-PH(−) group included HbA1c levels, BMI before pregnancy, and maternal age. Variables with high VIP in the GDM-PH(+) group included triglyceride levels, platelet count, and firstborn child’s birth year. SHAP (SHapley Additive exPlanation) summary plot (Mean (|SHAP Value|) is shown in Fig. 2a. Variables with high Mean (|SHAP Value|) in the GDM-PH(+) group included number of previous deliveries, 1st born child's birth year, and BMI before pregnancy. Figure 2b shows variables with a high Mean (|SHAP Value|) in the GDM-PH(−) group, including maternal age, HbA1c levels, and BMI before pregnancy.
Discussion
We compared four machine learning methods to improve GDM prediction models based on a large birth cohort. GBDT exhibited the highest accuracy, followed by LR, RF, and SVM. Without changing the sampling methods, overfitting occurred upon the use of all algorithms except for GBDT for GDM-PH(−). The accuracy for GDM prediction of all algorithms, except for SVM, improved without overfitting using undersampling or oversampling. Changing the risk thresholds improved the accuracy of GBDT.
Furthermore, GBDT results were more accurate than the existing method wherein LR used only maternal age, pre-pregnancy BMI, and laboratory results of specimens (see Supplementary Table S1 online). This could be because variables useful for GDM prediction can be increased using JCES data and GBDT can construct the boundary surface non-linearly.
There were some differences in variables important for predicting GDM in the GBDT model between the GDM-PH(+) (recurrent GDM) and GDM-PH(−) (new-onset GDM) groups. Thus, differences in VIP between recurrent GDM and new-onset GDM in JECS data were not based on parity.
The RF, GBDT, and SVM algorithms used are reportedly effective for structured data; thus, we compared them to determine the most appropriate one for the JECS data. For the GDM-PH(+) group, overfitting occurred in the data in the SVM model. Other algorithms yielded stable results without overfitting. For the GDM-PH(−) group, overfitting occurred in the data of all models, except for the GBDT model. Owing to the exploratory approach for predicting GDM, the data set used here was unique because it included many variables that do not affect GDM. Those noisy data cause compounding negative effects on generalizability and overfitting22. Imbalanced datasets often result in an overfitted model to achieve high classification accuracy23. The GDM-PH(+) group (N = 624) had a much smaller sample size than the GDM-PH(−) group (N = 82,074). Both groups included many variables that did not affect GDM. However, the ratio of the GDM and non-GDM groups were almost similar in the GDM-PH(+) group. Furthermore, the GDM-PH(−) group had a very low incidence of GDM (2.8%). In the SVM model, the problems related to many variables that do not improve the predictability negatively affect the analysis. SVM extracts records near the boundary surface as support vectors and creates a discrimination surface using only support vectors10. Thus, SVM can reduce the number of records used for analysis. However, SVM is not an algorithm to properly select variables from a large number of variables. Therefore, the choice of support vectors in our study was inappropriate, possibly leading to overfitting.
In contrast, RF and GBDT models use decision tree algorithms. The decision tree requires repeated binary decision-making. Therefore, variables that do not included the predictability of GDM are not included in the decision tree11,12. Therefore, decision tree algorithms are highly robust to data with many variables. In the RF algorithm, random sampling of the training dataset is performed as the first step to create multiple datasets11. Subsequently, the RF algorithm creates a decision tree model for each dataset to predict results by the majority rule. Sampling datasets from the GDM-PH(−) group using this particular algorithm may not ensure model diversity generated by random sampling, possibly leading to overfitting. In the GBDT model, hyperparameter optimization is performed using gradient descent before the start of each subsequent training session12. Therefore, unlike the RF model, the decision tree in the GBDT model may be trained while reducing the bias between the case and control groups. However, the TPR was not high even in the GBDT model (the only model without overfitting).
As in this study, the development of prediction models using data with a low case-to-control ratio requires adjustment of the sample size of the training data by changing the sampling method24. The TPRs of the RF, GBDT, and LR models were improved by changing the sampling method (Table 3). Undersampling could prevent overfitting with excessive control data. However, in the SVM model, the accuracy of GDM prediction has not improved, possibly because changes in sampling methods do not solve the problem of multidimensional data with many variables that do not improve the predictability. Considering oversampling, the TPR improved slightly in the GBDT and LR models, but overfitting occurred in the RF model. The oversampling technique randomly duplicates data until the case-to-control ratio reaches a specific value; thus, this technique may not solve the problem of the RF model (i.e., model diversity). In contrast, in the LR model, imbalance corrections, including changes in sampling methods may even worsen model performance25. In this study, the method of changing the risk threshold was used for imbalance corrections without changing the sampling method. The JECS data were not designed to estimate imbalance correction; thus, it was not possible to evaluate such effects in this study. However, our study demonstrated the potential to improve TPRs while maintaining the FPR low by changing the thresholds (Fig. 1). Generally, lowering the risk threshold increases both the TPR and FPR, but setting an appropriate risk threshold for LR and GBDT enables imbalance corrections without changing the sampling method. For setting the risk threshold, Goorbergh et al. used two fixed values—the prevalence of malignancy in the training dataset and the default risk threshold of 0.5. However, in this study, when the risk threshold was varied from 0 to 0.5 in steps of 0.005, the AUC reached its maximum value at 0.025, as did the GDM prevalence at 0.027.
We performed an exploratory analysis of factors contributing to GDM using AI. Typically, our exploratory methods have the following disadvantages: (1) the results may be inappropriate depending on the AI algorithms used, and (2) due to the cost, increasing the number of participants to obtain enough variables that are acceptable for the exploratory analysis was difficult. However, using sufficient data, selecting appropriate algorithms, and comparing VIPs, it was possible to identify variables previously not associated with GDM and verify previously reported associated factors.
In this study, we predicted the development of GDM based on information that could be collected in the early stages of pregnancy. Mothers are more likely to be diagnosed with GDM at 24–28 weeks of gestation. In this study, the average date of completion of the collected questionnaires was 14–15 weeks, which is considered early enough to predict the diagnosis, even if considering the time between the blood collection and the results of the tests. In meta-analysis, protective association of physical activity (21–46%) from GDM when comparing any type of physical activity to none in either the pre-pregnancy or early pregnancy period20. If a high-risk group near the 1st trimester can be extracted, it may lead to GDM prevention.
In this study, 775 questions were used to predict the incidence of GDM. Obviously, it would not be practical to build a prediction model using all of these questions, as it would take a lot of time to enter the predictors. Therefore, it is important to screen out as many variables as possible that are important for prediction. In this study, two evaluation criteria, VIP and Mean (|SHAP value|), were used to select predictors. High VIP variables identified in this study are listed in Table 4. Previous GDM studies identified a history of GDM in previous pregnancies, maternal age, and obesity as risk factors for GDM26. Additionally, the effect of GDM on the interpregnancy interval was reported27. The JECS data do not include the interpregnancy interval. Therefore, the firstborn child’s birth year was considered as an alternative variable. One study reported a significant difference in white blood cell count and platelet count between the GDM and non-GDM groups in the second trimester28. A meta-analysis showed a significant increase in lipid levels (e.g., triglyceride) in mothers with GDM in the first and second trimesters29. Variables that are reportedly associated with GDM in studies conducted before the JECS were also identified as factors with high VIP in this study. However, regarding urinary creatinine concentration, a study on the associations between urinary metals in early pregnancy and the subsequent risk of GDM reported no significant difference in urinary creatinine between GDM and non-GDM groups30. However, we revealed urinary creatinine concentration with a higher VIP from the GDM-PH(−) group, especially the nulliparous group. Although the reason for this is unknown, it may be a surrogate indicator for some other factor, such as physique.
The items in the questionnaire administered at enrollment in this study include the 8-item Short-Form Health Survey (SF-8) items for health-related quality of life (HRQOL)31. Physical component summary and mental component summary were variables with high VIP regardless of the presence or absence of a history of GDM. Regarding GDM and HRQOL, a systematic review examining the short- and long-term progression of HRQOL and their association with GDM diagnosis was reported; GDM does not directly lead to reduced QOL in mothers but causes some complicated interactions with psychological factors, resulting in reduced QOL32. The SF-8 data in this study were collected before 22 weeks of gestation. Our study results suggest that mothers’ HRQOLs are related to the risk of GDM; thus, GDM further reduces HRQOL.
Recent studies examining the association between mothers’ birth weights and GDM revealed that mothers with low birth weights or macrosomia were at higher risk of GDM33. We identified mothers’ birth weights as factors with high VIP in the GDM-PH(−) group. Hales et al. reported a correlation between low birth weight and subsequent glucose intolerance34. GDM is mild glucose intolerance; thus, mothers with low birth weights may have an increased risk of GDM.
High Mean|SHAP| variables identified in this study are shown Fig. 2a,b. Although similar variables to the VIPs were found in the top 20, SF-8 MCS and SF-8 PCS were absent from the top 20 for both GDM-PH(+) and GDM-PH(−).On the other hand, the GDM-PH(+) group showed a new variable, chocolate and vitamin D intake from the dietary questionnaire, and the GDM-PH(−) group showed a new variable, supplement intake (folic acid). Both vitamin D and folic acid have been reported to have an association with GDM35,36.
Although variables including those already reported to be associated with GDM, such as these, were detected in this study, the AUC score of gradient boosting for those with GDM history (0.67) was below the acceptable minimum for clinical implication (0.70). But in the JECS study, we are currently analyzing maternal genetic data, which will be provided in the future. By re-constructing the model after taking these genetic backgrounds into account, we expect to improve the prediction accuracy.
This study has some limitations. First, in Japan, the diagnostic criteria of the Japanese Society of Obstetrics and Gynecology are used to determine GDM. But the JECS is a multi-region, multi-medical institution cohort study; GDM data were obtained from medical record transcripts; thus, we could not review in detail the diagnostic criteria of GDM for the co-operating health care provider(s)21. Second, analysis in this study was performed considering information collected at the time of study enrollment. However, we did not consider the effects of other factors not identified in the JECS, especially genetic information and family history of diabetes mellitus. Third, information on diet was collected using self-administered questionnaires. Therefore, the results may not accurately reflect the actual food or nutrient intake. Fourth, the incidence of GDM in Japan is 7–13%37. However, the incidence of GDM in this study was 2.7%. This may indicate that the JECS included more health-conscious mothers or favored low enrollment for high-risk pregnancies, leading to sampling bias. Fifth, the JECS was conducted in Japan, and most participants were Japanese. Thus, generalization to populations from other countries may be inaccurate because the JECS results consider the unique living environment and lifestyle in Japan. Finally, with the size of the JECS data, it is difficult to obtain predictions by physicians as an external evaluation. Studies combining the findings of this study (mother's birth weight and psychological factors) with previously reported factors, including genes, are needed for more accurate prediction compared to prediction by other means, such as using genes.
In conclusion, we demonstrated that exploratory analysis using AI for a large birth cohort is possible through the appropriate use of algorithms. Algorithm comparison revealed high accuracy, interpretability, and superiority of decision tree-based algorithms, including GBDT considering datasets in this study. Further studies regarding GDM prediction using AI are needed to improve the TPR by collecting other variables, including genetic information and family history of diabetes mellitus. Using exploratory analysis of the JECS data, we identified the importance of previously reported variables related to GDM and new variables, such as HRQOL in early pregnancy and mothers’ birth weights related to GDM.
Material and methods
Data sources
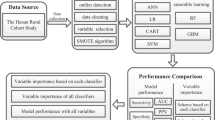
The JECS is a nationwide birth cohort study. The design of the JECS study is described elsewhere18. The eligibility criteria for participants in the JECS did not consider the presence or absence of disease. This study used the jecs-ta-20190930 dataset, which was released in October 2019. The following data were identified from the dataset and used for analysis: maternal questionnaire data and dietary data from the survey administered at study enrollment (T1), medical record transcripts during pregnancy, child’s sex determined after delivery, laboratory results of specimens collected by 21 weeks of gestation, parental education and household income data collected during mid-late pregnancy, and parents’ birth weights (reported by the mother) after delivery. For the study outcome, GDM cases were defined as GDM during pregnancy per medical record transcripts. The maternal questionnaire included the Kessler Psychological Distress Scale (K6) as an indicator of psychological distress38, a short version of the International Physical Activity Questionnaire as an indicator of physical activity39,40, SF-8 Health Survey (SF-8) as an indicator of health-related quality of life31, and environmental exposures41. Food-intake and nutrient-intake data were obtained from the food frequency questionnaires validated in the Japan Public Health Center-based Prospective Study for the Next Generation42.
Data preprocessing
The flow chart of the selection of participants is presented in Fig. 3. In this study, one pregnant woman with multiple fetuses is counted as one case. Of 103,060 pregnancies, the following were excluded from the present study: mothers with missing records on the diagnosis of GDM (N = 2375), mothers with a missing GDM history (N = 1960), and mothers who completed the T1 questionnaire at or after 22 weeks of gestation (N = 16,027). We followed 82,698 participants and observed 2253 pregnancies with GDM and 80,445 pregnancies without GDM. The recurrence rates of GDM were high43; thus, the remaining 82,698 pregnancies were divided as follows: (1) GDM in groups with a history of GDM (GDM-PH(+)) (N = 624) and (2) GDM in groups without a history of GDM (GDM-PH(−)) (N = 82,074).
As variables used to stratify this study, the history of GDM was reported in the T1 questionnaires. The qualitative text response data from open-ended questions and original variables summarized as separate variables (e.g., K6 and SF-8) were excluded from the analysis of the questionnaire data. Due to the very low number of triplets, information on multiple births was classified into singleton and multiple pregnancies. The variables used are shown in Supplemental Table S2. Non-ordinal categorical variables (e.g., marriage status, maternal occupation, infertility treatment status, and drugs used during pregnancy) were converted to dummy variables. BMI before pregnancy was calculated using weight before pregnancy as weight (kg)/height2 (m2). The variables reporting time of day with separate hours and minutes were converted to hours (i.e., hours + minutes/60). Continuous variables were normalized.
Statistical analysis
The machine learning methods used in this study included SVM10 (which uses radial basis function [Gaussian] kernel), RF11, and GBDT12. LR44 was used as a reference.
All analyses were performed using python 3.8.5 in Jupyter Nortbook (Project Jupyter). Among the python libraries, scikit-learn 1.0.2 was used in the LR, SVM, and RF models; lightgbm 3.1.1 was used in the GBDT model; and imbalanced-learn 0.9.0 was used in undersampling and oversampling. The settings of the hyperparameters of these algorithms are shown in Table 5. SVM and LR do not accept datasets with missing data; thus, single imputation using mean substitution was performed. For cross-validation, our data were randomly divided into two groups with a ratio of 4:1, a training set, and a test set. To prevent overestimation from the use of imbalanced data, we used undersampling and oversampling methods, and the ratio of the GDM group to the non-GDM group was set at 1:2. True-positive rate (TPR), calculated as true positive/(true positive + false negative), and false-positive rate (FPR), calculated as false positive/(false positive + true negative), were the parameters of GDM prediction across the models. The AUC was used for evaluating model accuracy. The risk threshold for GDM classification was set at 0.5. Changes in the TPR and FPR when the risk threshold was changed from 0 to 0.5 step by 0.005 were examined.
Ethical approval
The JECS protocol was reviewed and approved by the Ministry of the Environment's Institutional Review Board on Epidemiological Studies (approval no.: 100910001) and the Ethics Committees of all participating institutions; the Medical Support Centre (National Centre for Child Health and Development), and 15 Regional Centers (Hokkaido University, Tohoku University, Fukushima Medical University, Chiba University, Yokohama City University, University of Yamanashi, University of Toyama, Nagoya City University, Kyoto University, Osaka University, Hyogo College of Medicine, Tottori University, Kochi University, University of Occupational and Environmental Health, and Kumamoto University). This study was conducted in accordance with the Helsinki Declaration and other nationally valid regulations and guidelines. In the JECS, written informed consent was obtained from all participants.
Data availability
Data were unsuitable for public deposition due to ethical restrictions and the legal framework of Japan. It is prohibited by the Act on the Protection of Personal Information (Act No. 57 of May 30, 2003, amendment on September 9, 2015) to publicly deposit data containing personal information. Ethical Guidelines for Medical and Health Research Involving Human Subjects enforced by the Japan Ministry of Education, Culture, Sports, Science and Technology and the Ministry of Health, Labour and Welfare also restrict the open sharing of epidemiologic data. All inquiries about access to data should be sent to: jecs-en@nies.go.jp. The person responsible for handling inquiries sent to this e-mail address is Dr. Shoji F Nakayama, JECS Programme Office, National Institute for Environmental Studies.
References
Gabbe, S. G. Gestational diabetes mellitus. N. Engl. J. Med. 315, 1025–1026 (1986).
Ding, G. L. et al. Transgenerational glucose intolerance with Igf2/H19 epigenetic alterations in mouse islet induced by intrauterine hyperglycemia. Diabetes 61, 1133–1142 (2012).
Dabelea, D. et al. Intrauterine exposure to diabetes conveys risks for type 2 diabetes and obesity: A study of discordant sibships. Diabetes 49, 2208–2211 (2000).
Dabelea, D. et al. Association of intrauterine exposure to maternal diabetes and obesity with type 2 diabetes in youth: The SEARCH case–control study. Diabetes Care 31, 1422–1426 (2008).
Tam, W. H. et al. In utero exposure to maternal hyperglycemia increases childhood cardiometabolic risk in offspring. Diabetes Care 40, 679–686 (2017).
Koivusalo, S. B. et al. Gestational diabetes mellitus can be prevented by lifestyle intervention: The Finnish gestational diabetes prevention study (RADIEL): A randomized controlled trial. Diabetes Care 39, 24–30 (2016).
Shen, J. et al. Artificial intelligence versus clinicians in disease diagnosis: Systematic review. JMIR Med. Inform. 7, e10010 (2019).
Craik, A., He, Y. & Contreras-Vidal, J. L. Deep learning for electroencephalogram (EEG) classification tasks: A review. J. Neural. Eng. 16, 031001 (2019).
Grinsztajn, L., Oyallon, E. & Varoquaux, G. Why do tree-based models still outperform deep learning on typical tabular data?. Adv. Neural Inf. Process. Syst. 35, 507–520 (2022).
Cortes, C. & Vapnik, V. Support-vector networks. Mach. Learn. 20, 273–297 (1995).
Breiman, L. Random forests. Mach. Learn. 45, 5–32 (2001).
Ke, G. et al. Lightgbm: A highly efficient gradient boosting decision tree. In Advances in Neural Information Processing Systems, Vol. 30 (eds Guyon, I. et al.) (Curran Associates, 2017).
Artzi, N. S. et al. Prediction of gestational diabetes based on nationwide electronic health records. Nat. Med. 26, 71–76 (2020).
Wu, Y. T. et al. Early prediction of gestational diabetes mellitus in the chinese population via advanced machine learning. J. Clin. Endocrinol. Metab. 106, e1191–e1205 (2021).
Ye, Y. et al. Comparison of machine learning methods and conventional logistic regressions for predicting gestational diabetes using routine clinical data: A retrospective cohort study. J. Diabetes Res. 2020, 4168340 (2020).
Mennickent, D., Rodríguez, A., Farías-Jofré, M., Araya, J. & Guzmán-Gutiérrez, E. Machine learning-based models for gestational diabetes mellitus prediction before 24–28 weeks of pregnancy: A review. Artif. Intell. Med. 132, 102378 (2022).
Michikawa, T. et al. The Japan Environment and Children’s Study (JECS): A preliminary report on selected characteristics of approximately 10,000 pregnant women recruited during the first year of the study. J. Epidemiol. 25, 452–458 (2015).
Kawamoto, T. et al. Rationale and study design of the Japan environment and children’s study (JECS). BMC Public Health 14, 25 (2014).
Mizuno, S. et al. Association between social capital and the prevalence of gestational diabetes mellitus: An interim report of the Japan Environment and Children’s Study. Diabetes Res. Clin. Pract. 120, 132–141 (2016).
Mijatovic-Vukas, J. et al. Associations of diet and physical activity with risk for gestational diabetes mellitus: A systematic review and meta-analysis. Nutrients 10, 698 (2018).
Horsch, A. et al. Improving cardiometabolic and mental health in women with gestational diabetes mellitus and their offspring: Study protocol for MySweetHeart Trial, a randomised controlled trial. BMJ Open 8, e020462 (2018).
Berisha, V. et al. Digital medicine and the curse of dimensionality. NPJ Digit. Med. 4, 153 (2021).
van den Goorbergh, R., van Smeden, M., Timmerman, D. & Van Calster, B. The harm of class imbalance corrections for risk prediction models: Illustration and simulation using logistic regression. J. Am. Med. Inform. Assoc. 29, 1525–1534. https://doi.org/10.1093/jamia/ocac093 (2022).
Chen, Y. F. et al. Design of a clinical decision support system for fracture prediction using imbalanced dataset. J. Healthc. Eng. 2018, 9621640 (2018).
Zhu, Y. & Zhang, C. Prevalence of gestational diabetes and risk of progression to Type 2 diabetes: A global perspective. Curr. Diab. Rep. 16, 7 (2016).
Holmes, H. J., Lo, J. Y., McIntire, D. D. & Casey, B. M. Prediction of diabetes recurrence in women with class A1 (diet-treated) gestational diabetes. Am. J. Perinatol. 27, 47–52 (2010).
Sargın, M. A. et al. Neutrophil-to-lymphocyte and platelet-to-lymphocyte ratios: Are they useful for predicting gestational diabetes mellitus during pregnancy?. Ther. Clin. Risk. Manage. 12, 657–665 (2016).
Ryckman, K. K., Spracklen, C. N., Smith, C. J., Robinson, J. G. & Saftlas, A. F. Maternal lipid levels during pregnancy and gestational diabetes: A systematic review and meta-analysis. BJOG 122, 643–651 (2015).
Wang, X. et al. Exposure to multiple metals in early pregnancy and gestational diabetes mellitus: A prospective cohort study. Environ. Int. 135, 105370 (2020).
Tokuda, Y. et al. Assessing items on the SF-8 Japanese version for health-related quality of life: A psychometric analysis based on the nominal categories model of item response theory. Value Health 12, 568–573 (2009).
Fukuhara, S. & Suzukamo, Y. Manual of the SF-8 Japanese, 71–116 (Institute for Health Outcomes & Process Evaluation Research Kyoto, 2004).
Marchetti, D., Carrozzino, D., Fraticelli, F., Fulcheri, M. & Vitacolonna, E. Quality of life in women with gestational diabetes mellitus: A systematic review. J. Diabetes Res. 2017, 7058082 (2017).
Zhang, L., Zheng, W., Huang, W., Liang, X. & Li, G. Differing risk factors for new onset and recurrent gestational diabetes mellitus in multipara women: A cohort study. BMC Endocr. Disord. 22, 3 (2022).
Hales, C. N. et al. Fetal and infant growth and impaired glucose tolerance at age 64. BMJ. 303, 1019–1022 (1991).
Mahendra, A. & Fall, C. H. D. Maternal vitamin D deficiency and GDM risk: Evidence for the case of investing more attention in antenatal clinics. Proc. Nutr. Soc. 20, 1–7 (2021).
Chen, X. et al. Association of maternal folate and vitamin B12 in early pregnancy with gestational diabetes mellitus: A prospective cohort study. Diabetes Care 44, 217–223 (2021).
Morikawa, M. et al. Change in the number of patients after the adoption of IADPSG criteria for hyperglycemia during pregnancy in Japanese women. Diabetes Res. Clin. Pract. 90, 339–342 (2010).
Furukawa, T. A. et al. The performance of the Japanese version of the K6 and K10 in the World Mental Health Survey Japan. Int. J. Methods Psychiatr. Res. 17, 152–158 (2008).
Craig, C. L. et al. International physical activity questionnaire: 12-country reliability and validity. Med. Sci. Sports Exerc. 35, 1381–1395 (2003).
Murase, N., Katsumura, T., Ueda, C., Inoue, S. & Shimomitsu, T. International standardization of physical activity level: Reliability and validity study of the Japanese version of the International Physical Activity Questionnaire (IPAQ). J. Health Welf. Stat. (Kosei no Shihyo) 49, 1–9 (2003) (in Japanese).
Iwai-Shimada, M. et al. Questionnaire results on exposure characteristics of pregnant women participating in the Japan Environment and Children Study (JECS). Environ. Health Prev. Med. 23, 45 (2018).
Yokoyama, Y. et al. Validity of short and long self-administered food frequency questionnaires in ranking dietary intake in middle-aged and elderly Japanese in the Japan Public Health Center-Based Prospective Study for the Next Generation (JPHC-NEXT) Protocol Area. J. Epidemiol. 26, 420–432 (2016).
Kim, C., Berger, D. K. & Chamany, S. Recurrence of gestational diabetes mellitus: A systematic review. Diabetes Care 30, 1314–1319 (2007).
Cramer, J. S. The origins of logistic regression. Tinbergen Institute Working Paper No. 2002-119/4. SSRN: https://ssrn.com/abstract=360300 or https://doi.org/10.2139/ssrn.360300 (2002).
Acknowledgements
We would like to thank all participants and co-operating health care providers and staff involved in the Japan Environment and Children’s Study and Editage (www.editage.jp) for English language editing. We would also like to thank the members of the JECS Group as of 2023: Michihiro Kamijima (principal investigator, Nagoya City University, Nagoya, Japan), Shin Yamazaki (National Institute for Environmental Studies, Tsukuba, Japan), Yukihiro Ohya (National Center for Child Health and Development, Tokyo, Japan), Reiko Kishi (Hokkaido University, Sapporo, Japan), Nobuo Yaegashi (Tohoku University, Sendai, Japan), Koichi Hashimoto (Fukushima Medical University, Fukushima, Japan), Chisato Mori (Chiba University, Chiba, Japan), Shuichi Ito (Yokohama City University, Yokohama, Japan), Zentaro Yamagata (University of Yamanashi, Chuo, Japan), Hidekuni Inadera (University of Toyama, Toyama, Japan), Takeo Nakayama (Kyoto University, Kyoto, Japan), Tomotaka Sobue (Osaka University, Suita, Japan), Masayuki Shima (Hyogo Medical University, Nishinomiya, Japan), Seiji Kageyama (Tottori University, Yonago, Japan), Narufumi Suganuma (Kochi University, Nankoku, Japan), Shoichi Ohga (Kyushu University, Fukuoka, Japan), and Takahiko Katoh (Kumamoto University, Kumamoto, Japan).
Funding
This Japan Environment and Children’s Study was funded by the Ministry of the Environment, Japan. The funding source had no role in study design, data collection, and analysis, manuscript preparation, or the decision to publish. The findings and conclusions of this article are solely the responsibility of the authors and do not represent the official views of the government.
Author information
Authors and Affiliations
Consortia
Contributions
M.W. and K.S. designed the research. M.W., M.Y., C.M., and the JECS group collected data. M.W. and A.E. conducted data analysis. M.W., A.E., K.S., and M.Y. contributed to data interpretation. M.W. drafted the initial manuscript. C.M. made critical revisions. All authors reviewed and commented on the manuscript. All authors approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Watanabe, M., Eguchi, A., Sakurai, K. et al. Prediction of gestational diabetes mellitus using machine learning from birth cohort data of the Japan Environment and Children's Study. Sci Rep 13, 17419 (2023). https://doi.org/10.1038/s41598-023-44313-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-44313-1
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.