Abstract
Gut dysbiosis has been identified as a crucial factor of Alzheimer's disease (AD) development for apolipoprotein E4 (APOE4) carriers. Inulin has shown the potential to mitigate dysbiosis. However, it remains unclear whether the dietary response varies depending on sex. In the study, we fed 4-month-old APOE4 mice with inulin for 16 weeks and performed shotgun metagenomic sequencing to determine changes in microbiome diversity, taxonomy, and functional gene pathways. We also formed the same experiments with APOE3 mice to identify whether there are APOE-genotype dependent responses to inulin. We found that APOE4 female mice fed with inulin had restored alpha diversity, significantly reduced Escherichia coli and inflammation-associated pathway responses. However, compared with APOE4 male mice, they had less metabolic responses, including the levels of short-chain fatty acids-producing bacteria and the associated kinases, especially those related to acetate and Erysipelotrichaceae. These diet- and sex- effects were less pronounced in the APOE3 mice, indicating that different APOE variants also play a significant role. The findings provide insights into the higher susceptibility of APOE4 females to AD, potentially due to inefficient energy production, and imply the importance of considering precision nutrition for mitigating dysbiosis and AD risk in the future.
Similar content being viewed by others
Introduction
Apolipoprotein E4 (APOE4) is the strongest genetic risk factor for Alzheimer's disease (AD), the most common form of dementia characterized by extracellular beta-amyloid plaques, intraneuronal tau tangles, and neurodegeneration1,2. Recent evidence suggests that an imbalanced gut microbiome, or dysbiosis, may contribute to developing AD-like neuropathology3,4,5. Specifically, AD patients have an increased abundance of Escherichia coli (E. coli), decreased short-chain fatty acids (SCFAs) as assessed through fecal sampling, and increased systemic inflammation and microglia activation3. In addition to the APOE4 genetic factor, gender plays a significant role with females having a higher risk of AD than males6,7,8. Studies have found that APOE alleles and sex influence the gut microbiome structure9,10,11. Asymptomatic APOE4 carriers have more extensive gut dysbiosis than non-carriers, such as those with APOE e3 alleles (APOE3)12. In addition, females may have different gut microbiota composition than males, increasing the risk for AD pathology8. Thus, mitigating dysbiosis at an early stage may be crucial for preventing AD development.
Our prior research demonstrates that supplementing a diet with the prebiotic inulin, a fermentable prebiotic fiber, can positively impact gut microbiome composition, boost the production of SCFAs, enhance mitochondrial function, and decrease neuroinflammation in young, asymptomatic APOE4 mice13. Our results also demonstrate that these effects of dietary inulin supplementation revealed an APOE genotype-dependent response9. Despite these promising results, it remains unclear whether the response to the inulin diet varies based on sex and whether specific microbial metabolic pathways would be impacted. The present study aims to address this knowledge gap.
We had young, asymptomatic APOE4 male (E4-M) and female (E4-F) mice fed with either inulin or control diet for 16 weeks and collected their fecal samples pre and post the diet. We performed shotgun metagenomics sequencing and analyses on microbiome diversity, differential relative abundance of microbial taxonomy, and the Kyoto Encyclopedia of Genes and Genomes (KEGG) functional pathways. We found that E4-F and E4-M mice had distinctive responses to the diet. E4-F fed with inulin (E4-F-inulin) had altered β-diversity, and the impacts on the functional pathway were more on anti-inflammation, with significantly decreased E. coli abundance compared with the E4-F-control group. In addition, inulin normalized alpha (α)-diversity of E4-F mice compared with E4-M-inulin and E4-M-control mice, indicating restoration of evenness and richness of microbial community14 of the E4-F-inulin mice. Similarly, E4-M mice fed with inulin (E4-M-inulin) had altered beta (β)-diversity but had significantly increased SCFAs-producing bacteria, especially those related to acetate production, and decreased lactic acid bacteria (LAB). They also had increased gut abundance of SCFAs-related kinases, including acetate CoA-transferase, propionate kinase, and butyrate kinase, and family Erysipelotrichaceae contributed most to the abundance of acetate CoA-transferase and propionate kinase. The mitochondrial tricarboxylic acid (TCA) cycle level was also elevated in the E4-M-inulin mice.
We performed the same experiments with mice having the human APOE3 allele. Although inulin altered β-diversity in both sexes, and the E3-M-inulin mice also had positive responses on SCFAs-producing bacteria and TCA cycle elevation as the E4-M mice did, the overall sex effects were not as strong as those observed in the APOE4 groups. The findings indicate a significant role played by different APOE variants.
These results highlight the importance of both APOE genotype and sex on the gut microbiome changes in response to the inulin. In particular, sex difference plays a significant role in the APOE4 mice. The findings may shed light on the importance of considering precision nutrition for mitigating APOE4-related neurodegenerative disorders, such as AD.
Results
Inulin's effects on food intake and body weight in APOE3 and APOE4 Mice
We documented food intake and body weight changes throughout the study. Figure 1 shows the end-point results. Food intakes were increased in the APOE3 mice (p < 0.001) (Fig. 1a), both in the male and female mice (Fig. 1b). In contrast, APOE4 mice did not show changes in food intake. However, the food intake increases in the APOE3 mice did not alter the body weight (Fig. 1c,d). No body weight changes were observed in the APOE4 mice either.
Analysis of food intake and body weight. (a) Inulin increased the food intake in APOE3 mice compared to its control when stratifying mice by APOE genotype and diet. (b) When stratifying mice by genotype, sex, and diet, inulin increased the food intake in E3-M and E3-F mice compared to their controls. Inulin didn’t change the body weight of the mice when considering (c) genotype and diet or (d) genotype, diet, and sex. *p < 0.05. ***p < 0.001.
Inulin normalizes α-diversity of the APOE4 female mice
We analyzed the α-diversity using the Shannon index, which measures the richness or evenness of a microorganism within a sample14. Table 1 shows the changes in α-diversity caused by inulin in male and female mice with APOE3 and APOE4 genes (considering the interaction of Gene × Sex × Diet), reporting data from a generalized linear model (GLM) analysis and pairwise comparisons, including degrees of freedom, residual df, F value, and p-values.
The results are visualized in Fig. 2a. It shows that in the control groups, the α-diversity of E4-F was higher than that in E4-M (p = 0.027) and APOE3 female control (E3-F-control) mice (p = 0.002) with an overall sex effect among groups [F (1, 69) = 6.753, p = 0.012]; there were no differences between the E4-M or E3-M mice, either with the control or inulin diets. However, when the mice were given an inulin diet, the difference between E4-F and the other three groups diminished, indicating that the diet normalized the α-diversity of the E4-F mice.
Differences in α-diversity when stratifying mice by APOE genotype, sex, and diet and differences in β-diversity when stratifying mice by genotype, sex, and diet. (a) Inulin normalized the difference in the α-diversity between E4-F-control mice compared to E3-F-control and E4-M-control mice. However, there was no difference between E4-F-inulin, E3-F-inulin, and E4-M-inulin mice, indicating that inulin normalized the gut dysbacteriosis in E4-F mice. Inulin altered the β-diversity in (b) E3-inulin and E4-inulin mice compared to their controls in male and female mice. *p < 0.05. **p < 0.01.
Inulin alters β-diversity in both sexes
We analyzed the β-diversity using the Bray–Curtis index, which measures distinct differences in microbial composition between the control mice and those fed with inulin in our study14. Table 1 shows the data of changes in β-diversity due to inulin in APOE3 and APOE4 mice, considering both the effect of Gene × Diet and Gene × Sex × Diet, reporting ADONIS analysis and pairwise ANOSIM comparisons data including the sum of squares, mean squares, F value, R2, and respective p-values. E4-inulin mice experienced significant changes in β-diversity compared to its control (p = 0.001), with an overall diet effect among groups (ADONIS R2 = 0.436, F = 57.873, p = 0.001). E3-inulin mice showed a similar result, but a more pronounced impact was seen in E4 mice from the figure. Further stratifying by sex, we found that both male and female mice had significant changes in β-diversity when fed with inulin, regardless of their APOE genotype. Figure 2b show these results for males and females; an overall diet (ADONIS R2 = 0.020, F = 2.817, p = 0.036) and sex effect (ADONIS R2 = 0.436, F = 60.175, p = 0.001) followed by pairwise comparisons: E4-M-control vs. E4-M-inulin p = 0.001, E4-F-control vs. E4-F-inulin p = 0.001, E3-M-control vs. E3-M-inulin p = 0.001, and E3-F-control vs. E3-F-inulin p = 0.003.
Inulin increases the abundance of SCFAs-producing bacteria, primarily in acetate-producing bacteria in the APOE4 male mice
The β-diversity changes led us to analyze further the present taxa to understand the taxonomic changes induced by inulin. Table 2 shows that inulin significantly increased the abundance of bacteria that produce SCFAs, including acetate, propionate, and butyrate15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31. A log2 FC represents the ratio of the normalized mean abundance of the gut microbiota in the APOE3 and APOE4 inulin groups relative to their control groups for comparison. A positive log2 fold change (FC) indicates that the relative abundance of a bacteria species increases in the inulin group compared to the controls, while a negative log2 FC indicates the opposite. Inulin increased the abundance of SCFAs-producing bacteria in APOE4 mice compared to its control (Table 2). All the taxonomic changes with an FDR-corrected p-value (q) ≤ 0.05 were considered statistically significant in APOE4 mice; specifically, 16 different microbial taxa were more abundant. Although inulin similarly boosted the population of SCFAs-producing bacteria in APOE3 mice relative to its control group, the overall alteration in bacterial count (11) was less pronounced than in APOE4 mice (Table 2).
When stratified by sex, the one-sided volcano plot in Fig. 3a shows the log2 FC and − log (q value) in gene abundance in SCFAs-producing bacteria in all four groups receiving inulin compared to their controls. The radar chart in Fig. 3b illustrates the log2 FC in the 17 SCFAs-producing bacteria shown in Table 2. We found that E4-M-inulin vs. control mice showed more increases in SCFAs-producing bacteria compared to E4-F-inulin vs. control mice; 15 microbiota were increased in E4-M and 10 in E4-F. When we investigated specific gut microbiota, E4-M-inulin vs. control mice showed more increases in acetate-producing bacteria compared to E4-F-inulin vs. control mice; 12 microbiota were increased in E4-M, 7 in E4-F (Table 2, Fig. 3a,b). Inulin also modified the abundance of SCFAs-producing bacteria in E3-inulin vs. control group, but the extent of these changes was not as substantial as those observed in APOE4 mice. Specifically, there was an increase in the number of the abundance of SCFAs-producing bacteria by 10 in E3-M mice and 9 in E3-F mice. Furthermore, the number of the abundance of acetate-producing bacteria saw an uptick by 9 in E3-M mice and 8 in E3-F mice (Table 2, Fig. 3a,b). Overall, APOE4 mice and males had more changes than females, with E4-M showing the most significant changes.
Fold changes (FCs) in the gene abundance of the gut SCFAs producers in APOE-inulin mice stratified by sex compared to their controls. (a) A one-sided volcano plot shows the log2 FC and -log (Q value) in gene abundance of SCFAs producers in the gut in E4-inulin and E3-inulin mice stratified by sex compared to their controls. Plots with a greater fold change and/or a more significant Q value in E4-M mice were indicated. (b) A radar chart reveals the log2 FC with a Q value ≤ 0.05 in gene abundance of SCFAs producers in the gut in E4-inulin and APOE3-inulin mice stratified by sex compared to their controls.
Inulin reduces the abundance of lactic acid bacteria (LAB) more significantly in the APOE4 male mice while reducing E. coli more significantly in the APOE4 female mice
The differential analysis of the present taxa also revealed that inulin significantly reduced the abundance of LAB32,33,34,35,36,37,38,39,40,41,42,43 and the opportunistic pathogen E. coli (Table 3). All the taxa changes with a q ≤ 0.05 were considered statistical significance. Both APOE4 and APOE3 genotypes showed similar decreases in LAB and E. coli abundance. When further analyzed by sex, the radar chart in Fig. 4a illustrates the log2 FC, and the one-sided volcano plot in Fig. 4b depicts the log2 FC and q value in LAB among the four groups fed with inulin. We found that inulin led to a greater decrease in LAB in E4-M-inulin vs. control mice compared to E4-F-inulin vs. control mice, with a decrease of 16 species in E4-M and 11 in E4-F (Table 3, Fig. 4a,b). Inulin also decreased LAB in E3-inulin vs. control group, 7 in E3-M and 2 in E3-F (Table 3, Fig. 4a,b). Among those groups, APOE4 mice overall showed a more significant number of changes than APOE3 in the LAB decline regardless of sex.
Fold changes (FCs) in the gene abundance of gut lactic acid bacteria (LAB) and Escherichia coli (E. coli) in APOE-inulin mice stratified by sex compared to their controls. (a) A radar chart revealed the log2 FC with a Q value ≤ 0.05 in gene abundance of LAB in the gut in E4-inulin and E3-inulin mice stratified by sex compared to their controls. (b) A one-sided volcano plot showed the log2 FC and -log (Q value) in gene abundance of LAB in the gut in E4-inulin and E3-inulin mice stratified by sex compared to their controls. Plots with a greater fold change and/or a more significant Q value in E4-M and E4-F were indicated. (c) Inulin decreased E. coli in E4-M and E4-F mice compared to their controls. E3-F-inulin and E4-M-inulin showed less E. coli than E3-M-inulin, while E4-F-inulin mice exhibited less E. coli than E4-M-inulin mice. ***Q ≤ 0.001. (d) A bar graph showed the Log2 FC in the gut E. coli in E4-inulin and E3-inulin mice stratified by sex compared to their controls. Inulin declined the abundance of E. coli in E4-M-inulin and E4-F-inulin mice compared to their controls. ***Q ≤ 0.001.
In addition, the E. coli population was significantly reduced in E4-F-inulin (q = 2.00 × 10–6) and E4-M-inulin (q = 4.07 × 10–3) mice compared to their controls, with a greater fold change in E4-F mice (E4-F vs. E4-M: 12.62 folds vs. 4.58 folds, Table 3, Fig. 4c). The E. coli population did not change in E3-inulin mice compared to the controls when stratified by sex (Fig. 4c).
Inulin differentially impacts the abundance of gut microbiota in SCFAs-related and metabolism-related kinases and pathways in both the APOE4 and APOE3 mice
To gain further insight into how variations in the microbial taxa may affect metabolic pathways, we performed a 2-way ANOVA with experimental covariates (i.e., genotype, diet) followed by pairwise comparisons and a 3-way ANOVA with experimental covariates (i.e., genotype, diet, sex) followed by pairwise comparisons. We also performed a differential analysis of functional gene orthologs and higher level KEGG categories, i.e., pathways, modules, and BRITE levels, present. Results showed that inulin elevated the abundance of genes in propionate kinase in APOE4 (p < 0.001) mice compared to their controls with an overall diet effect [F (1, 69) = 31.63, p < 0.001] among groups (Fig. 5a). When stratified by sex, the relative abundance of propionate kinase genes was increased in E4-M (p < 0.001; Fig. 5b). Inulin also increased the abundance of acetate CoA-transferase [F (1, 69) = 19.84, p < 0.001], but only in the APOE4 group (p < 0.001, Fig. 5e) in male mice (p < 0.001; Fig. 5f). Investigation of the microbial taxa that contribute to these kinases revealed that the family Erysipelotrichaceae was a leading contributor. We found that inulin increased the abundance of family Erysipelotrichaceae, and that likely contributed to the change in acetate CoA transferase (Fig. 5g) and propionate kinase (Fig. 5h), especially in E4-M mice.
Changes in SCFAs-related kinases and fold changes (FCs) in functional gene abundance in APOE3 and APOE4 mice compared to their controls. Inulin increased the abundance of (a) propionate kinase and (c) butyrate kinase in E3-inulin mice and (a) propionate kinase and (e) acetate CoA-transferase in E4-inulin mice compared to their controls. When looking into the sex-dependent responses, inulin increased the abundance of (d) butyrate kinase in E3-M-inulin and (b) propionate kinase and (f) acetate CoA-transferase in E4-M-inulin mice compared to their controls. *p < 0.05. **p < 0.01. ***p < 0.001. Error bars show mean ± SD. Inulin increased the abundance of microbiota taxa that contributed to the abundance of (g) acetate CoA-transferase and (h) propionate kinase. (i) A one-sided volcano plot showed the log2 FC and -log (Q value) in gene abundance in metabolism- and inflammation-related kinases and pathways in E4-inulin and E3-inulin mice stratified by sex compared to their controls. Inulin changed the abundance of genes in the tricarboxylic acid (TCA) cycle in E4-M-inulin and E3-M-inulin mice compared to their controls. (j) A radar chart revealed the log2 FC with a Q value ≤ 0.05 in the gene abundance in metabolism- and inflammation-related kinases and pathways in E4-inulin and E3-inulin mice stratified by sex compared to their controls.
Table 4 shows the log2 FC in functional enzymes and pathways and their significance induced by inulin. We found enhancement of the abundance of the TCA cycle and suppression of glycolysis (glucose → pyruvate) and pyruvate oxidation (pyruvate → acetyl-CoA) in E4-inulin mice compared to its control. We observed that the abundance of the enzyme phosphate acetyltransferase (pta, K13788 in KEGG) and pta (K00625 in KEGG) increased in E4-inulin mice. We also observed a reduction in the abundance of the pyruvate dehydrogenase (PDH) E1 and E2 components in E4-inulin mice. Figure 5i,j display the log2 FC with q values and log2FC in functional enzymes and pathways found to be affected by inulin stratified by sex, as shown in Table 4. Inulin increased the abundance of orthologs in the TCA cycle in E4-M-inulin mice compared to its control.
Furthermore, inulin increased the abundance of propionate kinase in APOE3 mice (p = 0.034; Fig. 5a), yet this effect was absent when we stratified them by sex (Fig. 5b). Inulin also induced the butyrate kinase with an overall diet effect F (1, 69) = 21.40, p < 0.001. This impact, however, was specific to APOE3 mice (p < 0.001, Fig. 5c), particularly noticeable in E3-M mice (p < 0.001, Fig. 5d).
Inulin also stimulated an increase in the abundance of genes in the TCA cycle, pta (K00625 in KEGG), and PDH E1 and E2 components, while concurrently reducing the abundance of glycolysis and pyruvate oxidation in APOE3 mice (Table 4). When stratified by sex, inulin heightened the abundance of the TCA cycle in E3-M mice relative to its control group (Fig. 5i,j, Table 4).
Discussion
Our findings demonstrated that in E4-F and E4-M mice, the inulin diet elicited sex-dependent and distinct responses, with minimal impact on their food intake and body weight. E4-F control mice had increased α-diversity compared to the other groups, indicating gut microbiome dysbiosis. Previous studies have linked higher α-diversity to dysbiosis in conditions such as aging and stroke44,45. Interestingly, inulin was able to reduce and normalize the α-diversity of the E4-F mice to the level similar to other three groups. The finding aligns with previous studies demonstrating that inulin can reduce α-diversity in mice with APOE4 genotype and with traumatic brain injury (TBI)12,13,46.
Another major finding from the E4-F mice was the reduced in the abundance of E. coli. Elevated E. coli levels have also been linked to diseases such as stroke, TBI, and AD3,46,47. In this study, inulin decreased the abundance of E. coli, which might reduce the Lipopolysaccharide (LPS)-induced inflammatory pathways48,49,50. Therefore, our results imply that by reducing E. coli and potentially alleviating inflammation-related pathways, inulin may mitigate neurodegeneration by regulating inflammatory mediators, especially in E4-F-inulin mice. The decline in the abundance of E. coli may be due to the induced abundance of SCFAs-producing bacteria in inulin-fed APOE mice as the gut microbial ecosystem shifted51.
In contrast to the E4-F, E4-M mice exhibited greater abundance of SCFA-producing bacteria and genes in metabolic changes, particularly in the SCFAs- and TCA-related pathways. Importantly, these effects related to diet and sex were less prominent in APOE3 mice, indicating a significant role played by different APOE variants. Given that bioenergetic deficits are critical drivers of AD development52,53, these findings may provide insights into the higher susceptibility of APOE4 females to AD, potentially resulting from inefficient energy production. Our results showed that inulin increases the population of bacteria that produce SCFAs, mainly acetate, after fermentation in the gut, with more dramatic changes shown in E4-M mice. Acetate has been demonstrated to have various potentially advantageous effects on the brain. For instance, studies show that acetate can lower microglial activation and inflammatory markers in neuro-inflammation models, alleviate thalamic neurodegeneration, and increase cerebral blood flow in patients with Alcohol Use Disorders54,55,56,57,58,59,60.
In the E4-M mice, we also observed an increased abundance of SCFAs-associated enzymes, including acetate CoA-transferase and propionate kinase. Notably, inulin enhanced the abundance of the family Erysipelotrichaceae, which was inconspicuous in control groups. This included the species Faecalibaculum rodentium, whose increase likely played a crucial role in elevating the abundance of acetate CoA-transferase and propionate kinase, especially in E4-M mice. These observations suggest a correlation between the increased presence of the family Erysipelotrichaceae and the abundance of SCFAs-related kinases. Further, we noted an increase in the abundance of the Pta enzyme in E4-M-inulin mice compared to its control. This may imply a potential conversion of acetate to acetyl-CoA via the Pta-Ack pathway61, potentially enhancing mitochondrial function62.
Regardless of APOE genotype or sex, we saw a decrease in the abundance of the PDH enzyme, which may lead to a decline in pyruvate oxidation63. The reduced abundance in pyruvate oxidation and glycolysis may increase acetyl-CoA production, which may further increase the abundance in the TCA cycle64,65,66,67,68,69. This shift in energy sources within the gut in our results unveils that inulin may increase glucose availability for the brain and neurons by serving as a source of acetyl-CoA, thereby promoting proper neural function. The family Erysipelotrichaceae has been linked to inflammation following a high-fat or Western diet70. Interestingly, it has been found that Faecalibaculum rodentium, the species within this family, produces SCFAs and offers protective effects against cancer28. This aligns with our prior observation that inulin increases SCFAs-producing bacteria, potentially augmenting SCFAs production and improving mitochondrial function. These findings underscore the importance of studying specific species within the gut microbiota for their distinct roles.
Another key finding from the present study is the reduced LAB regardless of APOE genotype or sex. The increased SCFAs production due to inulin likely decreased the abundance of LAB, again, as the gut microbial ecosystem altered51. The decrease in LAB abundance may prevent excessive lactate production. The decreased LAB levels in APOE4 mice may point to a reduction in lactate synthesis in the gut, which lowers the risk of disease pathology, including inflammation and cancer71.
APOE4 is the strongest genetic risk factor for AD, and currently, there are no effective therapeutics to restore brain functionality after clinical symptoms have manifested. Accumulating evidence has shown that neurologically and systemically, metabolism may play a more critical role than amyloid beta plaques and tau tangles in AD progression52. In line with the findings in the present study, it has been shown that APOE3 and APOE4 carriers develop AD through different metabolic pathways72; therefore, it will be critical for future studies to identify precision nutrition approaches that tailored to different APOE variants to mitigate AD risk. In our prior work, we have demonstrated that inulin reduces neuroinflammatory gene expression and boosts SCFAs production in APOE4 mice, which mitigates their risk for AD development12,13. Our research suggest that inulin could be a preventative intervention. However, evidence is currently sparse regarding inulin's efficacy as a treatment after AD symptoms have manifested. This is a vital avenue for future exploration.
In summary, we investigated the effects of inulin on the potential gut microbial metabolism and revealed that treatment effectiveness was associated with sex in APOE4 carrier by analyzing the changes in gut microbiota and abundance of microbial metabolism in fecal samples. Our findings highlight the importance of considering sex in APOE4 carriers when exploring the link between diet, gut microbiome, and AD risk mitigation. It would be important to explore the development of gender-specific treatments, like acetate-producing bacteria supplementation for APOE4 females. It would also be beneficial to develop interventions tailored to different APOE variants. The findings from the study may provide insight for future precision nutrition applications to mitigate AD risk via gut-brain axis.
Methods
Animals and study design
We used a C57BL/6 mouse model with human-targeted replacement APOE (ε4 in homozygous APOE4 mice and ε3 in homozygous APOE3 mice) from Taconic (APOE4 model number: 1549-M and 1549-F and APOE3 model number: 1548-M and 1548-F). The mice were categorized into groups based on genotype and diet: E4-control (n = 18), E4-inulin (n = 19), E3-control (n = 17), and E3-inulin (n = 18). Additionally, groups were determined based on genotype, diet, and sex, resulting in the following categories: E4-M-control (n = 10), E4-F-control (n = 8), E4-M-inulin (n = 11), E4-F-inulin (n = 8), E3-M-control (n = 10), E3-F-control (n = 7), E3-M-inulin (n = 10), and E3-F-inulin (n = 8).
We fed mice a prebiotic inulin or control diet at four months and fed them for 16 weeks. Both diets were provided by TestDiet (control diet: 9GLK and inulin diet: 9GLL). The prebiotic inulin diet contained 8% fiber from inulin, and the control diet contained 8% fiber from cellulose, as we previously reported12,13,73. We fed the mice 8% of inulin because it has been shown that 8% of inulin increased cecal contents, produced more SCFA, and increased the amount of bacterial enzymes in the cecum compared to 4% of inulin74. Also, human studies showed that 8% of fiber (40 g of fiber per day) was considered the maximum amount for the western people to tolerate without side effects75. The detailed diet composition was provided in Table 5. We individually housed each mouse to avoid feces exchange with ad libitum access to food and water. We recorded the food intake and body weight biweekly. We collected the fecal samples when the mice reached eight months of age. The Institutional Animal Care and Use Committee (IACUC) at the University of Kentucky (UK) approved this study.
Fecal sample collection and gut microbiome analysis
Fecal samples were collected and frozen at − 80 °C until DNA extraction. Genomic DNA from feces was extracted using a ZymoBIOMICS DNA Mini Kit according to the manufacturer’s instructions. Shotgun metagenome libraries were prepared from fecal DNA using an Illumina DNA Prep kit according to the manufacturer’s instructions. The final library pool was sequenced on an Illumina NovaSeq6000 instrument using an S4 flow cell with paired-end 2 × 150 sequencing reads. Library preparation was performed at the Genomics and Microbiome Core Facility (GMCF) at Rush University, and sequencing was performed at the W.M. Keck Center for Comparative and Functional Genomics at the University of Illinois, Urbana-Champaign (UIUC). For taxonomic annotation, raw reads were mapped to the NCBI nucleotide database by Centrifuge for taxonomic annotation76. The least common ancestor algorithm was used for taxonomic annotations for each read. The annotations were then summarized across all reads to create counts per taxon. For functional gene annotations, raw reads were mapped to the Swissprot protein database using DIAMOND77,78. Gene orthologs annotations were then assigned using the consensus of aligned references and then summarized across all reads to create counts per ortholog for each sample. Higher-level summaries of orthologous functions are created using KEGG BRITE hierarchical annotations79. Raw counts were normalized to percentages for relative abundance.
Statistics and reproducibility
Sample size determination
A sample size was calculated for an anticipated 86% power to detect at least a 60% difference in comparing APOE4-control and APOE4-inulin, assuming a common standard deviation of 20 using a 2-way ANOVA considering all pairwise post-hoc comparisons (α = 0.05, β = 0.20).
Food intake and body weight
We monitored both food intake and body weight of the mice biweekly over a span of 16 weeks. To analyze the changes in food intake over this period and the body weight at its conclusion, we used a 2-way ANOVA with experimental covariates (i.e., genotype, diet) followed by pairwise comparisons and a 3-way ANOVA with experimental covariates (i.e., genotype, diet, sex) followed by pairwise comparisons.
Beta diversity/dissimilarity analyses
Before analysis, the normalized data were square root transformed. Using the vegan library, the Bray–Curtis index was calculated with default parameters in R (v3.6.2, https://www.r-project.org/)80. The resulting dissimilarity index was modeled and tested using the ADONIS test for significance with the sample covariates. Additional comparisons of the individual covariates were performed using ANSOIM. Plots were generated in R using the ggplot2 library81. A p-value less than 0.05 was considered statistically significant.
Alpha diversity analyses
Before analysis, the data were filtered as described above and rarefied to 1,000,000 counts per sample depth. The vegan library calculated the Shannon index with default parameters in R80. The resulting Shannon indices were then modeled with the sample covariates using a generalized linear model (GLM) assuming a Gaussian distribution. F-test was used to test the significance of the model (ANOVA). Post-hoc, pairwise tests were performed using the Mann–Whitney test. Plots were generated using the ggplot2 library in R81. A p-value less than 0.05 was considered statistically significant.
Differential relative abundance analysis of microbial taxonomy
Differential analyses of bacterial taxonomy compared with experimental covariates (i.e., genotype, diet, sex) were performed using the software package edgeR on raw sequence count82. The data were filtered to those taxa that were annotated as Bacteria but not annotated as chloroplast or mitochondria in origin, and removing taxa removed any taxon that had less than 1000 total counts across all samples and were present in less than 20% of the samples. Data were normalized as counts per million. Then normalized data were fit using a negative binomial GLM using experimental covariates (i.e., diet, sex, and genotype), and statistical tests were performed using a likelihood ratio test. A multi-factor comparison and repeated measures for the gut microbiome differential analysis were performed. Log2 fold-change (FC) and Q-value (false discovery rate-corrected P-value) for each taxon were reported. The Q value was calculated using the Benjamini–Hochberg false discovery rate (FDR) correction83. Significant taxa were determined based on an FDR threshold of 5% (0.05). The radar charts of log2 FC of SCFAs producers, LAB, and E. coli were created by Microsoft Office Excel (2022, https://www.microsoft.com/en-in/microsoft-365/excel). The one-sided volcano plots of log2 FC of SCFAs producers and LAB were created by GraphPad Prism 9 (https://www.graphpad.com/scientific-software/prism/).
Differential relative abundance analysis of microbial functional gene and pathway
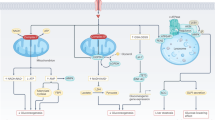
Before analyses, data were filtered to only include features with at least 100 counts across all samples and present in at least 30% of the samples for kinase level (KOs) analyses. For module and pathway analyses, data were filtered only to include a feature with at least 1000 counts across all samples and present in at least 30% of the samples. Differential analyses of functional genes compared with experimental covariates (i.e., genotype, diet, sex) were performed using GLM and various pairwise comparisons. Log2 FC and Q-value for each KO and pathway were reported. Significant taxa were determined based on an FDR threshold of 5% (0.05). For changes in SCFAs-related kinases gene abundance, we performed a 2-way ANOVA with experimental covariates (i.e., genotype, diet) followed by pairwise comparisons and a 3-way ANOVA with experimental covariates (i.e., genotype, diet, sex) followed by pairwise comparisons. A p-value less than 0.05 was considered statistically significant. To investigate the microbiota that contributed to the abundance of SCFAs-related kinases abundance, we determined the taxonomic annotations and total read counts for each KO, computed the fractional read counts for each taxon for a particular KO, and obtained normalized read counts for each taxon for each taxon for the particular KO by multiplying the computed fractional read counts by the previously normalized counts for the KO. The bar charts were generated in R. The radar chart of log2 FC of metabolism- and inflammation-related kinases and pathways was created by Microsoft Office Excel (2022, https://www.microsoft.com/en-in/microsoft-365/excel). The one-sided volcano plot of log2 FC of metabolism- and inflammation-related kinases and pathways was created by GraphPad Prism 9. The schematic diagram was created by Microsoft PowerPoint for Microsoft 365 MSO (v2210, https://www.microsoft.com/en-us/microsoft-365/powerpoint).
Data availability
The gene amplicon sequence data generated for this study have been submitted to the NCBI Bio-Project database (PRJNA880558). The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
References
Jack, C.R., et al. NIA-AA Research Framework: Toward a biological definition of Alzheimer's disease. (2018).
Jack, C. R. et al. A/T/N: An unbiased descriptive classification scheme for Alzheimer disease biomarkers. Neurology 87, 539–547 (2016).
Borsom, E.M., Lee, K. & Cope, E.K. Do the bugs in your gut eat your memories? Relationship between gut microbiota and Alzheimer's disease. Brain Sci. 10 (2020).
Vogt, N. M. et al. Gut microbiome alterations in Alzheimer’s disease. Sci. Rep. 7, 13537 (2017).
Fung, T. C., Olson, C. A. & Hsiao, E. Y. Interactions between the microbiota, immune and nervous systems in health and disease. Nat. Neurosci. 20, 145–155 (2017).
Hsu, M., Dedhia, M., Crusio, W. & Delprato, A. Sex differences in gene expression patterns associated with the APOE4 allele. (F1000Res, 2019).
Arnold, M. et al. Sex and APOE ε4 genotype modify the Alzheimer’s disease serum metabolome. Nat. Commun. 11, 1148 (2020).
Korf, J. M., Ganesh, B. P. & McCullough, L. D. Gut dysbiosis and age-related neurological diseases in females. Neurobiol. Dis. 168, 105695–105695 (2022).
Parikh, I.J., et al. Murine gut microbiome association with APOE alleles. Front. Immunol. 11 (2020).
Maldonado Weng, J. et al. Synergistic effects of APOE and sex on the gut microbiome of young EFAD transgenic mice. Mol. Neurodegener. 14, 47 (2019).
Zajac, D. J., Green, S. J., Johnson, L. A. & Estus, S. APOE genetics influence murine gut microbiome. Sci. Rep. 12, 1906 (2022).
Yanckello, L.M., et al. Apolipoprotein E genotype-dependent nutrigenetic effects to prebiotic inulin for modulating systemic metabolism and neuroprotection in mice via gut-brain axis. Nutr. Neurosci. 1–11 (2021).
Hoffman, J. D. et al. Dietary inulin alters the gut microbiome, enhances systemic metabolism and reduces neuroinflammation in an APOE4 mouse model. PLoS ONE 14, e0221828 (2019).
Walters, K. E. & Martiny, J. B. H. Alpha-, beta-, and gamma-diversity of bacteria varies across habitats. PLoS ONE 15, e0233872 (2020).
Parada Venegas, D. et al. Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front. Immunol. 10, 277–277 (2019).
Gronow, S. et al. Complete genome sequence of Bacteroides salanitronis type strain (BL78). Stand Genomic Sci. 4, 191–199 (2011).
Kralova, S., et al. gen. nov., sp. nov., Carrying two variants of. Microbiol Spectr 10, e0195421 (2022).
Lei, Y., et al. Parabacteroides produces acetate to alleviate heparanase-exacerbated acute pancreatitis through reducing neutrophil infiltration. Microbiome 9 (2021).
Nogal, A. et al. Circulating levels of the short-chain fatty acid acetate mediate the effect of the gut microbiome on visceral fat. Front. Microbiol. 12, 711359 (2021).
Louis, P. & Flint, H. J. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 19, 29–41 (2017).
Chia, L.W., et al. Bacteroides thetaiotaomicron fosters the growth of butyrate-producing Anaerostipes caccae in the presence of lactose and total human milk carbohydrates. Microorganisms 8 (2020).
Mirande, C. et al. Dietary fibre degradation and fermentation by two xylanolytic bacteria Bacteroides xylanisolvens XB1A T and Roseburia intestinalis XB6B4 from the human intestine. J. Appl. Microbiol. 109, 451–460 (2010).
Rios-Covian, D., et al. Bacteroides fragilis metabolises exopolysaccharides produced by bifidobacteria. BMC Microbiol. 16(2016).
Shimizu, J. et al. Propionate-producing bacteria in the intestine may associate with skewed responses of IL10-producing regulatory T cells in patients with relapsing polychondritis. PLoS ONE 13, e0203657 (2018).
Horvath, T.D., et al. Bacteroides ovatus colonization influences the abundance of intestinal short chain fatty acids and neurotransmitters. iScience 25, 104158 (2022).
Gurwara, S. et al. Dietary nutrients involved in one-carbon metabolism and colonic mucosa-associated gut microbiome in individuals with an endoscopically normal colon. Nutrients 11, 613 (2019).
Li, W. et al. Ecological and network analyses identify four microbial species with potential significance for the diagnosis/treatment of ulcerative colitis (UC). BMC Microbiol. 21, 1 (2021).
Zagato, E. et al. Endogenous murine microbiota member Faecalibaculum rodentium and its human homologue protect from intestinal tumour growth. Nat. Microbiol. 5, 511–524 (2020).
Dandachi, I. et al. Genome analysis of Lachnoclostridium phocaeense isolated from a patient after kidney transplantation in Marseille. New Microbes New Infect. 41, 100863 (2021).
Kanauchi, O., Taché, Y. & Larauche, M. Development of functional foods (enzyme-treated rice fiber) from rice by-products. 521–532 (2014).
Sundar, K. & Prabu, T.R. Chapter 12 - Microbial metabolites in nutrition and healthcare. in Volatiles and Metabolites of Microbes (eds. Kumar, A., Singh, J. & Samuel, J.) 235–256 (Academic Press, 2021).
Georgalaki, M. D. et al. Macedocin, a food-grade lantibiotic produced by Streptococcus macedonicus ACA-DC 198. Appl. Environ. Microbiol. 68, 5891–5903 (2002).
Song, A.A.-L., In, L. L. A., Lim, S. H. E. & Rahim, R. A. A review on Lactococcus lactis: From food to factory. Microb. Cell Fact. 16, 55–55 (2017).
Treu, L., et al. Whole-genome sequences of three Streptococcus macedonicus strains isolated from Italian cheeses in the Veneto region. Genome Announc. (Washington, DC) 5 (2017).
Alves-Barroco, C., et al. New insights on Streptococcus dysgalactiae subsp. dysgalactiae Isolates. Front. Microbiol. 12, 686413–686413 (2021).
Linares, D. M., O’Callaghan, T. F., O’Connor, P. M., Ross, R. P. & Stanton, C. Streptococcus thermophilus APC151 strain is suitable for the manufacture of naturally gaba-enriched bioactive yogurt. Front. Microbiol. 7, 1876–1876 (2016).
Giraffa, G. Lactobacillus helveticus: Importance in food and health. Front. Microbiol. 5, 338–338 (2014).
Rodríguez-Sojo, M. J., Ruiz-Malagón, A. J., Rodríguez-Cabezas, M. E., Gálvez, J. & Rodríguez-Nogales, A. Limosilactobacillus fermentum CECT5716: Mechanisms and therapeutic insights. Nutrients 13, 1–22 (2021).
Kaci, G. et al. Anti-inflammatory properties of Streptococcus salivarius, a commensal bacterium of the oral cavity and digestive tract. Appl. Environ. Microbiol. 80, 928–934 (2014).
Zheng, J. et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 70, 2782–2858 (2020).
Darby, T.M., et al. Lactococcus lactis Subsp. cremoris is an efficacious beneficial bacterium that limits tissue injury in the intestine. iScience 12, 356–367 (2019).
Drago, L. L. et al. Effect of Limosilactobacillus reuteri LRE02–Lacticaseibacillus rhamnosus LR04 combination on antibiotic-associated diarrhea in a pediatric population: A national survey. J. Clin. Med. 9, 3080 (2020).
Guerrero Sanchez, M., Passot, S., Campoy, S., Olivares, M. & Fonseca, F. Ligilactobacillus salivarius functionalities, applications, and manufacturing challenges. Appl. Microbiol. Biotechnol. 106, 57–80 (2022).
Hoffman, J. D. et al. Age drives distortion of brain metabolic, vascular and cognitive functions, and the gut microbiome. Front. Aging Neurosci. 9, 1 (2017).
Hammond, T. C. et al. Gut microbial dysbiosis correlates with stroke severity markers in aged rats. Front. Stroke 1, 1 (2022).
Yanckello, L. M. et al. Inulin supplementation prior to mild traumatic brain injury mitigates gut dysbiosis, and brain vascular and white matter deficits in mice. Front. Microbiomes 1, 1 (2022).
Hammond, T. C. et al. Functional recovery outcomes following acute stroke is associated with abundance of gut microbiota related to inflammation, butyrate and secondary bile acid. Front. Rehabil. Sci. 3, 1017180 (2022).
Zhang, W. & Liu, H. T. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 12, 9–18 (2002).
Sameer, A. S. & Nissar, S. Toll-like receptors (TLRs): Structure, functions, signaling, and role of their polymorphisms in colorectal cancer susceptibility. Biomed. Res. Int. 2021, 1157023 (2021).
Liu, T., Zhang, L., Joo, D. & Sun, S.-C. NF-κB signaling in inflammation. Signal Transduct. Target. Ther. 2, 17023 (2017).
Martín, R., Miquel, S., Ulmer, J., Langella, P. & Bermúdez-Humarán, L. G. Gut ecosystem: How microbes help us. Beneficial Microbes 5, 219–233 (2014).
Hammond, T. C. et al. beta-amyloid and tau drive early Alzheimer’s disease decline while glucose hypometabolism drives late decline. Commun. Biol. 3, 352 (2020).
Hammond, T. C. & Lin, A. L. Glucose metabolism is a better marker for predicting clinical Alzheimer’s disease than amyloid or tau. J. Cell Immunol. 4, 15–18 (2022).
Tanabe, J. et al. Effects of acetate on cerebral blood flow, systemic inflammation, and behavior in alcohol use disorder. Alcohol Clin. Exp. Res. 45, 922–933 (2021).
Brissette, C. A., Houdek, H. M., Floden, A. M. & Rosenberger, T. A. Acetate supplementation reduces microglia activation and brain interleukin-1β levels in a rat model of Lyme neuroborreliosis. J. Neuroinflam. 9, 249 (2012).
Reisenauer, C. J. et al. Acetate supplementation attenuates lipopolysaccharide-induced neuroinflammation. J. Neurochem. 117, 264–274 (2011).
Soliman, M. L., Smith, M. D., Houdek, H. M. & Rosenberger, T. A. Acetate supplementation modulates brain histone acetylation and decreases interleukin-1β expression in a rat model of neuroinflammation. J. Neuroinflam. 9, 51 (2012).
Qin, L. & Crews, F. T. Focal thalamic degeneration from ethanol and thiamine deficiency is associated with neuroimmune gene induction, microglial activation, and lack of monocarboxylic acid transporters. Alcohol Clin. Exp. Res. 38, 657–671 (2014).
Mews, P. et al. Alcohol metabolism contributes to brain histone acetylation. Nature 574, 717–721 (2019).
Sitkovsky, M. V. et al. Physiological control of immune response and inflammatory tissue damage by hypoxia-inducible factors and adenosine A2A receptors. Annu. Rev. Immunol. 22, 657–682 (2004).
Kim, J. N., Ahn, S. J. & Burne, R. A. Genetics and physiology of acetate metabolism by the Pta-Ack pathway of streptococcus mutans. Appl. Environ. Microbiol. 81, 5015–5025 (2015).
Magistretti, P. J. & Allaman, I. Lactate in the brain: From metabolic end-product to signalling molecule. Nat. Rev. Neurosci. 19, 235–249 (2018).
Bose, S., Ramesh, V. & Locasale, J. W. Acetate metabolism in physiology, cancer, and beyond. Trends Cell Biol. 29, 695–703 (2019).
Sgrignani, J., Chen, J., Alimonti, A. & Cavalli, A. How phosphorylation influences E1 subunit pyruvate dehydrogenase: A computational study. Sci. Rep. 8, 14683 (2018).
Mihaylova, M. M. & Shaw, R. J. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat. Cell Biol. 13, 1016–1023 (2011).
Han, Y. et al. Roles of KLF4 and AMPK in the inhibition of glycolysis by pulsatile shear stress in endothelial cells. Proc. Natl. Acad. Sci. 118, e2103982118 (2021).
Agarwal, S., Bell, C. M., Rothbart, S. B. & Moran, R. G. AMP-activated protein kinase (AMPK) control of mTORC1 Is p53- and TSC2-independent in pemetrexed-treated carcinoma cells. J. Biol. Chem. 290, 27473–27486 (2015).
Song, X. et al. Inulin can alleviate metabolism disorders in ob/ob mice by partially restoring leptin-related pathways mediated by gut microbiota. Genom. Proteom. Bioinf. 17, 64–75 (2019).
Li, Y. et al. Dietary fibers with different viscosity regulate lipid metabolism via ampk pathway: Roles of gut microbiota and short-chain fatty acid. Poult. Sci. 101, 101742 (2022).
Kaakoush, N.O. Insights into the role of Erysipelotrichaceae in the human host. Front. Cel. Infect. Microbiol. 5 (2015).
Li, X. et al. Lactate metabolism in human health and disease. Signal Transduct. Target. Ther. 7, 305 (2022).
Hammond, T. C. et al. Human gray and white matter metabolomics to differentiate APOE and stage dependent changes in Alzheimer’s disease. J. Cell Immunol. 3, 397–412 (2021).
Yanckello, L. M. et al. Inulin supplementation mitigates gut dysbiosis and brain impairment induced by mild traumatic brain injury during chronic phase. J. Cell Immunol. 4, 50–64 (2022).
Zduńczyk, Z., Juśkiewicz, J., Wróblewska, M. & Król, B. Physiological effects of lactulose and inulin in the caecum of rats. Arch. Anim. Nutr. 58, 89–98 (2004).
Coudray, C. et al. Effect of soluble or partly soluble dietary fibres supplementation on absorption and balance of calcium, magnesium, iron and zinc in healthy young men. Eur. J. Clin. Nutr. 51, 375–380 (1997).
Kim, D., Song, L., Breitwieser, F. P. & Salzberg, S. L. Centrifuge: Rapid and sensitive classification of metagenomic sequences. Genome Res. 26, 1721–1729 (2016).
Buchfink, B., Xie, C. & Huson, D. H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2015).
The UniProt Consortium. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 45, D158–D169 (2016).
Kanehisa, M., Furumichi, M., Tanabe, M., Sato, Y. & Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45, D353–D361 (2016).
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., Minchin, P. R., O’Hara, R. B., Simpson, G. L., Solymos, P., Stevens, M. H. H., Szoecs, E., & Wagner, H. vegan: Community Ecology Package. 2.4.0. (2018).
Wickham, H. Ggplot2: Elegant graphics for data analysis (Springer, 2009).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. Roy. Stat. Soc. 57, 289–300 (1995).
Acknowledgements
This research was supported by NIH grants RF1AG062480 (funded by NIA and ODS) to A-LL. Bioinformatics analysis in the project described was performed by the UIC Research Informatics Core, supported in part by NCATS through Grant UL1TR002003. The funders had no role in study design, data collection and analysis, decision to publish, or manuscript preparation. We thank the Division of Laboratory Animal Resources of UK for colony management.
Author information
Authors and Affiliations
Contributions
L.M.Y. and A-L.L. conceived the experiments. Y-H.C., L.M.Y., S.G., A.C., and A-L.L. conducted the experiments. Y-H.C., G.C., X.X., A.F., and A-L.L. analyzed the results. Y-H.C., G.C., A.R., A.C., and A-L.L. prepared the graphs and tables. Y-H.C., K.W., A.F., and A-L.L. searched for literature. Y-H.C., G.C., S.G., C.A., C.W., A.F., and A-L.L. wrote the paper. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chang, YH., Yanckello, L.M., Chlipala, G.E. et al. Prebiotic inulin enhances gut microbial metabolism and anti-inflammation in apolipoprotein E4 mice with sex-specific implications. Sci Rep 13, 15116 (2023). https://doi.org/10.1038/s41598-023-42381-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-42381-x
This article is cited by
-
Vivaria housing conditions expose sex differences in brain oxidation, microglial activation, and immune system states in aged hAPOE4 mice
Experimental Brain Research (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.