Abstract
Multidrug-resistant coagulase-negative staphylococci represent a real therapeutic challenge. The aim of the study was to emphasize the importance of heteroresistance to vancomycin presence in methicillin-resistant strains of S. epidermidis. The research comprised 65 strains of S. epidermidis. Heteroresistance to vancomycin was detected with the use of the agar screening method with Brain Heart Infusion and a population profile analysis (PAP test). In addition, types of cassettes and genes responsible for resistance to antibiotics for 22 multidrug resistant strains were determined. Our investigations showed that 56 of 65 S. epidermidis strains were phenotypically resistant to methicillin. The tested strains were mostly resistant to erythromycin, gentamicin, clindamycin, and ciprofloxacin. Six strains showed decreased susceptibility to vancomycin and their heterogeneous resistance profiles were confirmed with the PAP test. All tested multi-resistant strains exhibited the mecA gene. More than half of them possessed type IV cassettes. ant(4′)-Ia and aac(6′)/aph(2′′), ermC and tetK genes were most commonly found. The described phenomenon of heteroresistance to vancomycin in multidrug resistant bacteria of the Staphylococcus genus effectively inhibits a therapeutic effect of treatment with this antibiotic. That is why it is so important to search for markers that will enable to identify heteroresistance to vancomycin strains under laboratory conditions.
Similar content being viewed by others
Introduction
The role of coagulase-negative staphylococci (CoNS) as etiological agents of serious infection is increasing. They are one of the main causes of bacteremia in patients with indwelling medical devices, mainly induced by use of catheter and connected with vascular accesses1,2,3,4. It is often difficult to determine the clinical relevance of isolates obtained from patient samples because it requires differentiating between isolates being causative agents of infection and those that result from sample contamination. Recently, CoNS, mainly S. epidermidis, have been a real therapeutic challenge as the strains produce biofilm, which makes them even more resistant to many drugs5,6.
Occurrence of biofilm that adheres to surfaces and evades the host’s immune system is considered the primary cause of S. epidermidis virulence7,8. The ability to produce extracellular polysaccharide, called polysaccharide intercellular adhesin (PIA), is also regarded as one of most important factors9. It was reported that S. epidermidis also produces other proteins that may be crucial in biofilm formation, such as accumulation-associated protein (Aap) or Bhp protein, the latter being a homolog of biofilm-associated protein (Bap), found in Staphylococcus aureus10,11. Bacteria concentrated in biofilms are more resistant to antibiotics than their planktonic forms resulting in therapeutic failures.
Methicillin-resistant CoNS, characterized by clinical resistance to β-lactam antibiotics, are particularly dangerous12. This resistance is conditioned by the presence of the mecA gene encoding penicillin-binding protein 2a (PBP2a), which is located on mobile genetic elements, called the staphylococcal chromosome cassette mec (SCCmec)13. Among S. epidermidis strains, the SCCmec type IV is most frequently detected14,15. Isolates resistant to β-lactam antibiotics are often also resistant to other classes of antibiotics, most commonly to aminoglycosides, macrolides, and lincosamides16. This fact makes treatment of CoNS-induced infections increasingly difficult. Moreover, these bacteria, accumulating numerous antibiotic resistance genes, make up a reservoir of genes available for other bacterial species, including S. aureus13.
Glycopeptides, mainly vancomycin, are administered in treatment of methicillin-resistant CoNS-induced infections17. In recent years, resistance of gram-positive bacteria to vancomycin associated with transfer of van genes from enterococci has been reported18. Failure of vancomycin therapy may be also caused by heteroresistance of bacterial cells. The reason for this phenomenon are structural changes in cells, mainly their increased cell wall thickness. In addition, the bacteria produce peptidoglycan containing a large number of free D-Ala-D-Ala residues. Due to these changes fewer vancomycin molecules reach their target sites, which reduces the activity of the antibiotic19.
Considering the increasing resistance of coagulase-negative staphylococci to the majority of available antibiotics, and thus increasing difficulties in treatment of human infections induced by multi-resistant strains. It can be concluded that further studies are necessary to identify the mechanisms of transmission of antibiotic resistance genes in this group of bacteria. The aim of the study was to emphasize the importance of heteroresistance to vancomycin in methicillin-resistant S. epidermidis strains isolated hospitalized infected patients. An analysis of antibiotic susceptibility and selected resistance genotypes was also conducted.
Materials and methods
Tested strains
The research was conducted on 65 Staphylococcus epidermidis strains derived from patients of hospitals in Poland in 2018. The strains were isolated from blood, wounds and many different organs e.g. the eye, peritoneum, and urethra. The identification was performed with the use of PCR by amplification of species-specific DNA fragments20. Staphylococcus epidermidis ATCC 12228 was used as a control.
Detection of antibiotic resistance phenotype
Resistance to antibiotics was determined by the disc-diffusion method (using Becton Dickinson discs). The following antibiotics were applied: cefoxitin (FOX-30), clindamycin (CC-2), erythromycin (E-15), tetracycline (TE-30), gentamicin (GM-10), ciprofloxacin (CIP-5), tigecycline (TGC-15), linezolid (LZD-30), cotrimoxazole (SXT-1.25/23.75), rifampicin (RA-5), and fusidic acid (FA-10). Results were interpreted in accordance to EUCAST guidelines (The European Committee on Antimicrobial Susceptibility Testing)21. The authors applied the CLSI guidance (Clinical & Laboratory Standards Institute) for LZD-30. MLSB resistance mechanisms were detected by the D-test. Resistance to erythromycin and clindamycin was interpreted as a constitutive type of the MLSB mechanism (cMLSB), while resistance to erythromycin and existence of a flattening zone around the disc with clindamycin from the erythromycin side was interpreted as presence of inducible resistance (iMLSB). For methicillin-resistant Staphylococcus epidermidis (MRSE), the susceptibility to teicoplanin, vancomycin and daptomycin was determined with the broth microdillution method according to EUCAST guidelines. Dilutions of the antibiotics were applied at the following concentrations: 16–0.25 mg/L. The strains exhibited reduced susceptibility to vancomycin for the MIC value 1–2 mg/L. Staphylococcus aureus ATCC 29213 was used as a control strain.
Detection of vancomycin heteroresistance
In order to detect vancomycin heteroresistance in S. epidermidis strains, Brain Heart Infusion (BHI) screen agar was used22,23. The suspensions of 0.5 McFarland density were prepared with the use of night bacterial cultures cultivated on blood agar. Four drops of 10 μL from each suspension were put on a BHI agar plate (Oxoid) with 4 mg/L vancomycin (Sigma). The plates were incubated at 37 °C for 48 h. Mature colonies incubated in each drop were counted after the incubation. The strain was marked as heterogeneous vancomycin S. epidermidis (hVISE) if at least one drop contained minimum two colonies.
Population analysis profile (PAP) for vancomycin heteroresistance
Vancomycin heteroresistance in subpopulations was detected for the strains that grow on a BHI agar plate with 4 μg/mL vancomycin in accordance with the method described by Kim et al.24 For this purpose, serial tenfold dilutions of sterile saline bacterial suspensions of 0.5 McFarland density were prepared. Then, 100 μL was placed on BHI agar plates with vancomycin at concentrations of 2–16 mg/L. The plates were incubated at 35 °C for 48 h. Afterwards, the authors counted the colonies that grew in particular concentration of vancomycin. Heterogeneous vancomycin susceptible Staphylococcus aureus (hVSSA) strain ATCC 29213 was the negative control, while the heterogeneous vancomycin intermediate resistant Staphylococcus aureus (hVISA) strain Mu3 (ATCC 700698) was the positive control. The procedure was repeated three times.
DNA isolation for methicillin-resistant strains
Multi-resistant strains (resistant to at least four antibiotics) were selected for genetic studies. DNA was isolated with the use of the Genomic Micro AX Staphylococcus Gravity set (A&A Biotechnology) in accordance to the manufacturer’s protocol.
Detection of resistance genes by the PCR method
Genes conditioning the resistance to β-lactam antibiotics (mecA)25, aminoglycosides (aac(6′)/aph(2″), aph(3′)-IIIa, ant(4′)-Ia)26,27, macrolides and lincosamides (ermA, ermB, ermC, msrA, msrB, mphC, lnuA)28,29, as well as tetracyclines (tetK, tetL, tetM, tetO)30,31,32 were identified by PCR. The strains demonstrated decreased susceptibility to vancomycin (MIC = 1–2 mg/L) also vanA and vanB genes were detected33. Obtained PCR reaction products were split electrophoretically in 1% agarose gel with 1 µl Midori Green DNA Advance Stain (NIPPON Genetics Europe GmbH, Germany) supplement.
SCCmec cassette typing by PCR
For strains harboring the mecA gene, SCCmec cassette typing was performed according to the method described by Kondo et al. and Zhang et al. AB1, AB2, AB3 and C ccr gene complex types and A, B and C mec gene complex classes were identified34,35. The ccrAB4 gene was marked with the method described by Oliveira et al. with Zhang et al. with modification35,36. Cassettes with combination of ccr and/or mec genes undescribed before, in accordance to the rules set by International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements, were considered potentially new types of cassettes37.
Statistical analysis
A relationship between results obtained for the cassette type and phenotypic antibiotic resistance and gene presence was determined with the use of the chi-squared test. p < 0.05 was adopted as significant. STATISTICA 13.1PL software was used (StatSoft 2016, Poland) for the purpose of statistical analysis.
Ethics approval and consent to participate
Not applicable. In the study, informed consent was not required as the isolates included in the study were obtained as a result of standard medical care. Patients’ identity as well as all their personal information were confidential.
Results
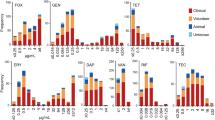
Our studies showed that 56 of 65 S. epidermidis strains were phenotypically resistant to methicillin, what suggests their resistance to all β-lactams. They were mostly resistant to erythromycin, gentamicin, clindamycin and ciprofloxacin. Single strains were resistant to fusidic acid, rifampicin and tigecycline. All of them were also methicillin-resistant. Resistance to linezolid was not observed. 22 of methicillin-resistant strains were multi-resistant and exhibited resistance to at least four of the antibiotics used in the studies. The resistance to antibiotics of all tested strains is presented in Table 1.
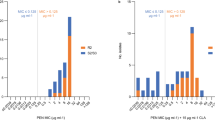
With regards to MRSE, MIC values for teicoplanin, vancomycin and daptomycin were determined with the microdillution broth method. Results are presented in Fig. 1.
Only one S. epidermidis strain was resistant to teicoplanin (MIC = 6 mg/L). According to EUCAST guidelines, all strains were sensitive to vancomycin and daptomycin. Vancomycin MIC for the majority of strains was below 1 mg/L. However the growth of three strains was inhibited with 2 mg/L vancomycin and three with 1 mg/L of this antibiotic. These strains were characterized by decreased susceptibility to vancomycin, and grew on a BHI agar plate with 4 mg/L of vancomycin. Their heterogeneous resistance profiles were confirmed in the PAP test. Subpopulations of four strains grew on BHI agar with 6 mg/L, while two grew with the application of 8 mg/L vancomycin. PAP test results for the tested strains of S. epidermidis and control strains (S. aureus ATCC 29,213, susceptible to vancomycin and Mu3, hVISA) are shown in Fig. 2.
Strains with reduced susceptibility to vancomycin were characterized by absence of van genes. Cassette types were determined and genes responsible for resistance to β-lactams, aminoglycosides, tetracyclines and the MLSB mechanism were identified in 22 multidrug-resistant strains. All tested strains, phenotypically resistant to methicillin, possessed the mecA gene. Type IV cassettes were detected in more than half of them, whereas type V cassettes were found in four strains. Presence of cassette types are shown in Table 2.
The phenotypic profile of antibiotic resistance and resistance genes, including the type of SCCmec cassette in 22 multi-resistant strains, is shown in Fig. 3.
Antibiotic resistance phenotypic profile and resistance genes in 22 multi-resistant strains of S. epidermidis. FOX-30—cefoxitin, E-15—erythromycin, CC-2—clindamycin, GM-10—gentamicin, TE-30—tetracycline, CIP-5—ciprofloxacin, TGC-15—tigecycline, LZD-30—linezolid, SXT-1.25/23.75—cotrimoxazole, RA-5—rifampicin, FA-10—fusidic acid; I—inductive mechanism MLSB (iMLSB), C—constitutive mechanism MLSB (cMLSB).
The following genes encoding resistance to aminoglycosides were the most common: ant(4')-Ia and aac(6')/aph(2''), observed respectively in 73% and 64% of the tested strains. The presence of the aac(6')/aph(2") gene was statistically significant for strains with type IV cassette (p = 0.03). Five strains with at least one of the genes, did not demonstrate phenotypic resistance to gentamicin.
The ermC gene was present in 16 strains, all of which were characterized by a constitutive or inductive MLSB mechanism. One cMLSB strain possessed only the ermA gene. This gene was present in three strains, but together with ermC. The mphC, msrA and msrB genes were present in three strains, and were always observed together. Two strains having these genes were erythromycin resistant, and in one strain, they occurred together with the ermC gene.
All strains with tet genes were resistant to tetracycline. The tetK gene was detected in 12 out of 13 resistant strains. Five strains were rifampicin resistant. All had type IV cassette, which was statistically significant (p = 0.02). There was also a statistically significant relationship between the presence of this cassette and clindamycin resistance (p = 0.02) and the presence of this cassette and the ermA gene (p = 0.04). Two strains were resistant to nine tested antibiotics, which is an alarming number from the therapeutic point of view. Both the strains had type IV cassette and at least five detectable genes. Besides, they demonstrated reduced sensitivity to vancomycin. Table 3 shows antibiotic resistance profiles and occurrence of detected genes in six strains characterized by vancomycin heteroresistance. The table also includes the origin of the isolate and the clinical department where the infected patient was hospitalized.
Discussion
Presently, more and more attention is being paid to the problem of increasing resistance in bacteria. Not only does it concern pathogenic bacteria, but also opportunistic pathogens that cause infections mainly in immunocompromised people. Such bacteria include S. epidermidis, which causes serious problems to hospitalized people who undergo invasive procedures, as well as newborns.
In our work, we studied 65 strains of S. epidermidis which induced infection in hospitalized people. 86% of these tested strains were methicillin-resistant, of which 39% were resistant to at least four antibiotics. This problem is also pointed out in works of other researchers in which the percentage of methicillin-resistant strains was comparable to that we observed or even higher38,39,40. The strains tested in our investigations were mostly resistant to erythromycin, cefoxitin, gentamicin, and clindamycin and the values were the following: 60%, 56%, 48%, 43% respectively. Macrolide resistance was the most common. A similar result was reported by Skiba-Kurek et al.38. Of the 100 of S. epidermidis strains they tested, 67% were resistant to erythromycin. The percentage of resistant strains observed in studies of Mirzaei et al. was even higher—as much as 80% of the tested strains appeared to be resistant to erythromycin39. Increased resistance to macrolides is a consequence of their widespread application in treatment of infections caused by gram-positive bacteria41. In our studies, almost half of the strains were resistant to gentamicin. In the studies presented by Wojtyczka et al., the resistance of tested S. epidermidis strains that were isolated from nosocomial infections, was below 4%. Nevertheless, in other studies this resistance was much higher42,43,44. Therefore, it is particularly important to emphasize that, according to EUCAST recommendations, gentamicin should be administered only in combination therapy21. Sensitivity to glycopeptides and daptomycin was determined for MRSE strains. Vancomycin is the first line antibiotic used to treat severe infections caused by methicillin-resistant staphylococci17. Daptomycin can be used in therapies in which vancomycin appears to be ineffective45. All the strains that we tested were susceptible to vancomycin and daptomycin according to EUCAST recommendations. However, six strains demonstrated reduced susceptibility to vancomycin. Moreover, these strains exhibited heterogeneous growth in the PAP test and could grow on a medium containing 6–8 mg/L vancomycin. Mashaly et al. or Nunes et al., who studied this mechanism in CoNS, showed that prior treatment with vancomycin may result in occurrence of a hetero-resistant phenotype19,23.
Its mechanism has not been fully recognised so far. It does not require the presence of the van genes responsible for the resistance to vancomycin46. It was confirmed by our studies as we didn’t detect such genes in the decreased susceptibility to vancomycin strains tested by us. For such strains, vancomycin therapy may turn out to be ineffective, although, according to recommendations, they are supposed to be susceptible to this antibiotic. As a result of increasing number of the infections caused by CoNS, decreased susceptibility of these bacteria to vancomycin may account for serious problem in hospital treatment that decreases therapeutic possibilities. This phenomenon was highlighted in many studies concerning severe infections caused by different species of CoNS23,47,48.
All tested strains, that were phenotypically resistant to methicillin, possessed the mecA gene. Type IV cassettes were detected in more than half of the strains. In the case of CoNS, the diversity of cassettes is very large and may result from sources of strain isolation49,50. The presence of type I and III cassettes is attributed to isolates of hospital origin, while the presence of type IV and V cassettes to isolates coming from CA-MRSE (community-associated methicillin-resistant S. epidermidis). Additionally, existence of these particular types of cassettes may be influenced by the species from which they are isolated. Type IV cassettes are often found in S. epidermidis. Moreover, this cassette has been isolated for the first time from these species51. The analysis of the cassettes existence in S. epidermidis over the recent years indicates that it was the type IV cassettes that were the most common isolated in such species14,15,48,52. Also types I, II, III or V cassettes were detected in S. epidermidis15,53,54. Studies of Havaei et al., based on S. epidermidis from hospitalized patients revealed the presence of type III cassettes in 36% of the tested strains15. Also Martins et al. most often identified type III cassettes (50%) in their studies on S. epidermidis. Moreover, 32.8% of the strains had type I cassettes55. In our studies, type III cassettes was detected in only two strains, while type I cassettes was not found in any of the tested strains. Type II and III cassettes often contain aminoglycoside, macrolide or tetracycline resistance genes in their structure, which was confirmed in studies of Szczuka et al. Strains of S. epidermidis that they tested possessed type III cassettes and were characterised by much greater resistance to non-β-lactam antibiotics than those with type IV cassettes56. In our studies, a correlation between type IV cassettes and resistance to clindamycin and rifampicin was observed. Moreover, there was also a correlation between the presence of such cassettes and existence of aac(6')/aph(2'') and ermA genes. In the tested methicillin-resistant S. epidermidis, described by Teodoro et al., the ermC gene was located mainly on SCCmec IV or non-typeable cassettes57.
Macrolides, lincosamides and streptogramins B have a similar mechanism of action leading to cross-resistance (MLSB), which is more and more frequently described in different groups of bacteria. The erm family genes is responsible for this mechanism. They encode one methylase which is responsible for the methylation of the adenine 23S rRNA ribosomal subunit. The ermC gene, being the dominant gene responsible for this mechanism in CoNS, was detected in 16 strains in our investigations. This gene was present in both constitutive and inductive MLSB mechanism strains. cMLSB determines resistance to all MLSB group of antibiotics, while iMLSB stands for clinical resistance to lincosamides and streptogramins B induced by 14- and 15-membered macrolides. The msrA and msrB genes encoding active efflux are responsible for resistance to macrolides and streptogramins B (MSB phenotype) in the case of the susceptibility to lincosamides. They were present in three strains and always together with the mphC gene encoding enzymes inactivating a specific substance28,58. Juda et al.59 in their studies described strains characterized by resistance to macrolides, lincosamides and streptogramins B possessed a more diverse genotype.
In the strains tested by us we were trying to identify aac(6′)/aph(2″), aph(3′)-IIIa, ant(4′)-Ia genes which are responsible for the most common aminoglycoside-modifying enzymes among the staphylococci genus. Resistance to aminoglycosides was encoded mainly by ant(4')-Ia and aac(6')/aph(2) genes. The aac(6')-Ie-aph(2'')-Ia gene dominated also in clinical isolates of staphylococci in studies conducted by Schmitz et al.26. Because of increased resistance to tetracycline we also attempted to find genes encoding the resistance to this group of antibiotics. 12 strains that we tested were characterized with presence of the tetK gene. This gene was also detected in nearly 60% of S. epidermidis strains tested by Chabi and Momtaz60. The authors also detected the tetM gene in more than 39% of strains. However, we did not such strain was detected in our studies. Two strains isolated from the blood of hospitalized patients and analyzed by us reveal how dangerous for therapeutic reasons, can be S. epidermidis-induced infections. They were resistant to as many as nine antibiotics and possessed at least five tested genes. Treatment of these strains with vancomycin appeared to be ineffective as they both of them demonstrated reduced susceptibility to this antibiotic.
The described phenomenon of heteroresistance to vancomycin in multidrug resistant bacteria of the Staphylococcus genus effectively inhibits a therapeutic effect of treatment with this antibiotic. It is difficult to determine this mechanism in a routine laboratory microbiology. Strains characterized by vancomycin heteroresistance are considered susceptible according to the method recommended by EUCAST. That is why it is so important to search for markers that will enable to identify vancomycin strains under laboratory conditions. To achieve this goal further studies are required.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Uckay, I. et al. Foreign body infections due to Staphylococcus epidermidis. Ann. Med. 41, 109–119 (2009).
Schoenfelder, S. M. K. et al. Success through diversity - how Staphylococcus epidermidis establishes as a nosocomial pathogen. Int. J. Med. Microbiol. 300, 380–386 (2010).
Wang, L. et al. Risk factors of nosocomial infection for infants in neonatal intensive care units: A systematic review and meta-analysis. Med. Sci. Monit. 25, 8213–8220 (2019).
Dong, Y., Speer, C. P. & Glaser, K. Beyond sepsis: Staphylococcus epidermidis is an underestimated but significant contributor to neonatal morbidity. Virulence 9, 621–633 (2018).
Sahal, G. & Bilkay, I. S. Multi drug resistance in strong biofilm forming clinical isolates of Staphylococcus epidermidis. Braz. J. Microbiol. 45, 539–544 (2014).
Pramodhini, S., Niveditha, S., Umadevi, S., Kumar, S. & Stephen, S. Antiobiotic resistance pattern of biofilm-forming uropathogens isolated from catheterised patients in Pondicherry, India. Aust. Med. J. 5, 344–348 (2012).
Otto, M. Staphylococcal infections: Mechanisms of biofilm maturation and detachment as critical determinants of pathogenicity. Annu. Rev. Med. 64, 175–188 (2013).
Wu, H., Moser, C., Wang, H. Z., Hoiby, N. & Song, Z. J. Strategies for combating bacterial biofilm infections. Int. J. Oral Sci. 7, 1–7 (2015).
Heilmann, C. et al. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20, 1083–1091 (1996).
Rohde, H. et al. Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol. Microbiol. 55, 1883–1895 (2005).
Bowden, M. G. et al. Identification and preliminary characterization of cell-wall-anchored proteins of Staphylococcus epidermidis. Microbiology-Sgm 151, 1453–1464 (2005).
Shore, A. C. & Coleman, D. C. Staphylococcal cassette chromosome mec: Recent advances and new insights. Int. J. Med. Microbiol. 303, 350–359 (2013).
Chon, J. W. et al. Virulence characteristics of mecA-positive multidrug-resistant clinical coagulase-negative staphylococci. Microorganisms 8, 19 (2020).
Martinez-Melendeza, A. et al. Molecular epidemiology of coagulase-negative bloodstream isolates: Detection of Staphylococcus epidermidis ST2, ST7 and linezolid-resistant ST23. Braz. J. Infect. Dis. 20, 419–428 (2016).
Havaei, S. A., Namvar, A. E., Moghim, S. & Lari, A. R. Evaluation of various staphylococcal cassette chromosome mec (SCCmec) types in Staphylococcus epidermidis invasive strains from hospitalised patients in Iran. Le Infezioni in Medicina (InfezMed) 23(1), 18–22 (2015).
Ma, X. X., Wang, E. H., Liu, Y. & Luo, E. J. Antibiotic susceptibility of coagulase-negative staphylococci (CoNS): Emergence of teicoplanin-non-susceptible CoNS strains with inducible resistance to vancomycin. J. Med. Microbiol. 60, 1661–1668 (2011).
Hope, R. et al. Non-susceptibility trends among staphylococci from bacteraemias in the UK and Ireland, 2001–2006. J. Antimicrob. Chemother. 62, II65–II74 (2008).
Navidinia, M. et al. Molecular characterization of resistance genes in MDR-ESKAPE pathogens. J. Pure Appl. Microbiol. 11, 779–792 (2017).
Nunes, A. P. F. et al. Heterogeneous resistance to vancomycin in Staphylococcus epidermidis, Staphylococcus haemolyticus and Staphylococcus warneri clinical strains: Characterisation of glycopeptide susceptibility profiles and cell wall thickening. Int. J. Antimicrob. Agents 27, 307–315 (2006).
Hirotaki, S., Sasaki, T., Kuwahara-Arai, K. & Hiramatsu, K. Rapid and accurate identification of human-associated staphylococci by use of multiplex PCR. J. Clin. Microbiol. 49, 3627–3631 (2011).
EUCAST, Breakpoint tables for interpretation of MICs and zone diameters, ver. 13.0. (https://www.eucast.org/clinical_breakpoints/: The European Committee on Antimicrobial Susceptibility Testing) (2023).
Satola, S. W., Farley, M. M., Anderson, K. F. & Patel, J. B. Comparison of detection methods for heteroresistant vancomycin-intermediate staphylococcus aureus, with the population analysis profile method as the reference method. J. Clin. Microbiol. 49, 177–183 (2011).
Mashaly, G. E. & El-Mahdy, R. H. Vancomycin heteroresistance in coagulase negative Staphylococcus blood stream infections from patients of intensive care units in Mansoura University Hospitals, Egypt. Ann. Clin. Microbiol. Antimicrob. 16(1), 1–5 (2017).
Kim, M. N., Pai, C. H., Woo, J. H., Ryu, J. S. & Hiramatsu, K. Vancomycin-intermediate Staphylococcus aureus in Korea. J. Clin. Microbiol. 38, 3879–3881 (2000).
Murakami, K. et al. Identification of methicillin-resistant strains of staphylococci by polymerase chain-reaction. J. Clin. Microbiol. 29, 2240–2244 (1991).
Schmitz, F. J. et al. The prevalence of aminoglycoside resistance and corresponding resistance genes in clinical isolates of staphylococci from 19 European hospitals. J. Antimicrob. Chemother. 43, 253–259 (1999).
Vakulenko, S. B. et al. Multiplex PCR for detection of aminoglycoside resistance genes in enterococci. Antimicrob. Agents Chemother. 47, 1423–1426 (2003).
Lina, G. et al. Distribution of genes encoding resistance to macrolides, lincosamides, and streptogramins among staphylococci. Antimicrob. Agents Chemother. 43, 1062–1066 (1999).
Sutcliffe, J., Grebe, T., TaitKamradt, A. & Wondrack, L. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 40, 2562–2566 (1996).
Argudin, M. A., Vanderhaeghen, W. & Butaye, P. Diversity of antimicrobial resistance and virulence genes in methicillin-resistant non-Staphylococcus aureus staphylococci from veal calves. Res. Vet. Sci. 99, 10–16 (2015).
Strommenger, B., Kettlitz, C., Werner, G. & Witte, W. Multiplex PCR assay for simultaneous detection of nine clinically relevant antibiotic resistance genes in Staphylococcus aureus. J. Clin. Microbiol. 41, 4089–4094 (2003).
Trzcinski, K., Cooper, B. S., Hryniewicz, W. & Dowson, C. G. Expression of resistance to tetracyclines in strains of methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 45, 763–770 (2000).
Saadat, S., Solhjoo, K., Norooz-Nejad, M.-J. & Kazemi, A. VanA and VanB positive vancomycin-resistant staphylococcus aureus among clinical isolates in, Shiraz South of Iran. Oman Med. J. 29, 335–339 (2014).
Kondo, Y. et al. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: Rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 51, 264–274 (2007).
Zhang, L. F. et al. Multilocus sequence typing and further genetic characterization of the enigmatic pathogen, Staphylococcus hominis. PLoS ONE 8, 9 (2013).
Oliveira, D. C., Milheirico, C. & de Lencastre, H. Redefining a structural variant of staphylococcal cassette chromosome mec, SCCmec type VI. Antimicrob. Agents Chemother. 50, 3457–3459 (2006).
Ito, T. et al. Classification of staphylococcal cassette chromosome mec (SCCmec): Guidelines for reporting novel SCCmec elements. Antimicrob. Agents Chemother. 53, 4961–4967 (2009).
Skiba-Kurek, I. Evaluation of biofilm formation and prevalence of multidrug-resistant strains of staphylococcus epidermidis isolated from neonates with sepsis in southern Poland. Pathogens 10, 16 (2021).
Mirzaei, R.A.-O., Yousefimashouf, R., Arabestani, M. R., Sedighi, I. & Alikhani, M. Y. The issue beyond resistance: Methicillin-resistant Staphylococcus epidermidis biofilm formation is induced by subinhibitory concentrations of cloxacillin, cefazolin, and clindamycin. PLoS ONE 17(11), e0277287 (2022).
Najar-Peerayeh, S., Moghadas, A. J. & Behmanesh, M. Antibiotic susceptibility and mecA frequency in staphylococcus epidermidis, isolated from intensive care unit patients. Jundishapur J. Microbiol. 7, 4 (2014).
Castro-Alarcon, N. et al. Molecular typing and characterization of macrolide, lincosamide and streptogramin resistance in Staphylococcus epidermidis strains isolated in a Mexican hospital. J. Med. Microbiol. 60, 730–736 (2011).
Wojtyczka, R. D. et al. Biofilm formation and antimicrobial susceptibility of staphylococcus epidermidis strains from a hospital environment. Int. J. Environ. Res. Public Health 11, 4619–4633 (2014).
van den Hoogen, A., Gerards, L. J., Verboon-Maciolek, M. A., Fleer, A. & Krediet, T. G. Long-term trends in the epidemiology of neonatal sepsis and antibiotic susceptibility of causative agents. Neonatology 97, 22–28 (2010).
Brzychczy-Wloch, M. Prevalence of antibiotic resistance in multi-drug resistant coagulase-negative staphylococci isolated from invasive infection in very low birth weight neonates in two Polish NICUs. Ann. Clin. Microbiol. Antimicrob. 12, 7 (2013).
Sader, H. S. & Jones, R. N. Antimicrobial activity of daptomycin in comparison to glycopeptides and other antimicrobials when tested against numerous species of coagulase-negative Staphylococcus. Diagn. Microbiol. Infect. Dis. 73, 212–214 (2012).
Srinivasan, A., Dick, J. D. & Perl, T. M. Vancomycin resistance in staphylococci. Clin. Microbiol. Rev. 15, 430–438 (2002).
Szemraj, M., Lisiecki, P., Glajzner, P. & Szewczyk, E. M. Vancomycin heteroresistance among methicillin-resistant clinical isolates S. haemolyticus, S. hominis, S. simulans, and S. warneri. Braz. J. Microbiol. 54, 159–167 (2023).
Peixoto, P. B. et al. Methicillin-resistant Staphylococcus epidermidis isolates with reduced vancomycin susceptibility from bloodstream infections in a neonatal intensive care unit. J. Med. Microbiol. 69, 41–45 (2020).
Chmielarczyk, A. et al. Molecular analysis of meticillin-resistant Staphylococcus aureus strains isolated from different types of infections from patients hospitalized in 12 regional, non-teaching hospitals in southern Poland. J. Hosp. Infect. 95, 259–267 (2017).
Xu, Z., Mkrtchyan, H. V. & Cutler, R. R. Antibiotic resistance and mecA characterization of coagulase-negative staphylococci isolated from three hotels in London, UK. Front. Microbiol. 6, 6 (2015).
Wisplinghoff, H. et al. Related clones containing SCCmec type IV predominate among clinically significant Staphylococcus epidermidis isolates. Antimicrob. Agents Chemother. 47, 3574–3579 (2003).
Ghaznavi-Rad, E., Fard-Mousavi, N., Shahsavari, A., Japoni-Nejad, A. & Van Belkum, A. Distribution of staphylococcal cassette chromosome mec types among methicillin-resistant coagulase negative staphylococci in central Iran. Iran. J. Microbiol. 10, 7–13 (2018).
Abadi, M. I. M. et al. Molecular characteristics of nasal carriage methicillin-resistant coagulase negative staphylococci in school students. Jundishapur J. Microbiol. 8, 7 (2015).
Noshak, M. A. et al. Molecular Detection and Characterization of the Staphylococcus epidermidis and Staphylococcus haemolyticus Isolated from Hospitalized Patients and Healthcare Workers in Iran. Biomed. Res. Int. 2023, 10 (2023).
Martins, A. et al. Antimicrobial resistance and persistence of Staphylococcus epidermidis clones in a Brazilian university hospital. Diagn. Microbiol. Infect. Dis. 77, 164–168 (2013).
Szczuka, E., Bosacka, K. & Kaznowski, A. Characterization of staphylococcal cassette chromosome mec (SCCmec) in methicillin-resistant staphylococcus epidermidis strains isolated from biomaterial-associated infections and their antibiotic resistance patterns. Pol. J. Microbiol. 65, 215–217 (2016).
Teodoro, C. R. S., Mattos, C. S., Cavalcante, F. S., Pereira, E. M. & dos Santos, K. R. N. Characterization of MLSb resistance among Staphylococcus aureus and Staphylococcus epidermidis isolates carrying different SCCmec types. Microbiol. Immunol. 56, 647–650 (2012).
Leclercq, R. Mechanisms of resistance to macrolides and lincosamides: Nature of the resistance elements and their clinical implications. Clin. Infect. Dis. 34, 482–492 (2002).
Juda, M., Chudzik-Rzad, B. & Malm, A. The prevalence of genotypes that determine resistance to macrolides, lincosamides, and streptogramins B compared with spiramycin susceptibility among erythromycin-resistant Staphylococcus epidermidis. Mem. Inst. Oswaldo Cruz 111, 155–160 (2016).
Chabi, R. & Momtaz, H. Virulence factors and antibiotic resistance properties of the Staphylococcus epidermidis strains isolated from hospital infections in Ahvaz. Iran. Trop. Med. Health 47, 9 (2019).
Acknowledgements
The authors would also like to thank Dorota Wawrzyniak, MA from the Foreign Language Centre of the Medical University of Lodz, Poland for a language consultation.
Funding
This study was supported by the statutory research funds (502–03/3–012-03/ 503–31-011) of the Medical University of Lodz.
Author information
Authors and Affiliations
Contributions
M.Sz., P.G., M.S. designed the experiments. M.Sz., P.G. performed the experiments. M.Sz., M.S. analyzed the data. M.Sz. and M.S. wrote the manuscript. All the authors read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Szemraj, M., Glajzner, P. & Sienkiewicz, M. Decreased susceptibility to vancomycin and other mechanisms of resistance to antibiotics in Staphylococcus epidermidis as a therapeutic problem in hospital treatment. Sci Rep 13, 13629 (2023). https://doi.org/10.1038/s41598-023-40866-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-40866-3
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.