Abstract
The positron emission tomography (PET) with 18F-flortaucipir can distinguish individuals with mild cognitive impairment (MCI) and Alzheimer’s disease (AD) from cognitively unimpaired (CU) individuals. This study aimed to evaluate the utility of 18F-flortaucipir-PET images and multimodal data integration in the differentiation of CU from MCI or AD through DL. We used cross-sectional data (18F-flortaucipir-PET images, demographic and neuropsychological score) from the ADNI. All data for subjects (138 CU, 75 MCI, 63 AD) were acquired at baseline. The 2D convolutional neural network (CNN)-long short-term memory (LSTM) and 3D CNN were conducted. Multimodal learning was conducted by adding the clinical data with imaging data. Transfer learning was performed for classification between CU and MCI. The AUC for AD classification from CU was 0.964 and 0.947 in 2D CNN-LSTM and multimodal learning. The AUC of 3D CNN showed 0.947, and 0.976 in multimodal learning. The AUC for MCI classification from CU had 0.840 and 0.923 in 2D CNN-LSTM and multimodal learning. The AUC of 3D CNN showed 0.845, and 0.850 in multimodal learning. The 18F-flortaucipir PET is effective for the classification of AD stage. Furthermore, the effect of combination images with clinical data increased the performance of AD classification.
Similar content being viewed by others
Introduction
Alzheimer’s dementia (AD) is the most common type of dementia among older adults1,2. In general, the progression of AD can be divided into three stages: cognitively unimpaired (CU), mild cognitive impairment (MCI), and AD. Although AD is characterized by the pathological hallmarks of β amyloid (Aβ) deposition and tau neurofibrillary tangles (NFTs), tau burden is known to be more strongly associated with cognitive dysfunction than Aβ accumulation3,4. Accordingly, imaging of Aβ deposition, pathologic tau, and neurodegeneration forms a research framework [AT(N)]. Imaging of AD biomarkers is accomplished using amyloid positron emission tomography (PET) for Aβ, tau-PET for NFTs, and 18F-fluoro-deoxyglucose (FDG) PET or magnetic resonance imaging (MRI) for neurodegeneration5. These imaging techniques can be utilized to classify and stage AD6,7. For example, MRI and FDG-PET can reflect neurodegeneration, and amyloid PET can provide pathological evidence of Aβ agglomeration8,9,10. However, MRI and FDG-PET cannot specifically reflect the molecular pathological hallmarks of AD such as Aβ or tau burden. In addition, amyloid PET could be accumulated 20 years before the diagnosis of AD. It means that it is difficult to visualize the progression of Aβ accumulation because it is saturated at the time of disease5. On the contrary, tau-PET scans can directly reflect the pathological changes of AD and have a high correlation with cognitive function and disease progression11. In addition, cerebral structural changes also reveal a close relationship with pathological tau deposition7,12. The strength of tau-PET images is their ability to reveal tau accumulation patterns and specific deposit sites in focal regions of the brain similar to that shown by tau histology performed through Braak staging7,13.
In response to the vast increase in the amount of medical imaging data, deep learning (DL) of medical images have been used for disease classification. Although many studies have used DL for classification of AD, they have mainly focused on MRI, amyloid PET, or co-registration of both types of images14,15,16. In addition, many previous studies on DL application focused on models using AD images limited to specific parts of the brain related to cognitive function17,18,19. Application of a convolutional neural network (CNN) to tau-PET scans is a novel approach, as the spatial characteristics and interpretation of this modality are quite different than amyloid PET, FDG-PET, or MRI. In particular, the PET signal highlights the specific region of tau molecular manifestation in the brain and is considered more informative than other imaging techniques. This can have implications for CNNs, which require processing of complex inputs as well as visualization of informative features.
In this study, we implemented a DL framework for the classification of AD stage using 18F-flortaucipir PET. Transfer learning (TL) for high classification performance was performed using the weight derived from CU versus AD classification for CU versus MCI classification. By identifying the phenotype of tau deposition through two-dimensional (2D) and three-dimensional (3D) 18F-flortaucipir-PET molecular imaging based on DL, the clinical usefulness of 18F-flortaucipir-PET is proposed.
Results
Subject characteristics
The characteristics of all subjects investigated in this study are presented in Table 1. The mean age was 71.4 years, with 70.0 years in the CU, 72.0 years in the MCI, and 73.7 years in the AD groups. One hundred thirty-six (49.3%) were female and 140 (50.7%) were male, with 47 (62.7%) and 40 (63.5%) males present in the MCI and AD groups, respectively. As a result of the normality test, all covariates had p > 0.05. The differences among the three groups for all variables showed p < 0.05 as a result of one-way ANOVA. The total Aβ positive was 141, with 43 of CU (22 of florbetapir and 21 of florbetaben), 46 of MCI (18 of florbetapir and 28 of florbetaben), and 52 of AD (23 of florbetapir and 29 of florbetaben).
Classification performance between CU and AD
The CU and AD classification results are shown in Table 2. Most of the result metrics in the 2D CNN-LSTM and 3D CNN models showed that the multimodal performance was slightly more significant compared to the image classification. For the 2D multimodal results, the receiver operating characteristic (ROC) area under the curve (AUC) was 0.947, accuracy 88.5%, precision 86.7%, recall 92.9%, F1 score 89.7% and specificity 84.6%. The 3D multimodal results were higher than those of image classification in all performance indicators, with AUC of 0.976, accuracy 92.3%, precision 92.9%, recall 92.9%, F1 score 92.9% and specificity 92.3% suggesting better performance than the 2D model.
Classification performance in between CU and MCI
The CU and MCI classification results are shown in Table 2. The multimodal performance was slightly higher than image classification result metrics of 2D CNN-LSTM and 3D CNN. The 2D multimodal results were the same or higher than those of image classification, with AUC of 0.923, accuracy 83.3%, precision 78.6%, recall 84.6%, F1 score 81.5% and specificity 82.4%. The 3D multimodal results show AUC of 0.850, and accuracy 82.8%, precision 86.7%, recall 81.3%, F1 score 83.9% and specificity 84.6%, which were higher than those of image classification in all performance indicators (Fig. 1).
Identification of informative features for AD classification
GRAD-CAM findings confirmed that 2D and 3D CNN learned through feature extraction from most areas in the image (Fig. 2). Figure 2A is the result of 3D CNN, and Fig. 2B is the result of 2D CNN-LSTM. The identification of informative features in the Grad-Cam results, the distinctive area extracted from the brain was an area associated with cognitive functions such as the hippocampus and the lateral and middle temporal regions. In addition, through 3D sagittal phase in AD group, it was able to observe some cingulate regions were included. As a result of 2D CNN-LSTM, the regions that appeared through GRAD-CAM via a single axial phase shows a lot of dependent parts of uptake region.
Discussion
In this study, DL was used to grading and differentiate syndromal cognitive stage between CU and AD, CU and MCI. In the MCI and AD groups used in this study, it was confirmed that there were some subjects who showed amyloid and flortaucipir positive or amyloid and flortaucipir negative at the same time. This suggests that this study was performed the syndromal cognitive stage grading of MCI and AD through flortaucipir PET. The classification among CU, MCI, and AD performed in this study is a syndromic cognitive stage grading, and AI modeling based on DL were performed with the goal of grading between CU and MCI, and between CU and AD. In addition, by applying the Tau PET image -based DL technique, the possibility of clinical syndromal grading was presented through CU VS MCI and CU VS AD comparison. It means that it is significance as a preliminary study to create a numeric staging model5. The results of 2D CNN-LSTM and 3D CNN proved the high performance of the classification ability of these imaging biomarkers. Moreover, multimodal data integration was performed by adding the demographic and neuropsychological variables into the CNN models as a method to use quantitative data which could be acquired at screening or baseline for disease classification. In 2D CNN-LSTM image classification for distinguishing between CU and AD has an AUC of 0.964 and accuracy of 88.5%. In addition, the results of 3D CNN image classification showed the AUC of 0.947 and accuracy of 88.5%. In multimodal classification, the results of 2D CNN-LSTM and 3D CNN showed the AUC of 0.947 and 0.976, respectively. For distinguishing between CU and MCI in image classification task has an AUC of 0.840 and accuracy of 80.0%. In addition, the results of 3D CNN image classification showed the AUC of 0.845 and accuracy of 83.3%. In multimodal classification, the results of 2D CNN-LSTM and 3D CNN showed the AUC of 0.923 and 0.850, respectively.
This study has several novel features. First, the classifiers generated in this study demonstrated that accumulated tau tangles may have an important role in AD pathogenesis based on the characteristics of their distribution. Previous studies using an ADNI database-driven approach have determined that the principal regions of tau pathology mainly overlap with the Braak stage III regions of interest (ROIs) (i.e., the amygdala, para-hippocampal gyrus, and spindle)6,17,20. It is generally known that stage III/IV ROIs could be observed in patients with CU as well as those with AD, whereas stage I/II is common in patients with CU and stage V/VI is common in those with AD21. In other words, it is difficult to classify tau deposition measurements as representative of cognitive decline including MCI and AD compared to CU through the ROIs of stage III/IV. We performed a systematic review of the existing literature to summarize the most common CU versus AD classification techniques that include comparison of CU versus MCI (Table 3)17,18,22,23,24,25,26,27. Notably, the classification between CU and MCI in this study showed better performance than other previously published methods. Significant accuracy was achieved for distinguishing both classifications based on regions with accumulated tau, which were set in the DL models. In addition, we generated regions with important identified features by GRAD-CAM in the DL process. The left and right amygdala, and left entorhinal, left para-hippocampal, inferior temporal, and right middle-temporal regions were identified as the main tau deposition regions. This suggests that tau deposition in the regions revealed by DL frameworks is similar to the regions of neurodegenerative and cognitive decline identified by Braak staging. Moreover, by including the entorhinal and inferior-temporal regions, which are known to be affected in early AD, among the Braak stage I/II regions and suggesting their importance, the classifiers generated in this study reflect the tau accumulation characteristics of AD and reinforce the suggestion of previous studies regarding their important role in early pathogenesis28,29. The results of our study also correspond well with the tau pattern and related regions as reported in previous studies 30,31,32. Second, we conducted TL by applying the weights of the classifier between CU and AD for maximized performance of classification between CU and MCI. The result for classification CU and MCI in this study was able to provide better performance than other previously published methods (Table 3). In particular, by presenting the results of CU and MCI classification with higher performance than other studies, we present the possibility of syndromal cognitive staging in early stages, which has recently attracted attention. In addition, it was confirmed that the DL based classification performance (2D; AUC of 0.840 and 0.923, 3D; AUC of 0.845 and 0.850) is superior to existing conventional ML model performance (Tau SUVR; AUC of 0.720, Tau SUVR with clinical variables; AUC of 0.800) of support vector machine (SVM) based classification and effective for classifying grade staging between CU and MCI which are relatively difficult to distinguish (Table 2). In the classification between CU and AD through ML, the difference in continuous numeric variables such as MMSE and Tau SUVR is stark, and it can be shown that the effect is better than that of DL (Supplementary Table S1). However, for the classification of CU vs MCI, which is relatively difficult to distinguish in terms of clinical symptoms, the DL-based classification performance was superior to staging. This suggests the possibility of clinically useful use through future research development. Although there might be some differences in the model structure and method of feature extraction, our results suggest that good performance of the classification between CU and MCI is presented through the application of weights of classification between CU and AD within the same data set. Third, the classifiers of this study could be applied to measurements that are easily obtained in clinical practice. In this study, we trained the 2D model using consecutive 2D slices by stacking two consecutive LSTM. Of a total of 144 slices, the model in this study used 72 consecutive even-numbered slices. In many clinical applications, brain PET scans for AD require fewer slices than the number of slices used in this study with 2-mm or 3-mm axial 2D slice thickness. The results of this study indicate that there is a possibility to learn all data at once without omitting the specific axial image information of each individual patient. In addition, multimodal layering was performed by concatenation of demographic and neuropsychological variables with the flattened layer of features extracted through CNN before entering the LSTM. The combined clinical variables used in this study were age, sex, education, and MMSE score, which are easily obtainable indicators at the screening stage for AD clinical trials or in hospital visits of outpatients. Through the results of our multimodal models, we demonstrated that the combination of clinical information with images could help to improve model performance slightly more than that of image DL.
However, there were some limitations in this study. First, we conducted 2D CNN-LSTM modeling utilized only consecutive even-numbered 72 slices in the axial direction among total of 144 consecutive slices of 3D PET data. The selected contiguous 72 slices were acquired after resampling the data from initial ADNI (96 slices per patient, 1.2 × 1.2 × 1.2 mm) to a voxel size of 1 × 1 × 1 mm. The method using consecutive even-numbered 72 slices was chosen as a way to overcome hardware limitations while maximally covering the entire brain volume area. As a result, we could present higher performance than other existing studies. However, these methods cannot be explained to completely cover the entire volume area, and some brain information is expected to be lost. If the hardware limitation is overcome in the future, the study could be conducted using total of 144 slices in the same process. Second, the small number of subjects was a problem. The data available in this study was less than that required for general DL because the ADNI 3 protocol was limited to 63 participants with AD. In DL training, if more samples are generally applied to the models, the better the results. Due to the small number of subjects, we allocated 20% of the training set for each cross-validation data set for validation. In addition, in the case of MCI subjects used in this study, as a late MCI, there was a limit for specific classification of syndromal cognitive staging with AD. In the future, when additional data is obtained using the model implemented in this study, it is possible to accurately grading for AD staging. In addition, the TL is used for applying a small data set through pre-trained models constructed from large data sets to obtain results with fine tuning. However, in this study, it was not possible to acquire many subjects; thus, the frozen layers method with feature extraction and cross-validation was performed to solve this problem and improve the reliability of the CU versus MCI classification model. Third, we use imperfect clinical diagnosis as the gold standard for modeling. As shown in Table 4, the clinical diagnosis presented in ADNI that we used is based on relatively objective criteria as a result of considering MMSE, CDR, logical memory test, and general cognition and function. In addition, it is being quality controlled by the ADNI clinical core, suggesting that many efforts are being made to compensate for incompleteness33. However, we need to conduct research using objective golden standards such as brain pathology or quantitative measures of biomarkers through study in the future. Lastly, the identification of extracted informational features for AD classification through GRAD-CAM shows a mixture of on-target binding and off-target binding. In particular, right off target binding is shown in the sagittal phase as a result of 3D CNN. This is seen as a limitation of flortaucirpir ligand, and effective research improvement can be presented through the second-generation tau ligand in the future. In addition, segmentation such as cortex, central structures and superior cerelleum before processing could be an alternative solution. In this study, we suggested that 18F-flortaucipir PET images could be a scalable biomarker by applying a DL framework for classification of AD stage. Our results show that the DL models using images in combination with clinical variables can effectively classify AD stages.
Methods
Subjects
A total of 271 subjects (138 CU, 75 MCI, and 63 AD) in the Alzheimer’s Disease Neuroimaging Initiative (ADNI3) for whom 18F-flortaucipir PET scans were performed at baseline were recruited. Age, sex, education, Mini-Mental State Examination (MMSE) score, 18F-flortaucipir-PET images, and diagnostic results were acquired (Fig. 3). All subjects were divided using criteria provided as clinical syndrome diagnoses within the ADNI cohort (Supplementary Table S2, S3). All subjects in the CU group had clinical dementia rating (CDR) scores of 0 or 0.5, which allowed them to be distinguished from participants with MCI and AD. The patients with MCI did not meet the dementia criteria and were evaluated based on an objective memory impairment determination. All participants with MCI had MMSE scores of 24 or higher up to 30 and CDR scores of 0.5, a CDR memory score of 0.5 or higher. In addition, d a score that indicated impairment on the delayed recall of Story A of the Wechsler Memory Scale-Revised (≥ 16 years of education: < 11; 8–15 years of education: ≤ 9; 0–7 years of education: ≤ 6) was applied34. All patients that met the criteria for AD had CDR scores of 0.5 of 1 and a score that indicated impairment on the delayed recall of Story A of the Wechsler Memory Scale-Revised (≥ 16 years of education: ≤ 8; 8–15 years of education: ≤ 4; 0–7 years of education: ≤ 2). A final total of 271 subjects from the ADNI3 cohort were selected for this study (Table 4).
Subjects flowchart through this study for the training and test datasets. Within the training data set, 20% was used as validation data and cross-validation was performed 5 times. Within the ADNI3 data set, 'AV1451 Coreg, Average, Standardized Image and Voxel Size' PET images were used, and each group (CU, MCI, AD) was classified based on clinical syndrome staging by registered in ADNI cohort. Aβ β-amyloid, PET positron emission tomography.
The study procedures were approved for all participating centers (https://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf), and written informed consent was obtained from all participants or their authorized representatives. A committee on human research at each participating institution approved the study protocol, and all participants or legal guardian(s)/legally authorized representatives gave their informed consent. In addition, all experiments were performed in accordance with the relevant guidelines and regulations outlined in the IRB.
Data acquisition and preprocessing
18F-flortaucipir 3D dynamic PET scan images were acquired for all individuals. All PET images were acquired by a 30-min scan, 75–105 min after intravenous (IV) injection of 18F radio isotope (RI) with 370 mBq (10.0 mCi) ± 10% radioactivity, considering the weight of each patient, and flortaucipir ligand. For this study, pre-processed PET images (AV1451 Coreg, Average, Standardized Image, and Voxel Size) provided and described were acquired from the ADNI3 cohort. As all images were preprocessed such as anterior–posterior axis fitting to the anterior commissure-posterior commissure line. Scans were normalized to Montreal Neurologic Institute (MNI) space using parameters generated from segmentation of the T1-weighted MRI scan in Statistical Parametric Mapping v12 (SPM12).
Intensity normalization was performed using a cerebellar gray matter as a reference region and standard uptake value ratio (SUVR) could be acquired for RI uptake calculation for each region in the brain35,36,37. More details of 18F-flortaucipir-PET preprocessing can be found in other related studies22,35,38. After acquisition images we converted the voxel size to 1 × 1 × 1 mm by resampling and resizing and acquired 3D PET images to use input data for the development of the DL framework. For 2D CNN long short-term memory (LSTM) DL framework development, we extracted 72 even-numbered sequential axial slice images from a total of 144 3D images per individual subject. The 3D CNN DL framework was performed using total image. All data such as demographic and clinical information, image voxel size was processed for min–max normalization for a multimodal framework.
The data for both frameworks were split as 80% of the total data for the training set and 20% of the total data for the test set. The validation set was 20% of the training set. Five-fold cross-validation was applied to derive stable performance (Fig. 3). The data ratio was maintained during five cross-validations, as one subset was selected for testing and the remaining four sets for validation.
Define of Aβ PET status
We downloaded the 18F-florbetapir and 18F-florbetaben analysis data from the ADNI. Moreover, we classified each participant as Aβ-positive PET scan on observing a global standardized uptake value ratio (SUVR) > 1.11 for the 18F-florbetapir 39. For 18F-florbetaben, tracer uptake was assessed according to the regional cortical tracer uptake system in four brain regions (frontal cortex, posterior cingulate cortex/precuneus, parietal cortex, and lateral temporal cortex) and the cut-off value was 1.140.
Classification for deep learning
TL was performed using the weight of both 2D CNN-LSTM and 3D CNN models built in classification between CU and AD to increase the classification between CU and MCI classification performance. In both 2D and 3D models, feature extraction methods similar to classification between CU and AD models was used by conducting a freezing technique to fix the feature extraction architecture for classification between CU and MCI by TL. From the first convolutional layer to the last layer (before the fully connected layer), which performs feature extraction within the image, it was frozen for TL, and DL was performed through a classifier composed of dense layers. The learning rate was changed to 0.00001, considering that it is more difficult to distinguish between CU and MCI. For each classification between CU and AD, CU and MCI, binary cross-entropy loss function was applied.
2D CNN-LSTM
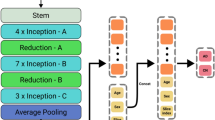
The 2D CNN model was prepared in conjunction with the LSTM (Fig. 4). Two LSTM algorithms were consecutively stacked after the 2D CNN to minimize the loss of brain information contained in the 72 axial images from the upper to the lower part of the head. All axial images were sequentially processed by LSTM configuration models after feature extraction from each slide through 2D CNN. Each slice index i and extracted features f were converted to the form of (i, f), and the model was constructed by stacking two LSTM layers consecutively. In the first layer of LSTM, the features of each slice are output in the form of (i, f) → (i, LSTM(output)). While maintaining the sequential slices information, the variable of f(Features) reduced by the size of the first LSTM output is input to the second LSTM layer, and finally output in the form of (i, LSTM(last output)). That is, the features corresponding to the entire slice information were sequentially extracted, and the model was constructed through two consecutive LSTM layers. To avoid excessive epochs that could lead to overfitting, early stopping was applied if the model did not show any improvement loss for ten iterations. The hyper-parameters for classification between CU and AD, adaptive moment estimation (Adam), a first-order gradient-based probability optimization algorithm with learning rate = 0.0001, decay rate = 0.96, and batch size of 1, was used (Fig. 5). The feature maps (8,16,32,64) were extracted from four hidden layers; kernel_size = 2, same padding, and Maxpool2D were applied to each layer to use the activation function of rectified linear unit (ReLU). Dropout (0.3) was applied to the third and fourth layers. Two LSTM (200,64) layers were applied, and dropout (0.25) was applied after the first layer.
3D CNN
The 3D CNN model was constructed more depth than 2D CNN-LSTM since the 3D images have volume including with height and width information (Fig. 4). To avoid overfitting, early stopping was applied if the model did not show any improvement loss for 15 iterations. Hyper-parameters for classification between CU and AD, such as optimization function, learning rate, decay rate, and batch size, were the same as for the 2D model. The feature maps (8,16,32,64,128) were extracted from four hidden layers; kernel_size = 3 and Maxpool3D were applied to each layer to use the activation function ReLU (Fig. 5). Features that had passed through the flattened layer were input into the three dense layers, and dropout (0.2) was subsequently applied.
Informative feature identification for AD classification
Gradient-weighted class activation mapping (GRAD-CAM) was used to identify informative features extracted through CNN models. The feature map could be visualized with the average pixel value up to final layers. We identified regions in the brain as the ReLU activation function was applied to visualize important parts in the model during the analysis process.
Evaluation performance
For the evaluation of the model performance, four metrics (accuracy, recall, precision, and F1 score) were used. Since this study focused on the accurate classification between CU and AD, CU and MCI, the metric of true positive was mainly established for overall performance evaluation of the classification model. The equations is Eqs. (1)–(4).
Statistical analysis
After performing the Shapiro–Wilk normality test on the variables from each group (CU, MCI, and AD), examination of significance differences among groups was conducted by one-way analysis of variance (ANOVA) and the chi-squared (\(\upchi \)2) test for continuous and categorical variables, respectively. After conducting Levene's test for checking the equality of variances, ANOVA was performed to test difference in the means among three groups. If the assumption of equal variance was not satisfied, Welch's ANOVA was performed to test the mean difference among groups. In addition, post-hoc analysis was performed using the Games-Howell test if the assumption of equal variance was established, and the Scheffe test if the assumption was not satisfied. Statistical significance was set at p < 0.05 and p < 0.001. All statistical analysis was performed in R (version 4.1.0).
Tools
Tensorflow 2.8.0 and Keras 2.8.0 were used to construct DL frameworks and performed by scratch on Python 3.7.0 for all processing.
Ethics approval and consent to participate
The study was approved by the institutional review boards of Kangwon National University Hospital (approval No. KNUH-2022-06-011) all participating institutions, and written informed consent was obtained from all participants or their authorized representatives.
Data availability
All ADNI data used in this study is available through the ADNI website (https://adni.loni.usc.edu/data-samples/access-data/). The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.
References
Wilson, H., Pagano, G. & Politis, M. Dementia spectrum disorders: Lessons learnt from decades with PET research. J. Neural Transm. 126, 233–251 (2019).
DeTure, M. A. & Dickson, D. W. The neuropathological diagnosis of Alzheimer’s disease. Mol. Neurodegener. 14, 32 (2019).
Braak, H. & Braak, E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. (Berl.) 82, 239–259 (1991).
Weiner, M. W. et al. The Alzheimer’s Disease Neuroimaging Initiative 3: Continued innovation for clinical trial improvement. Alzheimers Dement. 13, 561–571 (2017).
Jack, C. R. et al. NIA-AA Research Framework: Toward a biological definition of Alzheimer’s disease. Alzheimers Dement. 14, 535–562 (2018).
Márquez, F. & Yassa, M. A. Neuroimaging biomarkers for Alzheimer’s disease. Mol. Neurodegener. 14, 21 (2019).
Cho, H. et al. In vivo cortical spreading pattern of tau and amyloid in the Alzheimer disease spectrum. Ann. Neurol. 80, 247–258 (2016).
Politis, M. & Piccini, P. Positron emission tomography imaging in neurological disorders. J. Neurol. 259, 1769–1780 (2012).
Politis, M. Neuroimaging in Parkinson disease: From research setting to clinical practice. Nat. Rev. Neurol. 10, 708–722 (2014).
Kuznetsov, I. A. & Kuznetsov, A. V. Simulating the effect of formation of amyloid plaques on aggregation of tau protein. Proc. Math. Phys. Eng. Sci. 474, 20180511 (2018).
Baker, S. L., Maass, A. & Jagust, W. J. Considerations and code for partial volume correcting [18F]-AV-1451 tau PET data. Data Brief 15, 648–657 (2017).
Wang, L. et al. Evaluation of tau imaging in staging Alzheimer disease and revealing interactions between β-amyloid and tauopathy. JAMA Neurol. 73, 1070 (2016).
Villemagne, V. L., Fodero-Tavoletti, M. T., Masters, C. L. & Rowe, C. C. Tau imaging: Early progress and future directions. Lancet Neurol. 14, 114–124 (2015).
Litjens, G. et al. A survey on deep learning in medical image analysis. Med. Image Anal. 42, 60–88 (2017).
Shen, D., Wu, G. & Suk, H.-I. Deep learning in medical image analysis. Annu. Rev. Biomed. Eng. 19, 221–248 (2017).
Dimou, E. et al. Amyloid PET and MRI in Alzheimer’s disease and mild cognitive impairment. Curr. Alzheimer Res. 6, 312–319 (2009).
Jo, T., Nho, K., Risacher, S.L., Saykin, A.J. & Alzheimer’s Neuroimaging Initiative. Deep learning detection of informative features in tau PET for Alzheimer’s disease classification. BMC Bioinform. 21, 496 (2020).
Liu, M., Cheng, D., Yan, W. & Alzheimer’s Disease Neuroimaging Initiative. Classification of Alzheimer’s disease by combination of convolutional and recurrent neural networks using FDG-PET images. Front. Neuroinform. 12, 35 (2018).
Son, H. J. et al. The clinical feasibility of deep learning-based classification of amyloid PET images in visually equivocal cases. Eur. J. Nucl. Med. Mol. Imaging 47, 332–341 (2020).
Leuzy, A. et al. A multicenter comparison of [18F]flortaucipir, [18F]RO948, and [18F]MK6240 tau PET tracers to detect a common target ROI for differential diagnosis. Eur. J. Nucl. Med. Mol. Imaging 48, 2295–2305 (2021).
Schöll, M. et al. PET imaging of tau deposition in the aging human brain. Neuron 89, 971–982 (2016).
Cho, S. H. et al. Appropriate reference region selection of 18F-florbetaben and 18F-flutemetamol beta-amyloid PET expressed in centiloid. Sci. Rep. 10, 14950 (2020).
Lu, D. et al. Multimodal and multiscale deep neural networks for the early diagnosis of Alzheimer’s disease using structural MR and FDG-PET images. Sci. Rep. 8, 5697 (2018).
Choi, H., Jin, K. H., & Alzheimer’s Disease Neuroimaging Initiative. Predicting cognitive decline with deep learning of brain metabolism and amyloid imaging. Behav. Brain Res. 344, 103–109 (2018).
Li, R. et al. Deep learning based imaging data completion for improved brain disease diagnosis. Med. Image Comput. Comput. Assist. Interv. MICCAI Int. Conf. Med. Image Comput. Comput. Assist. Interv. 17, 305–312 (2014).
Wen, J. et al. Convolutional neural networks for classification of Alzheimer’s disease: Overview and reproducible evaluation. Med. Image Anal. 63, 101694 (2020).
Zou, J. et al. Deep learning improves utility of tau PET in the study of Alzheimer’s disease. Alzheimers Dement. Diagn. Assess. Dis. Monit. 13, e12264 (2021).
de Calignon, A. et al. Propagation of tau pathology in a model of early Alzheimer’s disease. Neuron 73, 685–697 (2012).
Hyman, B. T., Van Hoesen, G. W., Damasio, A. R. & Barnes, C. L. Alzheimer’s disease: Cell-specific pathology isolates the hippocampal formation. Science 225, 1168–1170 (1984).
Suk, H.-I. & Shen, D. Deep learning-based feature representation for AD/MCI classification. Med. Image Comput. Comput. Assist. Interv. MICCAI Int. Conf. Med. Image Comput. Comput. Assist. Interv. 16, 583–590 (2013).
Ossenkoppele, R. et al. Discriminative accuracy of [18F]flortaucipir positron emission tomography for Alzheimer disease vs other neurodegenerative disorders. JAMA 320, 1151–1162 (2018).
Jack, C. R. et al. Defining imaging biomarker cut points for brain aging and Alzheimer’s disease. Alzheimers Dement. J. Alzheimers Assoc. 13, 205–216 (2017).
Aisen, P. S., Petersen, R. C., Donohue, M., Weiner, M. W., & Alzheimer’s Disease Neuroimaging Initiative. Alzheimer’s disease neuroimaging initiative 2 clinical core: Progress and plans. Alzheimers Dement. 11, 734–739 (2015).
Wechsler, D. WMS-R: Wechsler Memory Scale-Revised : Manual (Psychological Corporation, 1987).
Maass, A. et al. Comparison of multiple tau-PET measures as biomarkers in aging and Alzheimer’s disease. Neuroimage 157, 448–463 (2017).
Landau, S. & Jagust, W. Flortaucipir (AV-1451) processing methods. Alzheimers Res. Ther. 14, 49 (2016).
Young, C. B., Landau, S. M., Harrison, T. M., Poston, K. L. & Mormino, E. C. Influence of common reference regions on regional tau patterns in cross-sectional and longitudinal [18F]-AV-1451 PET data. Neuroimage 243, 118553 (2021).
Baker, S. L. et al. Reference tissue-based kinetic evaluation of 18F-AV-1451 for tau imaging. J. Nucl. Med. Off. Publ. Soc. Nucl. Med. 58, 332–338 (2017).
Joshi, A. D. et al. Performance characteristics of amyloid PET with florbetapir F 18 in patients with Alzheimer’s disease and cognitively normal subjects. J. Nucl. Med. 53, 378–384 (2012).
Cho, S. H. et al. Concordance in detecting amyloid positivity between 18F-florbetaben and 18F-flutemetamol amyloid PET using quantitative and qualitative assessments. Sci. Rep. 10, 19576 (2020).
Acknowledgements
All data used in this study were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI, http://adni.loni.usc.edu/). The ADNI investigators contributed to the design and implementation of ADNI and/or provided data but did not participate in the working for manuscript. A complete listing of ADNI investigators can be found at https://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
Funding
This study was supported by Basic Science Research Program and Regional Innovation Strategy (RIS) through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (MOE) (NRF 2021R1I1A3059241, 2022RIS-005).
Author information
Authors and Affiliations
Contributions
S.W.P. designed the study, analyzed and interpreted the data, and wrote and revised the manuscript. N.Y.Y. analyzed the data and visualized the results. G.H.B. and Y.S.K. reviewed the manuscript. J.W.J. designed the study, supervised all work, interpreted the analysis results, and wrote and review the manuscript. All authors approved for the final manuscript. All participants provided written informed consent.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Park, S.W., Yeo, N.Y., Kim, Y. et al. Deep learning application for the classification of Alzheimer’s disease using 18F-flortaucipir (AV-1451) tau positron emission tomography. Sci Rep 13, 8096 (2023). https://doi.org/10.1038/s41598-023-35389-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-35389-w
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.