Abstract
Narrow band imaging (NBI) has been extensively utilized as a diagnostic tool for colorectal neoplastic lesions. This study aimed to develop a trial deep learning (DL) based four-class classification model for low-grade dysplasia (LGD); high-grade dysplasia or mucosal carcinoma (HGD); superficially invasive submucosal carcinoma (SMs) and deeply invasive submucosal carcinomas (SMd) and evaluate its potential as a diagnostic tool. We collected a total of 1,390 NBI images as the dataset, including 53 LGD, 120 HGD, 20 SMs and 17 SMd. A total of 598,801 patches were trimmed from the lesion and background. A patch-based classification model was built by employing a residual convolutional neural network (CNN) and validated by three-fold cross-validation. The patch-based validation accuracy was 0.876, 0.957, 0.907 and 0.929 in LGD, HGD, SMs and SMd, respectively. The image-level classification algorithm was derived from the patch-based mapping across the entire image domain, attaining accuracies of 0.983, 0.990, 0.964, and 0.992 in LGD, HGD, SMs, and SMd, respectively. Our CNN-based model demonstrated high performance for categorizing the histological grade of dysplasia as well as the depth of invasion in routine colonoscopy, suggesting a potential diagnostic tool with minimal human inputs.
Similar content being viewed by others
Introduction
Although it is relatively simple for human observers to recognize and describe the visual elements in empirical terms, it has been remarkably difficult to accurately define and analyze them with a computer. The electronic endoscope has allowed us to quantify any element making up a digitized endoscopic image through mathematical processes. Several studies have evaluated the effectiveness of feature extraction for computer-aided diagnosis (CAD) to classify the endoscopic severity of ulcerative colitis1,2 and to assess the risk of developing gastric cancer among Helicobacter pylori-positive patients3. However, the diagnostic accuracy of feature engineering was limited due to the challenges in extracting features for image analysis in gastrointestinal diseases.
With the proliferation of CNN, the task of classifying objects in natural images can be solved simply by presenting examples of images and the names of the objects to a neural network that acquired all its knowledge from the training data4. This groundbreaking technology has freed engineers from feature engineering and endoscopists from knowledge-based image interpretation. The CNN-based supervised learning has been applied in the automated localization of gastric cancer in routine gastroscopies5 and the automated detection of colon polyps6. Several studies have reported the utilization of a CNN-based model for distinguishing adenomatous from non-adenomatous polyps7, adenomatous from hyperplastic diminutive colorectal polyps 8, and neoplastic polyps from non-neoplastic polyps 9. The fine-tuning of a pre-trained CNN for gastric precancerous disease classification10 or the efficient channel attention deep dense CNN for the classification of esophageal disease11 has also been reported.
However, to the best of our knowledge, a multi-class model for evaluating the grade of histologic dysplasia along with the depth of invasion has not yet been developed. The NBI international colorectal endoscopic (NICE) classification applicable to with or without magnification was proposed for diagnosing submucosal invasive colon cancer12. However, criteria for classification described in empirical terms12 may inevitably suffer from a variety of biases in evaluation leading to different accuracy varying with endoscopists and disturb comparison of accuracy among different endoscopist communities. This study aimed to develop a trial CNN-based supervised learning model for evaluating histologic atypism or invading depth from NBI images of detected colonic neoplastic lesions and evaluate the potential as a diagnostic tool.
Methods
Preparation of endoscopic images
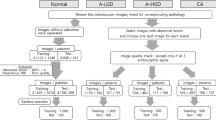
NBI images of neoplastic lesions from patients who underwent endoscopic or surgical resection at Sendai City Medical Center Sendai Open Hospital from April 2017 to December 2019 were used for this single center retrospective study. Characteristics of collected NBI images are summarized in Table 1. A total of 1390 NBI images were sampled from a total of 210 lesions with definite histologic diagnosis13: 53 low-grade dysplasia (LGD); 120 high grade dysplasia or mucosal carcinoma (HGD); 20 superficially invasive (the depth of the invasive front < 1000 µm) submucosal carcinoma (SMs) and 17 deeply invasive (the depth of the invasive front > 1000 µm) submucosal carcinomas (SMd). Pathological diagnosis was conducted by pathologists unaware of the study design in a blinded manner. The diagnosis of a mucosal lesion, LGD or HGD was assigned to the most severe grade regardless of the size of the component. Sampled picture number per lesion was 5.5 to 7 samples with an averaged image capturing conditions: no magnification 41.0%; low magnification 37.9%; high magnification 21.1%. The images of a solitary lesion at varying magnifications were carefully chosen to minimize potential bias in the selection process. The video endoscopes CF-HQ290ZI, PCF-H290ZI, PCF-H290TI and video endoscopy system EVIS LUCERA ELITE CV-290/CLV-290SL (Olympus Medical Systems, Co., Ltd., Tokyo, Japan) were used.
Preparation of dataset
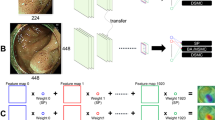
NBI images (Fig. 1a) were manually partitioned into the lesion (Fig. 1b) and background (Fig. 1c) from which the patch images (128 × 128 pixels) were cropped starting from the left upper corner (white dotted patch), rightwards (white solid patch), then downwards (red solid patch) at every 32-pixel-strides (white and red arrows) over the entire effective region of interest. The patches including blackouts with more than 10% of the effective region were automatically excluded from analysis. Blackouts were defined as regions with the intensity of red component lower than 50. Similarly, the patches with halations exceeding 5% of the effective region were also excluded. Halations were defined as regions with the intensity of green component higher than 250. In this study, the patches were further classified into in-focus patches and out-of-focus ones according to the amount of spatial high frequency area extracted by high pass filter with a cut-off of 6.25% Nyquist frequency. The in-focus patches were classified into (0) background (BG), (1) LGD, (2) HGD, (3) SMs and (4) SMd, and the out-of-focus ones into (5) background (BG-oof) and 6) lesion (L-oof). A total of 598,801 patches were classified into 7 categories (Table 2). The study did not have any inclusion or exclusion criteria for pictorial quality of the patches by endoscopists. As stated, the patches with excessive blackout or halation were automatically excluded before entry. The study aimed to establish an effective histologic classifier that can be used in any common shooting conditions of NBI.
Preparation of dataset. Original NBI image (a) were manually partitioned into the lesion (b) and background (c). The patch images (128 × 128 pixels) were trimmed from the lesion and background starting from the left upper corner (white dotted patch), rightwards (white solid patch), then downwards (red solid patch) at every 32-pixel-strides (white and red arrows) over the entire region of interest.
Evaluation method
We employed cross-validation to obtain more accurate results with less bias in the machine learning studies14. In this study, the dataset is randomly partitioned into three equal sized folds, one fold of which is for validation and the other folds are for training. The proportion of labels was equal in each fold. The training and validation processes were repeated three times using different folds each time. The three validation results could then be averaged to produce a single estimation.
Architecture of the CNN
ResNet50 (a CNN) proposed by He et al.15 and Pytorch were utilized. ResNet50 without pretraining was imported from Pytorch library (torchvision.models). The original patches with 128 × 128 pixels were converted into images with 224 × 224 pixels. We tuned hyper parameters, which were set by a human, as follows: optimizer, Adam; loss function, cross entropy loss; number of training epochs, 50; batch size, 256; learning rate, 0.00005 via trial and error; and number of the outer layers, 7 classes.
Image-level classification
An exemplification of SMd and the annotation mask without blackout or halation (denoted by X) are depicted in Fig. 2a,i, respectively. The patches classified into BG, LGD, HGD, SMs, SMd, BG-oof and L-oof, by the trained CNN, are illustrated by white (Fig. 2b), green (Fig. 2c), yellow (Fig. 2d), magenta (Fig. 2e), red (Fig. 2f), dark gray (Fig. 2g) and cyan (Fig. 2h) open squares, respectively, and the corresponding union masks in Fig. 2j–p, respectively. Classification algorithms must be developed by utilizing in-focus patches, without sacrificing pictorial information. Here, the union masks of the patches classified into labels BG, LGD, HGD, SMs and SMd are designated by M0, M1, M2, M3 and M4, respectively. Intersection over union between X and Mi (IoUi) are given by X ∩ Mi/ X ∪ Mi (i = 0, 1, 2, 3, 4). The lesion was classified into the argmax among IoUi (i = 0, 1, 2, 3, 4), with the IoUi values of 0.12, 0.05, 0.21, 0.04 and 0.57, respectively, leading to label 4 or histologic classification SMd.
The patch-level histological mapping predicted by the trained model, alongside the annotation mask. An example picture of SMd and the annotation mask without blackout or halation can be seen in (a) and (i), respectively. White (b), green (c), yellow (d), magenta (e), red (f), dark gray (g), and cyan (h) open squares indicate the patches classified as BG, LGD, HGD, SMs, SMd, BG-oof, and L-oof, respectively, with their corresponding union masks visible in (j–p), respectively. BG, background; HGD, high grade dysplasia; LGD, low grade dysplasia; SMs, superficially invasive submucosal carcinoma; SMd, deeply invasive submucosal carcinoma; BG-oof, out-of-focus background; L-oof, out-of-focus lesion.
Ethics approval and consent to participate
This study was approved by the Committee of Medical Ethics of Hirosaki University Graduate School of Medicine (Aomori, Japan; reference no. 2019–1099) and Sendai City Medical Center (Sendai, Japan: reference no. 2019–0029). Informed consent was obtained in the form of opt-out on our website (https://www.https://www.med.hirosaki-u.ac.jp/hospital/outline/resarch.html), with the approval of the Committee of Medical Ethics of Hirosaki University Graduate School of Medicine. This study was designed and carried out in accordance with the Declaration of Helsinki.
Results
Accuracy of patch-level and image-level classification
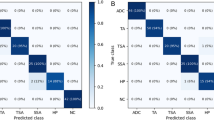
An averaged validation accuracy in the patches with label 0, 1, 2, 3, 4, 5 and 6 was 0.938, 0.876,0.957, 0.907, 0.929, 0.966 and 0.904, respectively (Table3). Table 4 displays the confusion matrix diagram depicting the outcomes of the image-level classification from the patch-based mapping across the entire image area using the trained CNN. The ground truth histology was located on the vertical axis, and the predicted histology was situated on the horizontal axis. Out of 1,390 pictures, 1,371 pictures were correctly classified into the correct histology with a total accuracy of 0.986. The precision and F1-scores were determined to be 0.973, 0.992, 1, 0.967 and 0.978, 0.991, 0.982, 0.980 for LGD, HGD, SMs, and SMd, respectively.
Examples of the patch-based mapping and image-level classification
Figure 3 illustrates the examples of input images, patch-level prediction map and bar graph of IoU. In cases 1, 2, 3 and 4, the ground truth histology was consistent with the predicted histology with the maximum intersection over union. In cases 5 and 6, HGD and SMd were misclassified as SMd and HGD, respectively. In these cases, misclassification of the surrounding background into the true lesion resulted in a lower intersection over union of the true lesion compared to the misclassified ones. A type of misclassification, stemming from an underestimation of the actual lesion compared to misclassified lesions across four SMs, has likely caused a decrease in accuracy relative to other lesions.
Discussion
In this study, we developed a trial CNN-based multi-class histology classifier model for detected colorectal neoplastic lesion in routine colonoscopy still images with NBI mode in common shooting condition. The NBI offers a significant advantage for CNN-based image classification thanks to its ability to provide high contrast or detailed pictorial information without requiring any pre-acquisition preparation. The diagnosis process includes patch-level histology mapping over the entire in-focus region of NBI image, trained on ResNet50 and the calculation of argmax among intersections over union between annotation mask and patch-level union masks for image-level histology. This model achieved an image-level accuracy of 0.986, suggesting its potential as a diagnostic tool.
The advancement of machine learning using CNN has enabled physicians to apply CAD of medical images in their specialized field. The American Society for Gastrointestinal Endoscopy AI Task Force16 stated that CAD plays a crucial role in screening and surveillance colonoscopy for colorectal cancer prevention. Similarly, a European Society of Gastrointestinal Endoscopy mentioned to the capability of AI for accurately predicting the histology of polyps from endoscopic images and improving the cost-efficiency and safety of colonoscopic colorectal cancer screening and surveillance17. The supervised learning of a CNN has enabled the development of a model for automated detection of colon polyps6, as well as binary classification models for distinguishing adenomatous from non-adenomatous polyps (with a ten-fold validation accuracy of 0.751)7, adenomatous from hyperplastic diminutive colorectal polyps (with an accuracy of 94%)8, and neoplastic polyps from non-neoplastic polyps (with a high confidence rate of 0.85)9.
However, a CNN model for multiclass differentiation among low-grade dysplasia, high-grade dysplasia, and carcinoma with superficial or deep submucosal invasion has not been developed. Although invading depth is determinant for therapeutic intervention (endoscopic resection or surgery), it has been evaluated so far by endoscopists with the use of knowledge-based criteria12,18 which inevitably suffers from a variety of biases in evaluation. One study has reported a CNN-based binary class prediction model for deeply submucosal invasive carcinoma with an overall accuracy of 85.5%, which is comparable to that of expert endoscopists19. Although it must be done with caution, comparison of model accuracy between studies with different designs revealed a prediction accuracy of 0.991 for carcinoma with deeply submucosal invasion in this study. To accurately compare the accuracy of ML models regardless of algorithm and class numbers, the promotion of a benchmark data set library with annotation masks20 is essential.
A patch-based CNN has been utilized for automated detection of a target area within a whole slide image in digital pathology21,22. This method has been recently applied for automated severity mapping along the entire colorectum in patients with ulcerative colitis from capsule endoscopy video files23, as it is considered to have an advantage when the object for classification is composed of topographically varying elements, such as severity or atypism. When reconstructing histologic maps in resected specimens, one often encounters topographic heterogeneity in the grade of dysplasia as well as invading depth. In this study, a single histology or label was assigned to a single lesion image, thus resulting in similar labels across the entire lesion area, which may have impacted the outcomes. None the less, a multi-class classification model developed from a trained patch-level classifier has achieved a high image-level accuracy of over 0.96, which may provide a potential diagnostic tool with minimal human input in routine colonoscopy.
The limitations of the study were: its single-center, retrospective nature, limited dataset size, and lack of external validation; moreover, other classification models including VGGNets, DenseNets and ViT were not explored; the applicability of the model to diagnosis in the subsequent endoscopy system is uncertain.
Data availability
The data generated or analyzed during this study are included in this published article. Some datasets generated and/or analyzed during the current study are not publicly available due to privacy but are available from the corresponding author on reasonable request.
References
Sasaki, Y., Hada, R. & Munakata, A. Computer-aided grading system for endoscopic severity in patients with ulcerative colitis. Dig. Endosc. 15, 206–209. https://doi.org/10.1046/j.1443-1661.2003.00246.x (2003).
Sasaki, Y., Fukuda, S., Mikami, T. & Hada, R. Endoscopic Quantification of Mucosal Surfaceroughness for grading severity of ulcerative colitis. Dig. Endosc. 20, 2891–2898. https://doi.org/10.1111/j.1443-1661.2008.00778.x (2008).
Sasaki, Y. et al. Computer-aided estimation for the risk of development of gastric cancer by image processing. In Artificial Intelligence in Theory and Practice III 197–204. https://doi.org/10.1007/978-3-642-15286-3_19 (2010).
Krizhevsky, A., Sutskever, I. & Hinton, G. E. ImageNet classification with deep convolutional neural networks. Commun. ACM 60, 84–90. https://doi.org/10.1145/3065386 (2017).
Hirasawa, T. et al. Application of artificial intelligence using a convolutional neural network for detecting gastric cancer in endoscopic images. Gastric Cancer 21, 653–660. https://doi.org/10.1007/s10120-018-0793-2 (2018).
Misawa, M. et al. Artificial intelligence-assisted polyp detection for colonoscopy: Initial experience. Gastroenterology 154, 2027–2029. https://doi.org/10.1053/j.gastro.2018.04.003 (2018).
Komeda, Y. et al. Computer-aided diagnosis based on convolutional neural network system for colorectal polyp classification: preliminary experience. Oncology 93, 30–34. https://doi.org/10.1159/000481227 (2017).
Byrne, M. F. et al. Real-time differentiation of adenomatous and hyperplastic diminutive colorectal polyps during analysis of unaltered videos of standard colonoscopy using a deep learning model. Gut 68, 94–100. https://doi.org/10.1136/gutjnl-2017-314547 (2019).
Rodriguez-Diaz, E. et al. Real-time artificial intelligence-based histologic classification of colorectal polyps with augmented visualization. Gastrointest Endosc. 93, 662–670. https://doi.org/10.1016/j.gie.2020.09.018 (2021).
Liu, X., Wang, C., Bai, J. & Liao, G. Fine-tuning pre-trained convolutional neural networks for gastric precancerous disease classification on magnification narrow-band imaging images. Neurocomputing 392, 253–267. https://doi.org/10.1016/j.neucom.2018.10.100 (2020).
Du, W. et al. Automatic classification of esophageal disease in gastroscopic images using an efficient channel attention deep dense convolutional neural network. Biomed. Opt. Express 12, 3066–3081. https://doi.org/10.1364/BOE.420935 (2021).
Hayashi, N. et al. Endoscopic prediction of deep submucosal invasive carcinoma: validation of the narrow-band imaging international colorectal endoscopic (NICE) classification. Gastrointest Endosc. 78, 625–632. https://doi.org/10.1016/j.gie.2013.04.185 (2013).
Bosman, F. T. et al. WHO Classification of Tumors of the Digestive System 160–165 (IARC Press, 2010).
Ojala, M. & Garriga, G. C. Permutation Tests for Studying Classifier Performance. J. Mach. Learn. Res. 11, 1833–1863 (2010).
He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition. In 2016 IEEE Conference on Computer Vision and Pattern Recognition. https://doi.org/10.48550/arXiv.1512.03385 (2016).
Berzin, T. M. et al. Position statement on priorities for artificial intelligence in GI endoscopy: A report by the ASGE Task Force. Gastrointest Endosc. 92, 951–959. https://doi.org/10.1016/j.gie.2020.06.035 (2020).
East, J. E. et al. Advanced endoscopic imaging: European Society of Gastrointestinal Endoscopy (ESGE) technology review. Endoscopy 48, 1029–1045. https://doi.org/10.1055/s-0042-118087 (2016).
Kobayashi, S. et al. Diagnostic yield of the Japan NBI Expert Team (JNET) classification for endoscopic diagnosis of superficial colorectal neoplasms in a large-scale clinical practice database. United Eur. Gastroenterol J. 7, 914–923. https://doi.org/10.1177/2050640619845987 (2019).
Lui, T. K. L. et al. Endoscopic prediction of deeply submucosal invasive carcinoma with use of artificial intelligence. Endosc. Int. Open 7, E514–E520. https://doi.org/10.1055/a-0849-9548 (2019).
Borgli, H. et al. HyperKvasir, a comprehensive multi-class image and video dataset for gastrointestinal endoscopy. Sci. Data 7, 283. https://doi.org/10.1038/s41597-020-00622-y (2020).
Hou, L., et al. Patch-Based Convolutional Neural Network for Whole Slide Tissue Image Classification. In 2016 IEEE Conference on Computer Vision and Pattern Recognition. 10.48550 /arXiv.1504.07947 (2016).
Roy, K., Bani, D., Bhattacharjee, D. & Nasipuri, M. Patch-based system for Classification of Breast Histology images using deep learning. Comput. Med. Imaging Graph. 71, 90–103. https://doi.org/10.1016/j.compmedimag.2018.11.003 (2018).
Higuchi, N. et al. Automated evaluation of colon capsule endoscopic severity of ulcerative colitis using ResNet50. PLoS ONE 17, e0269728. https://doi.org/10.1371/journal.pone.0269728 (2022).
Acknowledgements
We would like to thank Takashi Sawai, MD, PhD, Department of Pathology, Sendai City Medical Center, Toru Furukawa, MD, PhD, Fumiyoshi Fujishima, MD, PhD, Satoko Sato, MD, PhD, Department of Pathology, Tohoku University School of Medicine, and Miwa Uzuki, MD, PhD, Department of Nursing, Faculty of Medical Science and Welfare, for histologic evaluations.
Author information
Authors and Affiliations
Contributions
Y.S. and T.S. contributed to the study conception and design. T.S. and K.I. collected the clinical data and tumor annotation. Y.S. established the prediction model. Y.S. and T.S. wrote the first draft of the manuscript paper. M.M., H.S. and S.F. contributed to the review, and/or critical revision of the manuscript. All authors have approved the final article. Informed consent was obtained in the form of opt-out on our website (https://www.med.hirosaki-u.ac.jp/hospital/outline/resarch.html), with the approval of the Committee of Medical Ethics of Hirosaki University Graduate School of Medicine.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Shimizu, T., Sasaki, Y., Ito, K. et al. A trial deep learning-based model for four-class histologic classification of colonic tumor from narrow band imaging. Sci Rep 13, 7510 (2023). https://doi.org/10.1038/s41598-023-34750-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-34750-3
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.