Abstract
Lymph node metastasis is the most important prognostic factor in patients with lung squamous cell carcinoma. The current findings show that lymph node metastatic tumor cells can arise by programming metastasis in primary tumor cells. Thereby, the genetic alterations responsible for the metastasis could be detected in the primary tumors. This bioinformatic study aimed to determine novel potential prognostic biomarkers shared between primary lung squamous cell tumors (without lymph node metastasis) and lymphatic metastasis, using the Cancer Genome Atlas database. Differentially expressed genes were screened by limma statistical package in R environment. Gene ontology and biological pathways analyses were performed using Enrichr for up-regulated and down-regulated genes. Also, we selected lymph node metastasis related genes among DEGs using correlation analysis between DEGs and suitable references genes for metastasis. Receiver operating characteristic curves was applied using pROC and R package ggplot2 to evaluate diagnostic value of differentially expressed genes. In addition, survival and drug resistance analyses were performed for differentially expressed genes. The miRNA-mRNA interaction networks were predicted by miRwalk and TargetScan databases and expression levels analysis of the miRNAs which were mainly targeting mRNAs was performed using UALCAN database. Protein–protein interaction network analysis and hub genes identification were performed using FunRich and Cytoscape plugin cytoHubba. In this study, a total of 397 genes were differentially expressed not only with a significant difference between N + vs. normal and N0 vs. normal but also with significant difference between N + vs. N0. Identified GO terms and biological pathways were consistent with DEGs role in the lung squamous cell carcinoma and lymph node metastasis. A significant correlation between 56 genes out of 397 differentially expressed genes with reference genes prompted them being considered for identifying lymph node metastasis of lung squamous cell carcinoma. In addition, SLC46A2, ZNF367, AC107214.1 and NCBP1 genes were identified as survival-related genes of patients with lung squamous cell carcinoma. Moreover, NEDD9, MRPL21, SNRPF, and SCLT1 genes were identified to be involved in lung squamous cell carcinoma drug sensitivity/resistance. We have identified several numbers of miRNAs and their related target genes which could emerge as potential diagnostic biomarkers. Finally, CDK1, PLK1, PCNA, ZWINT and NDC80 identified as hub genes for underlying molecular mechanisms of lung squamous cell carcinoma and lymphatic metastasis. Our study highlights new target genes according to their relation to lymph node metastasis, whose expressions in the primary lung squamous cell carcinoma are able to accurately assess the presence of lymphatic metastasis.
Similar content being viewed by others
Introduction
Lung Squamous cell carcinoma (LUSC) is a common subtype of non-small cell lung cancer (NSCLC), accounting for 20% of all cases and causing more than 400,000 new cases worldwide each year1,2. In general, LUSC tends to be aggressive, and 60% of patients are diagnosed with local and distant invasion3,4,5. Metastasis of LUSC is a major cause of mortality, often presenting diagnostic challenges and is the main drawback of successful treatments6,7.
During metastasis, malignant cells may invade through the blood or lymphatic vessels, however, spread to the hilar or interlobar lymph nodes (N1), then to the ipsilateral (N2), or contralateral mediastinal lymph nodes (N3) is the primary route for LUSC and other NSCLC histologic subtypes metastasis, which facilitate tumor distant metastasis6,8,9. Lymphatic vessels with slower flow rate along with reduce shear stress due to impaired lymphatic vessels contraction increase the tumor cells survival more than systematic vessels. In addition, stagnation lymph flow can directly connect cancer cells to each other, as well as activate the integrin-mediated signaling pathway and increase tumor growth in the vessel10. Also, there are many gap junctions in the discontinuous basement membrane, which causes leakage of lymphatic vessels and metastatic spread of tumor cells11. Accordingly, the prognosis of LUSC greatly depends on the presence of lymph node metastasis (LNM)6. It has recently been suggested that although the metastasis appears in the later stages of cancer, the initiation of metastatic programming occurs much earlier in the primary tumor cells12,13.
Genetic alterations in some of the tumor cells in the primary tumor give them advantages that enable them to form large populations in the primary tumor mass. These beneficial traits include acquisition of mitogenic signal propagation, capability to resist growth inhibiting signals, the ability to prevent apoptosis or programmed cell death, and lymphangiogenesis growth induction. A subset of these genetic changes, acquired by cells in the early stages of tumorigenesis confer the ability to metastasis later. It seems that there is a genetic programing to enable cancer cells in the primary tumors to metastasize14,15,16. Interestingly, some of the genomic aberrations required for progression and metastatic spread identified in LUSC patients with lymph node metastasis correspond to aberrations that impair the function of growth regulatory proteins and inactive growth inhibitory pathways in primary LUSC17. Therefore, the identification of molecular biomarkers in the early stages of tumor, whose expressions are able to accurately predict the progression to lymph node metastasis, is strongly needed18,19.
Recent advances in molecular oncology have described some coding and non-coding genes including micro RNAs (miRNAs), as molecular biomarkers in the early cancer detection and prognostic markers20. Uncovering biomarkers whose differential expression in primary tumor is strongly associated with the potential LUSC tumor progression could improve the prognosis by developing novel and groundbreaking therapeutic approaches that target specific genetic alterations in primary tumors.
Identification of key genes involved in molecular mechanism of cancer, especially for metastasis and lymph node metastasis, has been addressed in certain genetic studies21,22, and in some of them it may even have been shown that the same genes are shared between the primary and metastatic lesions. However, to our knowledge, there are no studies that identify genes involved not only in growth and development of tumor but also in lymph node metastasis in LUSC. Hence, the present bioinformatic study aimed to determine new molecular pathways and potential novel prognostic biomarkers expressed differentially between N + and N0 compared to normal, which were also significantly expressed differentially in N + compared to N0 in LUSC using The Cancer Genome Atlas (TCGA) database.
Methods
Data collection and preparation
Approval for this bioinformatics-based study was obtained from the Islamic Azad University- Kazerun Branch Ethics Committee (IR.IAU.KAU.REC.1400.141). All methods were performed in accordance with the guidelines and regulations of the Islamic Azad University- Kazerun Branch.
In this study, the Cancer Genome Atlas Lung Squamous Cell Carcinoma (TCGA-LUSC) data (Accession number: caArray_EXP-592) were downloaded from LinkedOmics (http://linkedomics.org/). All genes with insignificant and close to zero expression levels were removed from the matrix using the counts per million (CPM) method. Also, normalization was performed using the trimmed mean of the m-values (TMM) method, and all values were converted to log2 scale. The flowchart is presented in Fig. 1.
Differential gene expression analysis
To perform differential gene expression (DGE) analysis, linear model of the limma package in the R environment was used. All samples were divided into three groups: patients with N + (including N1, N2 and N3) metastasis (n = 176), patients without lymph node metastasis or N0 condition (n = 320) and normal tissue samples (n = 49). DGE was performed between N + vs. normal, N0 vs. normal and N + vs. N0. The selection criteria were the adj. p value < 0.05 and log fold change was not considered. Genes were selected as DEGs in final that were not only significantly deregulated in N + and N0 compared to normal, but also deregulated in N + compared to N0.
Signaling pathway and gene ontology analyses
Pathway analysis was done using Enrichr online web tool, comprising analyses of REACTOME pathways and Wikipathways (https://amp.pharm.mssm.edu/Enrichr). Furthermore, we performed Gene ontology (GO) function analysis including the cellular component (CC), molecular function (MF), and biological process (BP), and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis on the differentially expressed genes.
Genes selection for lymph node metastasis
First, we conducted a literature search to find suitable reference genes for lymph node metastasis in lung cancer by searching PubMed, Google Scholar, Embase, ScienceDirect and Cochrane Library with the keywords: “Metastasis”, “Gene Expression”, “Lung cancer”and “Lymph node metastasis”. For example, the search query in PubMed was (Metastasis [MeSH Terms]) AND (Gene Expression [MeSH Terms]) AND (Lung cancer [MeSH Terms]).Then determined the correlation coefficient (CC) between all the differentially expressed genes with the reference genes. Differentially expressed genes with a correlation coefficient range of > 0.4 or < − 0.4 with the reference genes were selected as lung cancer progression and lymph node metastasis.
Clustering and ROC analysis
DE genes presented a positive correlation with the reference genes were clustered through the kmeans method to draw a heat map. We divided candidate genes into the number of groups equal to the number of clusters and applied Receiver Operating Characteristic (ROC) curve analysis using the pROC package to examine how each gene in each cluster could distinguish cancer from normal samples. In addition, the calculated sensitivities and specificities were used to draw ROC curves through the ggplot2 R package.
Survival analysis
First, the clinical data including T-stage, N-stage, pathological-stage, gender, tobacco smoking history, age, state of living, and follow-up time were added to the expression matrix. Then, all expression values of each gene were scaled between zero and one, and these scaled values were rounded into 1–10 to have ten gene expression levels. Next, the univariate Cox regression analysis was performed on each gene to evaluate their role in the patient survival (log rank test < 0.05) also, the same analysis was performed on them for survival-related clinical data.
Selected genes from univariate Cox regression analysis of identified DE genes were used to compute a risk score model to assess the patients' survival rate with the following formula:
where Wj is the multivariate coefficient for gene j, Expij is the expression value of gene j in patient i, and n is the number of testing genes.
In addition, multivariate cox regression analysis was performed considering significant clinical parameters and risk scores. Patients were divided into high and low-risk groups based on the average risk score as the cut-off value and finally the survival plots were depicted based on each gene and model genes.
Drug resistance/sensitivity analysis
Drug resistance analysis was performed using the PharmacoGX package in R which is connected to various public databases to find out which candidate genes are related to cell drug response. We used lung samples from Cancer Cell Line Encyclopedia (CCLE) to perform our analysis. The association between expression of each gene and drugs was calculated based on IC50 and other parameters such as estimate, p.value, and the number of samples was also calculated. The estimate measures the correlation of each gene to sensitivity or resistance to each drug. The number is between − 1 and 1; − 1 means that the higher gene expression causes the higher sensitivity to the specific drug, while 1 indicates the highest resistance to the drug based on the increased gene expression. Only Food and Drug Administration (FDA)-approved drugs were checked in this study (p value less than 0.05).
Analysis of mRNA- miRNAs interaction
For all identified DE genes, possible interactions with miRNAs were checked through TargetScan (https://www.targetscan.org/) and miRWalk (http://mirwalk.umm.uni-heidelberg.de/) databases and those miRNAs that could pass double-checking process in both databases, were selected. Cytoscape v 3.7 was applied to visualize miRNA-mRNA interaction networks. In addition, we used the UALCAN database (http://ualcan.path.uab.edu)23 to analyze expression levels of the miRNAs which were targeting any more mRNAs in LUSC.
Identification of hub genes
First, FunRich tool version 3.1.324 was applied to illustrate the interaction of differentially expressed genes. Then, hub genes were selected using the Cytoscape plugin cytoHubba. Protein–protein interaction network of these hub genes were constructed by STRING (http://string-db.org; version 10.5) and visualized by Cytoscape.
Moreover, to identify critical upstream signals that lead to lymph node metastasis we expanded the network by adding Human Protein Reference Database (HPRD)-derived direct interacting partners.
Soft wares and statistics
All statistical and in-silico analyses were performed in R environment version 4.0.1 and 0.05 was considered as cut-off p.value in all steps of the study. Cytoscape version 3.9.1, GraphPad Prism version 9, FunRich version 3.1.3 and cytoHubba, were used for network analysis and visualization.
Informed consent
This work is not involving “human participants” because in this bioinformatics-based study, as a Secondary Research collected information for another primary activity has been used. Generally speaking, all samples in TCGA have been collected and utilized following strict human subjects protection guidelines, informed consent and IRB review of protocols.
Results
Identification of DEGs
A total of 397 genes were significantly showed expressed differentially between N + and N0 compared to normal, which were also significantly expressed differentially in N + compared to N0 (including 281 upregulated (Fig. 2, Supplementary File 1) and 116 genes downregulated (Fig. 3, Supplementary File 2)).
Reactome /GO/KEGG analysis for all identified DEGs
We utilized Enrichr web tools to identifying possible signaling pathways for all 397 DE genes. In REACTOME analysis (Table 1), the most outstanding pathway for down-regulated DEGs was “surfactant metabolism pathway” which provides valuable insights into the pathways implicated in lung cancer tumor-infiltrating lymphocytes. From the results of Wikipathway analysis for down-regulated DEGs (Table 2), the most highly correlated pathway was “Lung fibrosis”. There is only one cellular component ontology of down-regulated DEGs enriched categories, which were associated with alveolar lamellar body (Table 3).
Gene enrichment analysis results of up-regulated DEGs using the Reactome Pathway (Table 4), Wikipathway (Table 5), KEGG Pathway (Table 6), Gene ontology Biological Process, Gene ontology Cellular Component and Gene ontology Molecular Function (Table 7), mainly concentrated in “mitotic division,” and “cell cycle”.
Identification of lymph node metastasis diver genes among all DEGs
We retrieved 143 articles related to lung cancer lymph node metastasis following strictly filtering based on exclusion criteria (conference abstracts, letters, and animal model studies were excluded) by searching PubMed, Google Scholar, Embase, Sci-enceDirect and Cochrane Library. Eventually, a total of four genes including, CD151, MMP1, PVT1, and SKP2 genes were selected as reference genes based on their known functions in lung cancer progression and lymph node metastasis which have been pointed out in the found articles. CD151 is a member of the transmembrane 4 superfamily, also known as the tetraspanin family. It is involved in cellular processes including cell adhesion and enhances cell motility, invasion and metastasis of cancer cells. MMP1 is a member of zinc-dependent endopeptide proteases family, which is prominently associated with extracellular matrix destruction (a genetic alteration responsible for programming metastasis in primary tumors) that is critical to the development of a primary tumor and its metastatic progeny. The PVT1 gene is known as an oncogene, and its overexpression is associated with many types of cancers, including breast and ovarian cancers. The SKP2 gene encodes a member of the F-box protein family, this gene is established as a protooncogene involved in the pathogenesis of lymphomas (Supplementary File 3).
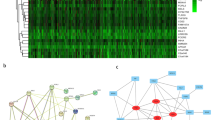
Correlation results showed that out of 397 differentially expressed genes in our study, 157 were highly correlated to CD151, 77 with MMP1, 160 with PVT1, and 227 with SKP2 (CC > 0.4 or < − 0.4). 56 of them were shared between all four reference genes (Supplementary Table 1, Fig. 4A) and selected as lymph node metastasis driver genes.
Genes that are significantly expressed differentially between N + and N0 compared to normal, which were also significantly expressed differentially in N + compared to N0. (A) Venn diagram of the significantly correlated DEGs with the reference genes, (B) 56 candidate genes are significantly correlated with CD151, MMP1, PVT1, and SKP2 and divided into two clusters.
In addition, Kmeans clustering results indicated that these 56 genes could be divided into two up-regulated (with 50 genes) and down-regulated (with 6 genes) clusters, and the correlation between genes and clusters was shown by heatmap (Fig. 4B).
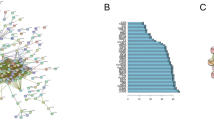
The ROC analysis was performed for each cluster (Fig. 5) and have shown that these genes could be used as excellent potential biomarkers (AUC > 0.9) (Supplementary Table 2).
A four‐gene‐based prognostic model may influence patient survival
The univariate cox regression results indicated that 45 of 397 DE genes might affect patients' survival. On the other hand, only T-stage, N-stage, tobacco smoking history, and pathological-stage showed a significant association with survival among clinical parameters (Table 8). Also, multivariate CoxPH analysis was performed and results showed that SLC46A2, ZNF367, AC107214.1 and NCBP1 genes can significantly affect survival independent of clinical parameters (Table 9). Also, T-stage and N-stage and pTNM showed p value < 0.05 in multivariate results (Table 9).
The risk score models for these four genes were calculated with the following formula:
And, was performed for each patient to obtain another multivariate Cox regression analysis to see whether it can independently impact survival. The results revealed that the risk score could significantly affect the survival of patients (p value < 0.0001). Moreover, we tested the model by dividing patients into high and low-risk groups and drawing their survival plots (Fig. 6A). Also, the survival plot for each gene in two conditions of high expression and low expression is shown in Fig. 6B.
Classification of patients into two high and low-risk groups based on risk score model consisting of four genes and their survival rate. (A) The survival plot of high and low-risk patients based on median risk score and the risk score distribution of patients as a scatter plot. (B) The survival plot of each gene applied to compute the risk score based on each gene's high and low expression levels, using their median as the cut-off value.
Mainly, survival analyses showed that the same expression of SLC46A2, ZNF367, AC107214.1 and NCBP1 genes in the primary and LNM lesions can provides more potent prognostic information when no analyses of the primary tumor have been done. This prediction gene signature of prognosis in early LUSC will support treatment decision-making.
Contribution of five crucial genes to Dovitinib and Paclitaxel resistance/sensitivity in LUSC
We used the CCLE database to assess the association of 397 DE genes with drugs used for lung cancer chemotherapy applying PharmacoGX package in the R environment. IC50 was used to compare the expression of each gene for each drug and calculate a False Discovery Rate (FDR). Our parameter called estimate shows the association of each drug and gene with a number in the range of − 1 and 1 (1 shows the highest resistance and − 1 shows the highest sensitivity). We identified abnormal expression NEDD9 gene confer resistance to Dovitinib with an estimate value of 0.77. On the other hand, high expression of SNRPF and SCLT1 genes cause sensitivity to Paclitaxel with estimate values of around − 0.7. However, MRPL21 might cause sensitivity to both drugs, with the estimate value around − 0.7 (Table 10).
Construction of mRNAs-miRNAs interaction network
Our results revealed that there are probably 526 miRNA interactions with candidate genes. Among downregulated mRNAs, KCND3, SCN1A, NFIA, and CYB561D1 were targeted by the highest number of miRNAs. However, hsa-miR-526b-3p and hsa-let-7e-5p target the most genes in this group. On the other hand, among overexpressed mRNAs group, TLK2, CDC25A, FAM104A, and HNRNPU were targeted by more than 10 miRNAs. The number of miRNAs in this group is much more; among them, hsa-let-7b-5p, hsa-miR-129-5p, hsa-miR-3681-3p, hsa-miR-520d-3p, hsa-miR-497-5p, hsa-miR-216a-3p revealed the highest number of mRNAs target (Fig. 7A,B). NEDD9 was the only gene among the drug resistance mRNAs that was targeted by 6 miRNAs. Then, expression levels of the miRNAs which were targeting more mRNAs including, hsa-miR-526b-3p, hsa-let-7e-5p, hsa-let-7b-5p, hsa-miR-129-5p, hsa-miR-3681-3p, hsa-miR-520d-3p, hsa-miR-497-5p, hsa-miR-216a-3p were analyzed by the UALCAN website. As shown in the Fig. 8, the miRNA expression of hsa-miR-526b-3p, hsa-let-7b-5p, hsa-miR-129-5p, hsa-miR-3681-3p, hsa-miR-497-5p, hsa-miR-216a-3p were differ significantly between the tumor tissues and normal (p value < 0.05). However, the expression of hsa-let-7e-5p and hsa-miR-520d-3p in tumor tissues were not significant.
mRNA-miRNA interactions. (A) is the overexpressed group interaction, and (B) is the down-expressed group. The nodes represent genes, and miRNAs and edges represent interactions. The bigger nodes in the middle of the figure depict mRNAs, while the smaller ones are miRNAs. The light green nodes are survival-related mRNAs.
Hub genes selection
The Protein–protein interaction (PPI) network of the 397 differentially expressed genes created by the FunRich software. CDK1, PLK1, PCNA, ZWINT, NDC8 were the important nodes (genes) with many edges with at least 10 proteins (Fig. 9A), and all of which selected and ranked by at least one of the methods available in the cytoHubba, one of the Cytoscape plugin. CytoHubba provides a simple interface to analyze a network based on shortest paths by computing eleven topological analysis methods including Degree, Edge Percolated Component, Maximum Neighborhood Component, Density of Maximum Neighborhood Component, Maximal Clique Centrality and six centralities (Bottleneck, EcCentricity, Closeness, Radiality, Betweenness, and Stress) in one stop shopping way25. Among them, ZWINT and NDC80 were identified in this study for the first time as our knowledge as hub genes for lymph node metastasis in LUSC. CDK1 and ZWINT were as 10 top genes in more than four cytoHubba methods, but NDC80 was only in the bottleneck cytoHubba method. Interestingly, CDK1, ZWINT and NDC80 were commonly as 10 top genes in bottleneck method (Fig. 9B). In this work, a directed statistically significant reliable Protein–protein interaction network was constructed from the hub genes (Fig. 10). In the present study we displayed (Supplementary File 4) direct and indirect interactions to point out signaling pathways which involved in a lymph node metastasis upstream of hub genes.
Discussion
Given the importance of lymph node metastasis in the process of metastasis in LUSC, in this study we compared the gene-expression profiles of patients with N + metastasis, without lymph node metastasis and normal tissue samples to find genes involved in lymph node metastasis encoded in the primary tumor.
Our results showed that a total of 281 and 116 genes were significantly up- and down- regulated, respectively, not only in the N0 and N + compared with normal but also in N + compared to N0. Among the 397 identified DE genes, 56 genes were significantly correlated with CD151, MMP1, PVT1, and SKP2 as reference genes for lymph node metastasis, and due to meeting best criteria in the ROC analysis were known be excellent for the distinguish lymph node metastasis cancer from primary LUSC without metastasis (N0). Takada M et al. in 200426 achieved diagnostic genes which predicted lymph node metastasis in lung squamous cell carcinomas and lung adenocarcinomas patients, compared to our study, they used microarray data and lower samples. Importantly, we analyzed genes that were not only deregulated significantly in N + and N0 rather than normal, but also in N + compared to N0, which could have greater predictive value for metastatic status.
The results of functional enrichment analysis showed that the DEGs were related to pathways such as “surfactant metabolism pathway” (which has a role in the regulation of the cancer microenvironment and has been suggested as a target in cancer immunotherapy27),“Lung fibrosis” (Several studies have provided histopathological evidence of an increased incidence of lung cancer in pulmonary fibrosis28), “mitotic division,” and “cell cycle” which are consistent with DEGs role in LUSC lymph node metastasis.
In addition, in our study among the total differentially expressed genes, SLC46A2, ZNF367, AC107214.1 and NCBP1 were significantly associated with overall survival in multivariate Cox regression analyses. SLC46A2 belongs to the solute carrier proteins (SLC) superfamily, which includes more than 400 transport proteins in 65 families and transport a wide variety of substances across the cell membrane and they also transport drugs across the lipid bilayer, so they play a role in carcinogenesis29. Consistent with our study, previously published article also described SLC46A2 as a prognostic biomarker for lung squamous cell carcinomas and lung adenocarcinomas patients30. Interestingly, our results showed that the SLC46A2 gene was significantly associated with metastasis and survival, indicating the importance of this gene in LUSC.
Zinc finger protein 367 (ZNF367) belongs to the zinc finger protein family and functions as a transcriptional factor with a unique zinc finger motif. Several studies have shown that this gene involved in overall survival and prognosis of breast cancer31,32. Nuclear cap-binding protein 1 (NCBP1) is require for capped RNA synthesis and is able to mediated proliferation, migration and invasion by participate in transcriptional and post-transcriptional processes and, it has been shown that NCBP1 to be associated with survival in lung cancer patients33,34. According to our results and previous studies, SLC46A2, ZNF367 and NCBP genes could be introduced as important factors for survival and clinical management in LUSC. In this study, for the first time to our knowledge, we report ZNF367 and AC107214.1 genes in LUSC as an important factor in survival.
In addition, we have identified NEDD9, MRPL21, SNRPF, and SCLT1 genes that can predict the response of LUSC patients to chemotherapy. Previous studies have shown that NEDD9 plays a key role in tumor initiation, progression, and metastasis35, and Kondo et al. found that NEDD9 could confer resistance to the chemotherapeutic agents in lung cancer 293 T, A549, PC-9 and PC-14 cell lines36. The mitochondrial proteins which are important for various mitochondrial functions, such as ribosomal subunit proteins (MRPL21), may contribute to drug resistance, for example, Huang et al. in 2020 showed that increased levels of MRPL21 is responsible for treatment strategies to reduce the chemotherapeutic resistance to recurrent acute myeloid leukemia37,38. It has been reported that SCLT1 by inducing apoptosis combined with chemotherapy drugs enhanced cancer cell death. SCLT1- induced apoptosis might be related to DNA damage response39 and Liu et al. have shown SCLT1 to be useful biomarker to predict response to chemotherapy in patients with hepatocellular carcinoma39. SNRPF encoded spliceosome small nuclear ribonucleoproteins. SNRPF dysregulation has been reported in some cancers, including colorectal, laryngeal squamous cell carcinoma cells and renal cell carcinoma, but not in lung cancer40. Therefore, the SNRPF, SCLT1 and MRPL21 genes presented in this article are considered as novel biomarkers for predicting the response of LUSC patients to chemotherapy, as they have not been published before.
Further, thorough the analysis of mRNA-miRNA interaction networks, we found DEGs which were targeted by more miRNAs, including KCND3, SCN1A, NFIA, CYB561D1, TLK2, CDC25A, FAM104A, and HNRNPU. As a member of the nuclear factor I family, NFIA can lead to uncontrolled cell proliferation and tumor initiation and progression and has been reported by Zhao et al. In 2017 in patients with squamous cell cancer, adenocarcinoma and large cell carcinoma41. So far, there have been no reports on other three down regulated mRNAs (KCND3, SCN1A and CYB561D1) in lung cancer targeted by miRNAs. Among up regulated mRNAs found in this study, the association of mutant TLK2 (A member of Tousled-like kinases family) has been reported in breast cancer42. However, there are no reports on the function of TLK2 in the context of lung cancer. Overexpression of cell division cycle 25A (CDC25A), a member of Cdc25 family, has also been reported as a poor prognosis marker in NSCLC. Li et al. in 2020 has been reported that miR-365 target CDC25A mRNA and reduce the expression of CDC25A in lung cancer43. Interestingly, so far, there are no report in the literature on the FAM104A and HNRNPU.
Moreover, we found various miRNAs including hsa-miR-526b-3p and hsa-let-7e-5p, hsa-let-7b-5p, hsa-miR-129-5p, hsa-miR-3681-3p, hsa-miR-520d-3p, hsa-miR-497-5p and hsa-miR-216a-3p could target the most candidate differentially expressed genes. Interestingly, some of these miRNAs such as hsa-miR-526b-3p was also detected as hub miRNAs associated with colorectal cancer progression by Motieghader et al.44. Cell migration mediated by let-7e-5p has been confirmed in the colon carcinoma. Although its underlying mechanism is unclear, modulation of MYC pathways assumed in this study45. Gharib et al. suggested that miR-497-5p overexpression affect the development of colorectal cancer by regulating cell proliferation. Therefore, miR-497-5p upregulation could be considered as a potential therapeutic target46. The functional molecular of hsa-let-7b-5p, hsa-miR-129-5p, hsa-miR-3681-3p, hsa-miR-520d-3p, hsa-miR-216a-3p and their targets in lymph node metastasis have not been reported previously. Also, the miRNA expression of hsa-miR-526b-3p, hsa-let-7b-5p, hsa-miR-129-5p, hsa-miR-3681-3p, hsa-miR-497-5p, hsa-miR-216a-3p were found differ significantly between the tumor tissues and normal in further analysis.
Generally speaking, identified miRNAs and target mRNAs show various expression patterns that are proving to be clinically relevant to LUSC's lymph node metastasis. Such observations indicate that a subgroup of miRNAs play an important role in lung squamous cell carcinoma lymphatic progression.
Finally, CDK1, PLK1, PCNA, ZWINT and NDC80 identified as hub genes for underlying molecular mechanisms of lung squamous cell carcinoma lymphatic metastasis. Among them, CDK1, ZWINT and NDC80 genes were identified for the first time to our knowledge in lymph node metastasis of LUSC. The ZW10 interacting kinetochore protein (ZWINT), encodes a protein involved in kinetochore function during mitotic cycle. Overexpression of ZWINT often resulted in abnormal mitosis in human cancers, which is a common feature of most malignancies. In addition, nuclear division cycle 80 (NDC80) is another mitotic regulator highly expressed in various human malignancies. NDC80, participate in regulation of mitosis by spindle assembly checkpoint47. NDC80 may interact with a kinase NEK2 and one of the centrosome proteins CEP250 to play a role in lymph node metastasis in cancers48,49.
Cyclin dependent kinase 1 (CDK1) as a serine/threonine kinase, regulate the cell cycle by promoting the G2/M as well as G1/S transitions. Alteration in CDK1 activity due to upregulation of CDK1 is closely related to cell proliferation50. In line with our results, CDK1 was one of the 20 key hub genes related to pancreatic cancer metastasis and prognosis. CDK1 and MYC can promote the formation of metastasis niches by regulating the activity of CD4 + T cells51. Also, Chen et al. found that CDK1 promotes the EMT and migration of head and neck squamous cell carcinomas (HNSCCs) cells by inhibiting ∆Np63α52. In addition, CDK1 can bind to FGFR1, which leads to cell proliferation, invasion and migration and affects lymph node metastasis48,53.
PCNA plays an important role in DNA replication, but is also associated with other functions such as chromatin remodeling, DNA repair, sister-chromatid cohesion and cell cycle control54,55. It has been shown taht PCNA was significantly related to lymph node metastasis in gastric cancer56. Interestingly, a preveoius study has shwon that PCNA as an oncogene can be involved in NSCLC progression through up-regulation of STAT357.
Polo-like kinase 1 (PLK1) is a highly conserved serine/threonine kinase with important roles in mitosis and cell cycle regulation58. PLK1 is highly expressed in multiple tumors and promotes tumor cell proliferation and cell transformation, and is associated with clinical stages and invasion59. Previous studies have shown that PLK1 can play a role invasion and metastasis through beta-1 integrin by vimentin phosphorylation in breast cancer60 or through CD44v6, matrix metalloproteinase (MMP)-2, and MMP-9 in thyroid cancer61. Interestingly, consistent with our results, in NSCLC, active PLK1 has been shown to upregulate the levels of p-Smad2, a TGF-β effector, leading to the promote metastasis and invasion thorough upregulates TNFAIP662. Thus, it is reasonable that deregulation of identified hub genes is associated not only with tumorigenesis but also with lymph node metastasis.
Generally speaking, in the present work ROC curve analysis was used to evaluate diagnostic ability of DEGs which could be useful at the time of diagnosis of the initial stage in determining the course of a LUSC lesion (from tumor promotion, malignant conversion to lymphatic metastasis). The prognostic importance of DEGs is also assessed that provides information on the likely patient health outcome. Since the decision about treatment choice in the early-stage of cancer may be difficult for clinicians, LUSC drug resistance/sensitivity associated DEGs were also confirmed. Simply put, we performed a series of analyses to ensure whether our new expression profile (including, differentially expressed genes with significant differences not only between N + vs. normal and N0 vs. normal but also between N + vs. N0), could be functional at different phases of patient management. Besides, five identified hub genes were herald as underpin molecular mechanisms of lung squamous cell carcinoma lymphatic metastasis. Although, in the present study lymph node metastasis has been speculated as a result of disruption of normal regulation of the cell cycle caused by deregulation of these hub genes, it needs to be credible.
Conclusion
In summary, this study, by identifying lymph node metastasis predicting biomarkers and improving understanding of the less well-known genes of LUSC, hopes to address problems with poor prognosis of this subset of patients due to delayed diagnosis. The results of the present study should be interpreted with caution because only bioinformatics techniques were used to identify biomarkers, and the results were not confirmed through in vitro study.
Data availability
Dataset analyzed during the current study (TCGA-LUSC dataset) were previously generated and are available from LinkedOmics (http://linkedomics.org/). Also, this study includes research data from UALCAN database (http://ualcan.path.uab.edu) available in web link.
Abbreviations
- LUSC:
-
Lung squamous cell carcinoma
- NSCLC:
-
Non-small cell lung cancer
- LNM:
-
Lymph node metastasis
- miRNA:
-
Micro RNA
- TCGA:
-
The cancer genome atlas
- CPM:
-
Counts per million
- TMM:
-
Trimmed mean of the M-values
- DGE:
-
Differential gene expression analysis
- GO:
-
Gene ontology
- CC:
-
Cellular component
- MF:
-
Molecular function
- BP:
-
Biological process
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- CC:
-
Correlation coefficients
- ROC:
-
Receiver operating characteristic
- CCLE:
-
Cancer cell line encyclopedia
- FDA:
-
Food and drug administration
- HPRD:
-
Human protein reference database
- FDR:
-
False discovery rate
- AUC:
-
Area under curve
- PPI network:
-
Protein–protein interaction network
- SLC:
-
Solute carrier proteins
- ZNF367:
-
Zinc finger protein 367
- NCBP1:
-
Nuclear cap-binding protein 1
- CDC25A:
-
Cell division cycle 25A
- ZWINT:
-
ZW10 interacting kinetochore protein
- NDC80:
-
Nuclear division cycle 80
- CDK1:
-
Cyclin dependent kinase 1
References
Jiang, W. et al. The efficacy and safety of albumin-bound paclitaxel plus carboplatin as neoadjuvant therapy for potentially resectable lung squamous cell carcinoma: a real-world retrospective cohort study. Transl Lung Cancer Res. 11(4), 647–655. https://doi.org/10.21037/tlcr-22-252 (2022).
Lu, J. et al. Identification of tumour-infiltrating myeloid subsets associated with overall survival in lung squamous cell carcinoma. J Pathol. https://doi.org/10.1002/path.6015 (2022).
Savini, A. et al. Squamous cell carcinoma of the lung: clinical criteria for treatment strategy. J. Cancer Metastasis Treat. 1, 90–93. https://doi.org/10.4103/2394-4722.157974 (2015).
Liu, Y., Yang, J., Sun, X. J., Lu, S. N. & Liu, S. Positive therapy outcome of lung squamous cell cancer: A case report. Exp. Ther. Med. 23(6), 433. https://doi.org/10.3892/etm.2022.11360 (2022).
Langer, C. J. et al. Incremental innovation and progress in advanced squamous cell lung cancer: Current status and future impact of treatment. J. Thorac. Oncol. 11(12), 2066–2081. https://doi.org/10.1016/j.jtho.2016.08.138 (2016).
Dong, S., Zhu, P. & Zhang, S. Expression of collagen type 1 alpha 1 indicates lymph node metastasis and poor outcomes in squamous cell carcinomas of the lung. PeerJ 8, e10089. https://doi.org/10.7717/peerj.10089 (2020).
Rydzanicz, M. et al. Nonrandom DNA copy number changes related to lymph node metastases in squamous cell carcinoma of the lung. Neoplasma 55(6), 493–500 (2008).
Podgrabinska, S. & Skobe, M. Role of lymphatic vasculature in regional and distant metastases. Microvasc. Res. 95, 46–52. https://doi.org/10.1016/j.mvr.2014.07.004 (2014).
Abdul Pari, A. A., Singhal, M. & Augustin, H. G. Emerging paradigms in metastasis research. J. Exp. Med. 218(1), e20190218. https://doi.org/10.1084/jem.20190218 (2021).
Zhou, H., Lei, P. J. & Padera, T. P. Progression of metastasis through lymphatic system. Cells 10(3), 627. https://doi.org/10.3390/cells10030627 (2021).
Paduch, R. The role of lymphangiogenesis and angiogenesis in tumor metastasis. Cell Oncol. (Dordr.) 39(5), 397–410. https://doi.org/10.1007/s13402-016-0281-9 (2016).
Gui, P. & Bivona, T. G. Evolution of metastasis: New tools and insights. Trends Cancer 8(2), 98–109 (2022).
Yamamoto, K. N., Nakamura, A. & Haeno, H. The evolution of tumor metastasis during clonal expansion with alterations in metastasis driver genes. Sci. Rep. 5, 15886. https://doi.org/10.1038/srep15886 (2015).
Bernards, R. & Weinberg, R. A. A progression puzzle. Nature 418(6900), 823. https://doi.org/10.1038/418823a (2002).
Gupta, G. P. & Massagué, J. Cancer metastasis: Building a framework. Cell 127(4), 679–695. https://doi.org/10.1016/j.cell.2006.11.001 (2006).
Chaffer, C. L. & Weinberg, R. A. A perspective on cancer cell metastasis. Science 331(6024), 1559–1564. https://doi.org/10.1126/science.1203543 (2011).
Johnson, J. L., Pillai, S. & Chellappan, S. P. Genetic and biochemical alterations in non-small cell lung cancer. Biochem. Res. Int. 2012, 940405. https://doi.org/10.1155/2012/940405 (2012).
Zhao, J., Li, J., Li, N. & Gao, S. Clinical significance of skipping mediastinal lymph node metastasis in N2 non-small cell lung cancer. J. Thorac. Dis. 10(3), 1683–1688. https://doi.org/10.21037/jtd.2018.01.176 (2018).
Fazi, F. & Fontemaggi, G. MicroRNAs and lymph node metastatic disease in lung cancer. Thorac. Surg. Clin. 22(2), 167–175. https://doi.org/10.1016/j.thorsurg.2011.11.001 (2012).
Alarcón-Zendejas, A. P. et al. The promising role of new molecular biomarkers in prostate cancer: From coding and non-coding genes to artificial intelligence approaches. Prostate Cancer Prostatic Dis. 25(3), 431–443. https://doi.org/10.1038/s41391-022-00537-2 (2022).
Euer, N. et al. Identification of genes associated with metastasis of mammary carcinoma in metastatic versus non-metastatic cell lines. Anticancer Res. 22(2A), 733–740 (2002).
Schwirzke, M. et al. Identification of metastasis-associated genes by transcriptional profiling of a pair of metastatic versus non-metastatic human mammary carcinoma cell lines. Anticancer Res. 21(3B), 1771–6 (2001).
Chandrashekar, D. S. et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 25, 18–27. https://doi.org/10.1016/j.neo.2022.01.001 (2022).
Fonseka, P., Pathan, M., Chitti, S. V., Kang, T. & Mathivanan, S. FunRich enables enrichment analysis of OMICs datasets. J. Mol. Biol. 433(11), 166747. https://doi.org/10.1016/j.jmb.2020.166747 (2021).
Chin, C. H. et al. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 8(4), S11. https://doi.org/10.1186/1752-0509-8-S4-S11 (2014).
Takada, M. et al. Prediction of lymph node metastasis by analysis of gene expression profiles in non-small cell lung cancer. J. Surg. Res. 122(1), 61–69. https://doi.org/10.1016/j.jss.2004.06.002 (2004).
Pocha, K. et al. Surfactant expression defines an inflamed subtype of lung adenocarcinoma brain metastases that correlates with prolonged survival. Clin. Cancer Res. 26(9), 2231–2243. https://doi.org/10.1158/1078-0432.CCR-19-2184 (2020).
Königshoff, M. Lung cancer in pulmonary fibrosis: Tales of epithelial cell plasticity. Respiration 81(5), 353–358. https://doi.org/10.1159/000326299 (2011).
Pizzagalli, M. D., Bensimon, A. & Superti-Furga, G. A guide to plasma membrane solute carrier proteins. FEBS J. 288(9), 2784–2835. https://doi.org/10.1111/febs.15531 (2021).
Alcaraz-Sanabria, A. et al. Transcriptomic mapping of non-small cell lung cancer K-RAS p.G12C mutated tumors: Identification of surfaceome targets and immunologic correlates. Front. Immunol. 12, 786069. https://doi.org/10.3389/fimmu.2021.786069 (2022).
Wu, X. et al. Zinc finger protein 367 promotes metastasis by inhibiting the Hippo pathway in breast cancer. Oncogene 39(12), 2568–2582. https://doi.org/10.1038/s41388-020-1166-y (2020).
Zeng, H. et al. ZNF367-induced transcriptional activation of KIF15 accelerates the progression of breast cancer. Int. J. Biol. Sci. 16(12), 2084–2093. https://doi.org/10.7150/ijbs.44204 (2020).
Zhang, H. et al. NCBP1 promotes the development of lung adenocarcinoma through up-regulation of CUL4B. J. Cell Mol. Med. 23(10), 6965–6977. https://doi.org/10.1111/jcmm.14581 (2019).
Lu, F. et al. Construction of a novel prognostic model in lung adenocarcinoma based on 7-methylguanosine-related gene signatures. Front. Oncol. 12, 876360. https://doi.org/10.3389/fonc.2022.876360 (2022).
Guerrero, M. S., Parsons, J. T. & Bouton, A. H. Cas and NEDD9 contribute to tumor progression through dynamic regulation of the cytoskeleton. Genes Cancer 3(5–6), 371–381. https://doi.org/10.1177/1947601912458585 (2012).
Kondo, S. et al. Impact of the integrin signaling adaptor protein NEDD9 on prognosis and metastatic behavior of human lung cancer. Clin. Cancer Res. 18(22), 6326–6338. https://doi.org/10.1158/1078-0432.CCR-11-2162 (2012).
Aasebø, E. et al. The progression of acute myeloid leukemia from first diagnosis to chemoresistant relapse: A comparison of proteomic and phosphoproteomic profiles. Cancers (Basel) 12(6), 1466. https://doi.org/10.3390/cancers12061466 (2020).
Huang, G., Li, H. & Zhang, H. Abnormal expression of mitochondrial ribosomal proteins and their encoding genes with cell apoptosis and diseases. Int. J. Mol. Sci. 21(22), 8879. https://doi.org/10.3390/ijms21228879 (2020).
Liu, Y. & Gong, Y. SCLT1 depletion impair hepatoma cell survival by suppressing cell proliferation and promoting apoptosis. Int. J. Sci. 8(4), 114–120 (2021).
Li, F. P. et al. Overexpressed SNRPB/D1/D3/E/F/G correlate with poor survival and immune infiltration in hepatocellular carcinoma. Am. J. Transl. Res. 14(6), 4207–4228 (2022).
Zhao, J. et al. A novel pathway in NSCLC cells: miR-191, targeting NFIA, is induced by chronic hypoxia, and promotes cell proliferation and migration. Mol. Med. Rep. 15(3), 1319–1325. https://doi.org/10.3892/mmr.2017.6100 (2017).
Lin, M., Yao, Z., Zhao, N. & Zhang, C. TLK2 enhances aggressive phenotypes of glioblastoma cells through the activation of SRC signaling pathway. Cancer Biol. Ther. 20(1), 101–108. https://doi.org/10.1080/15384047.2018.1507257 (2019).
Li, H. et al. MiR-365 enhances the radiosensitivity of non-small cell lung cancer cells through targeting CDC25A. Biochem. Biophys. Res. Commun. 512(2), 392–398. https://doi.org/10.1016/j.bbrc.2019.03.082 (2019).
Motieghader, H., Kouhsar, M., Najafi, A., Sadeghi, B. & Masoudi-Nejad, A. mRNA-miRNA bipartite network reconstruction to predict prognostic module biomarkers in colorectal cancer stage differentiation. Mol. Biosyst. 13(10), 2168–2180. https://doi.org/10.1039/c7mb00400a (2017).
Silva, C. M. S. et al. Circulating let-7e-5p, miR-106a-5p, miR-28-3p, and miR-542-5p as a promising microRNA signature for the detection of colorectal cancer. Cancers (Basel) 13(7), 1493. https://doi.org/10.3390/cancers13071493 (2021).
Gharib, E., Nasri Nasrabadi, P. & Reza, Z. M. miR-497-5p mediates starvation-induced death in colon cancer cells by targeting acyl-CoA synthetase-5 and modulation of lipid metabolism. J. Cell Physiol. 235(7–8), 5570–5589. https://doi.org/10.1002/jcp.29488 (2020).
Yang, L. et al. ZWINT: A potential therapeutic biomarker in patients with glioblastoma correlates with cell proliferation and invasion. Oncol. Rep. 43(6), 1831–1844. https://doi.org/10.3892/or.2020.7573 (2020).
Wang, C., Xie, X., Li, W. & Jiang, D. Expression of KIF2A, NDC80, CDK1, and CCNB1 in breast cancer patients: Their interaction and linkage with tumor features and prognosis. J. Clin. Lab. Anal. 36(9), e24647. https://doi.org/10.1002/jcla.24647 (2022).
Zeng, L. et al. Involvement of NEK2 and its interaction with NDC80 and CEP250 in hepatocellular carcinoma. BMC Med. Genom. 13(1), 158. https://doi.org/10.1186/s12920-020-00812-y (2020).
Li, M., He, F., Zhang, Z., Xiang, Z. & Hu, D. CDK1 serves as a potential prognostic biomarker and target for lung cancer. J. Int. Med. Res. 48(2), 300060519897508. https://doi.org/10.1177/0300060519897508 (2020).
Chai, C., Yang, Z., Huang, M., Xu, J. & Lu, X. CDK1/TTR/MYC promote the formation of metastatic niches and affect the prognosis of pancreatic cancer by participating in immune CD4+ T cell infiltration as indicated by an integrated bioinformatics analysis. Dig. Med. Res. 3, 40. https://doi.org/10.21037/dmr-20-98 (2020).
Chen, H. et al. CDK1 promotes epithelial-mesenchymal transition and migration of head and neck squamous carcinoma cells by repressing ∆Np63α-mediated transcriptional regulation. Int. J. Mol. Sci. 23(13), 7385. https://doi.org/10.3390/ijms23137385 (2022).
Zhou, W., Zhu, Y., Chen, S., Xu, R. & Wang, K. Fibroblast growth factor receptor 1 promotes MG63 cell proliferation and is associated with increased expression of cyclin-dependent kinase 1 in osteosarcoma. Mol. Med. Rep. 13(1), 713–719. https://doi.org/10.3892/mmr.2015.4597 (2016).
Maga, G. & Hubscher, U. Proliferating cell nuclear antigen (PCNA): A dancer with many partners. J. Cell Sci. 116(Pt 15), 3051–3060. https://doi.org/10.1242/jcs.00653 (2003).
Strzalka, W. & Ziemienowicz, A. Proliferating cell nuclear antigen (PCNA): A key factor in DNA replication and cell cycle regulation. Ann. Bot. 107(7), 1127–1140. https://doi.org/10.1093/aob/mcq243 (2011).
Isozaki, H. et al. Significance of proliferating cell nuclear antigen (PCNA) expression in gastric cancer in relation to lymph node metastasis. J. Surg. Oncol. 61(2), 106–110. https://doi.org/10.1002/(SICI)1096-9098(199602)61:2%3c106::AID-JSO4%3e3.0.CO;2-G (1996).
Wang, L., Kong, W., Liu, B. & Zhang, X. Proliferating cell nuclear antigen promotes cell proliferation and tumorigenesis by up-regulating STAT3 in non-small cell lung cancer. Biomed. Pharmacother. 104, 595–602. https://doi.org/10.1016/j.biopha.2018.05.071 (2018).
van de Weerdt, B. C. & Medema, R. H. Polo-like kinases: A team in control of the division. Cell Cycle 5(8), 853–864. https://doi.org/10.4161/cc.5.8.2692 (2006).
Degenhardt, Y. & Lampkin, T. Targeting Polo-like kinase in cancer therapy. Clin. Cancer Res. 16(2), 384–389. https://doi.org/10.1158/1078-0432.CCR-09-1380 (2010).
Rizki, A., Mott, J. D. & Bissell, M. J. Polo-like kinase 1 is involved in invasion through extracellular matrix. Cancer Res. 67(23), 11106–11110. https://doi.org/10.1158/0008-5472.CAN-07-2348 (2007).
Zhang, X. G., Lu, X. F., Jiao, X. M., Chen, B. & Wu, J. X. PLK1 gene suppresses cell invasion of undifferentiated thyroid carcinoma through the inhibition of CD44v6, MMP-2 and MMP-9. Exp. Ther. Med. 4(6), 1005–1009. https://doi.org/10.3892/etm.2012.729 (2012).
Shin, S. B., Jang, H. R., Xu, R., Won, J. Y. & Yim, H. Active PLK1-driven metastasis is amplified by TGF-β signaling that forms a positive feedback loop in non-small cell lung cancer. Oncogene 39(4), 767–785. https://doi.org/10.1038/s41388-019-1023-z (2020).
Acknowledgements
Special thanks go to the scientists who took time for consulting in the field of Bioinformatics.
Funding
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
M.M. designed the study and critically reviewed the manuscript. K.K.V. performed formal analysis. M.M. and S.N. adminstrated project. The final manuscript has been approved by all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Khashei Varnamkhasti, K., Moghanibashi, M. & Naeimi, S. Genes whose expressions in the primary lung squamous cell carcinoma are able to accurately predict the progression of metastasis through lymphatic system, inferred from a bioinformatics analyses. Sci Rep 13, 6733 (2023). https://doi.org/10.1038/s41598-023-33897-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-33897-3
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.