Abstract
This study aimed to explore how kinesin family member 2C (KIF2C) influences the progression of non-small cell lung cancer (NSCLC). The levels of KIF2C and microRNA-186-3p (miR-186-3p) were examined by quantitative real-time polymerase chain reaction (qRT-PCR). Through the utilization of cell counting kit-8 (CCK-8) assay, colony formation assay, wound closure assay, and Transwell assay, NSCLC cell proliferation, migration, and invasion were identified, respectively. NSCLC cell apoptosis was assessed using the TUNEL assay and flow cytometry (FCM) assay. Luciferase reporter analysis was used to investigate the relationship between KIF2C and miR-186-3p. Western blot assays were conducted to investigate the influence of KIF2C on the AKT-GSK3β-β-catenin pathway. The results showed that KIF2C was up‐regulated in NSCLC cells, which predicted poor prognosis. KIF2C overexpression promoted the proliferation, migration, and invasion of NSCLC cells as well as inhibited NSCLC cell apoptosis. KIF2C was as a key target of miR-186-3p. High expression of KIF2C, meanwhile, increased the levels of β-catenin, p-GSK-3β and phosphorylated protein kinase B (p-AKT). KIF2C downregulation and miR-186-3p upregulation reversed these outcomes. As an oncogenic factor, KIF2C is negatively regulated by miR-186-3p and participates in the progression of NSCLC through the AKT-GSK3β-β-catenin pathway.
Similar content being viewed by others
Introduction
Around 18% of cancer cases globally are lung cancer. The 5-year survival rate for non-small cell lung cancer (NSCLC), which makes up about 85% of all lung malignancies, is just 19.7%1,2.
As a member of the kinesin 13 family member, kinesin 2C (KIF2C) has been demonstrated to be important for cell mitosis and to play a role in spindle assembly, chromosomal aggregation and segregation, and centromere-microtubule junctions. Although KIF2C is substantially expressed in a number of tumor types, its exact mode of action is yet unknown in lung cancer3. Study has found that KIF2C participated in the modification of the cytoskeleton during cell invasion and migration, which are important steps in tumor cell proliferation4.
Numerous cancers have been shown to have abnormally active Wnt/β-catenin signaling pathways with a wide range of causes. It participates in a range of physiological and pathological processes related to carcinogenesis and embryonic development5,6.
Researchers found that miRNA modulated tumor angiogenesis, cell proliferation, differentiation, apoptosis, metastasis, and metabolism, all of which were involved in the incidence and development of tumors7,8. Studies have shown that miR-186-3p suppressed the progression of tumors by directly targeting epithelial regulatory proteins in breast cancer cells that were estrogen receptor positive9.
The regulatory involvement of miR-186-3p in NSCLC, however, remains unknown.
The aim of this study was to clarify the ability of KIF2C to enhance cancer cell viability, proliferation and invasion in NSCLC by activating the AKT-GSK3β-β-catenin pathway. At the same time, miR-186-3p can negatively regulate the expression of KIF2C, thereby exerting a tumor suppressor effect10. Our research revealed the mechanism of KIF2C on NSCLC and the regulatory effect of miR-186-3p on KIF2C, which provides a novel therapeutic target for NSCLC.
Materials and methods
Cell culture and transfection
Human immortalized normal lung epithelial cell line BEAS-2B and lung cancer cell lines, including A549, A549-DDP, LLC, PC-9, SHP-77, NCI-H1703 and human embryonic kidney cell line 293T were purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). Cisplatin (DDP) was first synthesized by M. Peyrone in 1845, and later proved to have a killing effect on NSCLC cells. It is now recommended for adjuvant chemotherapy in NSCLC patients after tumor resection. A549 cells are a human adenocarcinoma cell line, while A549-DDP refers to a human adenocarcinoma cell line resistant to DDP. A549-DDP cells were cultured in RPMI-1640 medium (Gibco, BRL, USA). Others were maintained in DMEM medium (Gibco, BRL, USA) at 37 °C in the humidified air with 5% CO2.
These media were supplemented with 10% fetal bovine serum (FBS; Invitrogen, CA, USA) and penicillin–streptomycin liquid (100 U/ml penicillin and100 mg/ml streptomycin), and replaced every two to three days. Using 0.25% trypsin (Roche, Basel, Switzerland), cells were passaged once the cell confluence reached 80%. Empty vector (pcDNA-NC), KIF2C overexpression plasmid (PCDNA-KIF2C), small interfering RNA targeting KIF2C (si-KIF2C) and its negative control (si-NC), and miR-186-3p inhibitor and its control (inh-NC), miR-186-3p mimics and their control (miR-NC) were constructed by GenePharma (Shanghai, China). A549 and A549/DDP cells were transfected with these vectors and oligonucleotides using Lipofectamine® 3000 (Invitrogen, CA, USA). Quantitative real-time polymerase chain reaction was carried out after 24 h.
Quantitative real-time polymerase chain reaction (qRT-PCR)
The total RNA was extracted from these cells utilizing TRIzol reagent (Invitrogen, MA, USA) and complementary DNA (cDNA) synthesis was accomplished using the Prime Script kit (Takara, Beijing, China). The qRT-PCR was performed using SYBR Green (Takara, Beijing, China) in an ABI-7300 RT-PCR system (Applied Biosystems, CA, USA). The ratio to GAPDH was used as an internal control for mRNA, and miRNA expression was normalized utilizing U6. The sequences of the following primers were made:
KIF2C-F: 5′-CAGTGGAATGGGCAGAAGGA-3′
KIF2C-R: 5′-CGGGCAAATTCCCAGTTTGG-3′
miR-186-3p-F: 5′-GCCCAAAGGTGAATTTTTTGGG-3′
miR-186-3p-R: 5′-CAGTGCGTGTCGTGGAGT-3′
GAPDH-F: 5′-GGACTGACCTGCCGTCTAG-3′
GAPDH-R: 5′- TAGCCCAGGATGCCCTTGAG-3′
U6-F: 5′-CTCGCTTCGGCAGCACA-3′
U6-R: 5′-AACGCTTCACGAATTTGCGT-3′
Cell counting kit-8 (CCK-8) assay
Cells were planted on 96-well plates following transfection at a starting density of 3000 cells per well.
Each well received 10 μL CCK-8 (Dojindo Molecular Technologies, Kumamoto, Japan) at a different monitored time point and incubated with the cells according to the manufacturer's instructions. A multifunctional microplate reader (Tecan Sunrise, Switzerland) was used to detect the absorbance at the wavelength of 450 nm.
Apoptosis assay
Apoptosis in cells was measured using the Annexin-V-FITC/PI apoptosis detection kit (BD Biosciences, San Jose, CA, USA) in accordance with the manufacturer's instructions. Then the cell suspension was shaken thoroughly for 15 min at room temperature in darkness and supplemented with 10 μL propidium iodide (Sigma Aldrich, St. Louis, USA). Flow cytometry (FCM) was then used to analyze apoptotic cells. Three different runs of each experiment were conducted. A TUNEL assay kit (cat. no. MK1015; Wuhan Boster Biological Technology, Ltd.) was used to detect apoptotic cells according to the manufacturer's instructions. Cells were incubated overnight at 4 °C with anti-neuronal nuclei antibody (cat. no. BM4354; 1:100; Wuhan Boster Biological Technology, Ltd.) according to the manufacturer's instructions. The cells were incubated with TUNEL reaction mixture for 1 h at 37 °C before being washed three times with PBS. Images were captured using an inverted fluorescence microscope at high magnification (400×). Images in three randomly selected areas were quantitated, and the positive cells were measured using Image J software.
Transwell assay
Transwell migration chambers (8‐μm pore size; Millipore, Billerica, MA, USA) were used to measure cell migration. A total of 2 × 105 cells were suspended in the serum-free DMEM media and planted into the top chamber together. In the lower chamber, DMEM medium with 10% FBS was added. The chambers were incubated for 48 h before removal, and crystal violet was used to stain these cells from the lower chamber following 4% paraformaldehyde for 15 min. Random six fields at high magnification (Olympus, Tokyo, Japan) were chosen, and cell migration was calculated. Additionally, the invasion assays were carried out in Transwell chambers covered with matrigel (Clontech, CA, USA).
Colony formation assay
At a cell density of 1000 cells/dish, the transfected cells were seeded in triplicate into 60 mm cell culture plates. After 14 days of growth in DMEM medium containing 10% FBS, cells were fixed and stained with 0.1% crystal violet for 30 min. After that, the colonies were meticulously cleaned with PBS until the background was completely clear. The colony formation rate was calculated as follows:
Wound healing assay
Transfected cells were planted in six-well plates for 24 h to form a monolayer. After that, each well was scratched with a cross using a sterile pipette tip held vertically. Subsequently, cells were washed with phosphate-buffered saline (PBS) and shaken for 5 min to remove the detached cells. Then the diluted samples and fresh medium were incubated for 72 h. After washing cells with PBS, add the preheated medium or a fresh sample, and take images. A Keyence BZ-9000 microscope (Keyence, Neu-Isenburg, Germany) was used to observe and capture the scratch closure at intervals of 24 h at low magnification.
Western blot
After cells were collected and fixed, the protein was extracted and examined according to the manufacturer's instructions. Put simply, the lysates were prepared with RIPA lysis buffer (Beyotime Biotechnology, Beijing, China) and centrifuged for 10 min at 4 °C at 12,000 rpm, and the BCA protein assay kit (Pierce, WI, USA) was applied to determine protein concentration in the supernatants. The sample mixed with loading buffer was added to 10% SDS-PAGE and then transferred to a nitrocellulose membrane. The primary antibodies in this part include the Anti-KIF2C antibody, Anti-p-AKT antibody, Anti-AKT antibody, Anti-p-GSK-3β antibody, Anti-GSK-3β antibody, Anti-β-actinin antibody, and Anti-GAPDH antibody. The above antibodies were purchased separately from Cell Signaling Technology (CST, MA, USA). The membranes were washed after being incubated with the primary antibodies overnight and treated for an hour with a secondary antibody labeled with horseradish peroxidase (HRP). The signal was detected with chemiluminescence phototope-HRP kit (Pierce, Rockford, USA USA) after washing the membrane.
Luciferase reporter assay
In a nutshell, the binding sequences of KIF2C 3'UTR of miR-186-3p, as predicted by the Target Scan Human 7.2 database, were amplified and put into the pmirGLO vector (Promega, WI, USA). The primers were as follows, KIF2C-WT, 5′-GAGGAGTTGTGTTTGTGGAC-3′ (forward), 5′-TGTAAAACGACGGCCAGT-3′ (reverse), and KIF2C-mut (mutated has-miR-186-3p binding site), 5′-GAGGAGTTGTGTTTGTGGAC-3′ (forward), 5′-TGTAAAACGACGGCCAGT-3′ (reverse). A549 cells were co-transfected with the mixture of pluc3′UTR (WT or mut), Renilla luciferase (Promega, Madison, WI) and Lipofectamine 3000 (Invitrogen, CA, USA). After transfection, luciferase activity was measured 48 h later using a dual-luciferase assay system (Promega, WI, USA).
Statistical analysis
The results were presented as means and standard deviation. Statistics were analyzed using SPSS 20.0 (Chicago, USA). Differences between the groups were compared using the student t-test.
Results
NSCLC with a higher expression of KIF2C has a poor outcome
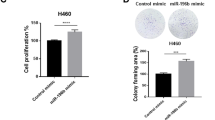
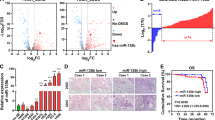
The purpose of this study was to determine KIF2C expression levels in human lung epithelial cells BEAS-2B and different types of NSCLC cell lines, such as mouse lung adenocarcinoma LLC, human lung adenocarcinoma cells A549, PC-9, SHP-77, A549-DDP, NCI-H1703, through qRT-PCR. In comparison with BEAS-2B, KIF2C expression was significantly higher in NSCLC cells (Fig. 1a). At the same time, KIF2C mRNA levels have been analyzed in human lung adenocarcinomas (LUAD), lung squamous cell carcinomas(LUSC), and normal lung tissue in the Cancer Genome Atlas (TCGA) dataset. According to the results, tumor tissues had significantly higher levels of KIF2C mRNA than normal tissues (Fig. 1b,c). Both high KIF2C expression in LUAD and LUSC tissues were associated with worse prognosis according to the TCGA database (Fig. 1d,e).
Expression of KIF2C in NSCLC cell lines. (a) KIF2C expression is significantly high-expressed in NSCLC cell lines; (b,c) KIF2C expression was increased in human lung adenocarcinoma(LUAD) and human lung squamous cell carcinoma(LUSC) tissues; (d,e) High KIF2C expression in human lung adenocarcinoma(LUAD) and human lung squamous cell carcinoma(LUSC) tissues were associated with worse prognosis (*P < 0.05, **P < 0.01, and ***P < 0.001).
KIF2C affects biologically malignant behaviors of NSCLC cellsWith the popularization of computed tomography and other imaging screening methods, adenocarcinoma incidence has increased significantly and has surpassed squamous carcinoma as the most common type. Adenocarcinoma is an invasive carcinoma, which is more likely to relapse and metastasize than squamous carcinoma and difficult to treat11. Therefore, we selected LUAD in NSCLC as the research object. Results have showed that A549-DDP cells showed the highest KIF2C expression among NSCLC cells, whereas its corresponding A549 cells showed a relatively low KIF2C expression. As a result, we used plasmid to overexpress KIF2C in low-expressing A549 cells and used small interfering RNA (siRNA) to knock down KIF2C in high-expressing A549-DDP cells. The transfection efficiencies of KIF2C overexpression and silencing were determined by qRT-PCR (Fig. 2a,b). In addition, CCK8 and flow cytometry (FCM) assays were performed to assess the proliferative ability of NSCLC cells transfected with KIF2C and KIF2C siRNA. The results showed that overexpression of KIF2C significantly promoted proliferation and inhibited apoptosis, while silencing of KIF2C inhibited proliferation and promoted apoptosis (Fig. 2c–f). Cancer cell proliferation, migration and invasion are a significant part of cancer progression, so we investigated the effect of KIF2C on these features in NSCLC by performing transwell, colony formation and wound healing assays. We performed cell proliferation, migration, and invasion assays to investigate the biological function of KIF2C in controlling the biologically malignant behaviors of NSCLC cells. The results revealed that, in comparison to the control group, KIF2C overexpression significantly promoted cell proliferation, migration, and invasion, whereas KIF2C knockdown significantly inhibited above cell behavior (Fig. 2g–i).
The effects of KIF2C on the proliferation, migration, invasion and apoptosis of NSCLC cells. (a,b) Overexpression and silence efficiency of KIF2C evaluated by real-time PCR analysis; (c.d) CCK8 and Colony formation assays were performed to assess the proliferative ability transfected with KIF2C and KIF2C siRNA plasmids; (e,f) Flowcytometry assays was used to detect cell apoptosis of overexpression and silence of KIF2C; (g,h,i) The effects of KIF2C on migration and invasion were evaluated by the wound healing and transwell assays (*P < 0.05, **P < 0.01, and ***P < 0.001).
KIF2C activated AKT-GSK3β-β-catenin pathway
Further studies were conducted to investigate how KIF2C influences proliferation, migration, and invasion in NSCLC cells. Wntβ–catenin pathways are broadly involved in promoting tumorigenicity, stemness, and EMT induction12,13.Therefore, we investigated if KIF2C could impact the AKT-GSK3β-β-catenin pathway via Western blotting assays. The results demonstrated that overexpression of KIF2C increased β-catenin, p-GSK-3β and p-AKT levels, while the result of KIF2C knockdown was the opposite (Fig. 3).
miR-186-3p targeted KIF2C regulation
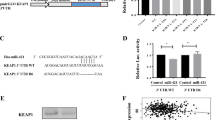
The StarBase database was chosen to predict the miRNAs that could control KIF2C for a further understanding of the upstream regulatory mechanisms of KIF2C. It was demonstrated that miR-186-3p and KIF2C 3'UTR have binding sites (Fig. 4a, Supplementary file). To verify the targeting relationship between KIF2C 3'UTR and miR-186-3p, we performed a dual luciferase reporter assay. According to the results, overexpression of miR-186-3p reduced the luciferase activity in KIF2C WT cells, but had no discernible effect on KIF2C MUT cells (Fig. 4b). Compared with BEAS-2B cells, miR-186-3p expression was lower in lung cancer cells (Fig. 4c).
miR-186-3p target KIF2C and expression of miR-186‐3p in NSCLC Cell Lines. (a) To further explore the upstream mechanism of KIF2C, the StarBase database (starbase.sysu.edu.cn) was used to predict miRNAs that may regulate KIF2C. It was displayed that there existed binding sites between miR-186-3p and KIF2C 3'UTR; (b) subsequently, a dual-luciferase reporter gene assay was conducted for verifying the targeted relationships between KIF2C 3'UTR and miR-186-3p, and it was shown that miR-186-3p overexpression inhibited KIF2C WT's luciferase activity, yet did not significantly affect KIF2C MUT's luciferase activity. (c) The expression of miR-186-3p was lower in lung cancer cells.
The miR-186-3p/KIF2C axis regulated the biological behaviors in NSCLC cells
An analysis of qRT-PCR results showed that miR-186-3p had been successfully silenced and overexpressed after transfection (Fig. 5a,b). In order to confirm if miR-186-3p contributed to the NSCLC progression by altering KIF2C, A549-DDP cells were co-transfected with pcDNA-KIF2C and miR-186-3p mimics, and A549 cells were co-transfected with si-KIF2C and miR-186-3p inhibitors.
Effects of miR-186-3p/KIF2C axis on NSCLC cell proliferation, migration, and invasion. (a,b); The effects of transfection of miR‐186-3p and si- miR‐186-3p on A549 and A549-DDP cell proliferation were determined by qRT-PCR; (c,d) the effects of miR‐186-3p silencing and overexpression on A549-DDP and A549 cell proliferation were detected by CCK‐8 assays; (e,f) tunel assays were used to detect the cell proliferation and apoptosi; (g,h) A549-DDP cells were co-transfected with pcDNA-KIF2C and miR-186-3p mimics, and A549 cells were co-transfected with si-KIF2C and miR-186-3p inhibitors. After transfection, Western blot were used to detect the changes of β-catenin, p-GSK-3β and p-AKT; (i,l,m) the effects of KIF2C on migration and invasion were evaluated by the wound healing and transwell assays (*P < 0.05, **P < 0.01, and ***P < 0.001).
A549-DDP cells were co-transfected with pcDNA-KIF2C and miR-186-3p mimics, whereas A549 cells were co-transfected with si-KIF2C and miR-186-3p inhibitors using CCK8 assays (Fig. 5c,d). To determine whether the proliferation of NSCLC cells was influenced by apoptosis, we performed TUNEL staining.
The results showed that A549-DDP cells transfected with miR-186-3p mimics and A549 cells transfected with si-KIF2C exhibited a higher rate of apoptosis than control cells (Fig. 5e,f). After transfection, western blot was conducted to investigate the effect of miR-186-3p/KIF2C axis on AKT-GSK3β-β-catenin pathway. We found that the levels of β-catenin, p-GSK-3β and p-AKT decreased in A549-DDP cells transfected with miR-186-3p mimics and A549 cells transfected with si-KIF2C compared with those of controls, which indicated that the AKT-GSK3β-β-catenin pathway was inhibited by miR-186-3p/KIF2C axis (Fig. 5g,h). Subsequently, we investigated the effect of miR-186-3p/KIF2C axis on cell proliferation, migration and invasion in NSCLC by performing transwell, colony formation and wound healing assays. The findings revealed that miR-186-3p inhibitors promoted proliferation, migration and invasion of A549 cell compared with the control group, whereas miR-186-3p mimics impeded above features of A549-DDP cells (Fig. 5i,l,m). Overall, these findings suggest that miR-186-3p/KIF2C axis play an important role in biologically malignant behaviors of NSCLC cells.
Discussion
Incidence and mortality from cancer are most frequently caused by lung cancer worldwide14. NSCLC is distinguished by slow cancer cell growth, metastasis, and development15. One of the most important symptoms of NSCLC is the metastasis of tumor cells16. Therefore, inhibiting tumor cell proliferation and metastasis has a huge auxiliary effect on patients with lung cancer15. Researchers found that KIF2C was highly expressed in liver cancer and thyroid cancer, and was a landmark tumor-promoting factor3,17. However, the activation mechanism of KIF2C in NSCLC has not been investigated18. Therefore, we designed this experiment to explore a new mechanism for restricting the proliferation and metastasis of NSCLC. The study of cancer uses three main approaches: fresh tumor tissue, animal models, and cell cultures. They all have advantages and disadvantages. However, each tumor tissue contains varying amounts of non-malignant cells, and there are restrictions on the acquisition and use of these cells. Animal experiments need to consider species differences. These reasons have made cancer cell lines an important tool for studying lung cancer. Currently an estimated 300–400 human lung cancer cell lines have been established, including small cell (SCLC) and NSCLC. These cell lines have been widely used in the scientific community all over the world and their research has generated more than a thousand citations, and the research results have played a crucial role in revealing the pathogenesis of lung cancer19. We found that KIF2C was closely related to chemotherapy resistance of breast cancer through the literature20. The latest research has found that KIF2C, as a stemness related genes, may affect the efficacy of chemotherapy and immunotherapy by regulating the tumor microenvironment in LUAD21. This also reflects the innovation of our research. In this paper, we presented experimental proof that, in comparison to BEAS-2B, KIF2C was significantly expressed in various kinds of lung cancer cell lines such as LUAD and LUSC. The results of cell experiments showed that KIF2C overexpression significantly enhanced cell proliferation, migration, and invasion while inhibiting cell apoptosis when compared to the control group, whereas KIF2C knockdown prevented cell proliferation, migration and invasion, and enhanced cell apoptosis. The above results illustrated the critical role of KIF2C in the proliferation and metastasis of NSCLC. Meanwhile, according to Western blotting results, overexpression of KIF2C raised the levels of β-catenin, p-GSK-3β, and p-AKT, but downregulation of the protein had the reverse effect. The results demonstrated that KIF2C may contribute to NSCLC growth and metastasis by triggering the AKT-GSK3β-β-catenin signaling pathway.
Many miRNAs act as tumor suppressors or oncogenes in most cancers, and some have tissue-specific expression patterns8,22. Researchers found that miR-186 suppressed growth, migration, and invasion of NSCLC23,24,25. MiR-186-3p is involved in a variety of cancer pathogenesis functions26. Lu et al. found that miR-186-3p can reduce tamoxifen resistance in breast cancer cells through the EREG axis and can prevent the onset and progression of cervical cancer via targeting IGF127. There are also reports that miR-186-3p can block tumor growth in lung cancer28.
We experimentally verified the targeting link between miR-186-3p and KIF2C 3'UTR, and the results suggested that miR-186-3p may control KIF2C expression. In addition to directly inhibiting NSCLC growth and metastasis, miR-186-3p can also reverse the NSCLC-promoting effects of KIF2C. The relationship between KIF2C and miR-186-3p has been shown in the schematic diagram (Fig. 6).
In conclusion, KIF2C is widely expressed in NSCLC, which can promote the proliferation and metastasis of NSCLC. Mir-186-3p can slow down the deterioration of NSCLC by inhibiting the activation of AKT-GSK3β-β-catenin signaling pathway by KIF2C. As a result, we believe KIF2C could be a possible target for NSCLC treatment (Supplementary Information).
References
Hirsch, F. R. et al. Lung cancer: Current therapies and new targeted treatments. Lancet 389(10066), 299–311. https://doi.org/10.1016/S0140-6736(16)30958-8 (2017).
Choi, W. I., Park, S. H., Park, B. J. & Lee, C. W. Interstitial lung disease and lung cancer development: A 5-year nationwide population-based study. Cancer Res. Treat. 50(2), 374–381. https://doi.org/10.4143/crt.2017.119 (2018).
Wei, S. et al. KIF2C: A novel link between Wnt/β-catenin and mTORC1 signaling in the pathogenesis of hepatocellular carcinoma. Protein Cell. 12(10), 788–809. https://doi.org/10.1007/s13238-020-00766-y (2021).
Moon, H. H. et al. Mitotic centromere-associated kinesin (MCAK/KIF2C) regulates cell migration and invasion by modulating microtubule dynamics and focal adhesion turnover. Cancers (Basel). 13(22), 5673. https://doi.org/10.3390/cancers13225673 (2021).
Hussain, M. et al. Wnt/β-catenin signaling links embryonic lung development and asthmatic airway remodeling. Biochim. Biophys. Acta Mol. Basis Dis. 1863(12), 3226–3242. https://doi.org/10.1016/j.bbadis.2017.08.031 (2017).
Zhong, Z. & Virshup, D. M. Wnt signaling and drug resistance in cancer. Mol. Pharmacol. 97(2), 72–89. https://doi.org/10.1124/mol.119.117978 (2020).
Konishi, H., Sato, H., Takahashi, K. & Fujiya, M. Tumor-progressive mechanisms mediating miRNA–protein interaction. Int. J. Mol. Sci. 22(22), 12303. https://doi.org/10.3390/ijms222212303 (2021).
Lee, Y. S. & Dutta, A. MicroRNAs in cancer. Annu. Rev. Pathol. 4, 199–227. https://doi.org/10.1146/annurev.pathol.4.110807.092222 (2009).
He, M. et al. The miR-186-3p/EREG axis orchestrates tamoxifen resistance and aerobic glycolysis in breast cancer cells. Oncogene 38(28), 5551–5565. https://doi.org/10.1038/s41388-019-0817-3 (2019).
Xia, T., Zhang, Z., Zhang, X. & Li, Q. Hsa-miR-186-3p suppresses colon cancer progression by inhibiting KRT18/MAPK signaling pathway. Cell Cycle 21(7), 741–753. https://doi.org/10.1080/15384101.2021.2023305 (2022).
Succony, L., Rassl, D. M., Barker, A. P., McCaughan, F. M. & Rintoul, R. C. Adenocarcinoma spectrum lesions of the lung: Detection, pathology and treatment strategies. Cancer Treat. Rev. 99, 102237. https://doi.org/10.1016/j.ctrv.2021.102237 (2021).
Krishnamurthy, N. & Kurzrock, R. Targeting the Wnt/beta-catenin pathway in cancer: Update on effectors and inhibitors. Cancer Treat. Rev. 62, 50–60. https://doi.org/10.1016/j.ctrv.2017.11.002 (2018).
Koni, M., Pinnarò, V. & Brizzi, M. F. The Wnt signalling pathway: A tailored target in cancer. Int. J. Mol. Sci. 21(20), 7697. https://doi.org/10.3390/ijms21207697 (2020).
Nasim, F., Sabath, B. F. & Eapen, G. A. Lung cancer. Med. Clin. N. Am. 103(3), 463–473. https://doi.org/10.1016/j.mcna.2018.12.006 (2019).
Herbst, R. S., Morgensztern, D. & Boshoff, C. The biology and management of non-small cell lung cancer. Nature 553(7689), 446–454. https://doi.org/10.1038/nature25183 (2018).
Chen, Z. et al. Up-regulated LINC01234 promotes non-small-cell lung cancer cell metastasis by activating VAV3 and repressing BTG2 expression. J. Hematol. Oncol. 13(1), 7. https://doi.org/10.1186/s13045-019-0842-2 (2020).
Lin, Q. et al. Activation of the TGF-β1/Smad signaling by KIF2C contributes to the malignant phenotype of thyroid carcinoma cells. Tissue Cell. 73, 101655. https://doi.org/10.1016/j.tice.2021.101655 (2021).
Gan, H. et al. KIF2C exerts an oncogenic role in nonsmall cell lung cancer and is negatively regulated by miR-325-3p. Cell Biochem. Funct. 37(6), 424–431. https://doi.org/10.1002/cbf.3420 (2019).
Gazdar, A. F., Gao, B. & Minna, J. D. Lung cancer cell lines: Useless artifacts or invaluable tools for medical science?. Lung Cancer 68(3), 309–318 (2010).
Gluz, O. et al. Efficacy of deescalated chemotherapy according to PAM50 subtypes, immune and proliferation genes in triple-negative early breast cancer: Primary translational analysis of the WSG-ADAPT-TN trial. Int. J. Cancer 146(1), 262–271 (2020).
Zeng, H. et al. Stemness related genes revealed by network analysis associated with tumor immune microenvironment and the clinical outcome in lung adenocarcinoma. Front. Genet. 16(11), 549213 (2020).
Yoshida, K., Yokoi, A., Yamamoto, Y. & Kajiyama, H. ChrXq27.3 miRNA cluster functions in cancer development. J. Exp. Clin. Cancer Res. 40(1), 112. https://doi.org/10.1186/s13046-021-01910-0 (2021).
Wang, J., Zhang, Y. & Ge, F. MiR-186 suppressed growth, migration, and invasion of lung adenocarcinoma cells via targeting Dicer1. J. Oncol. 11(2021), 6217469. https://doi.org/10.1155/2021/6217469.PMID:34804161;PMCID:PMC8601821 (2021).
Huang, T. et al. MiR-186 inhibits proliferation, migration, and invasion of non-small cell lung cancer cells by downregulating Yin Yang 1. Cancer Biomark. 21(1), 221–228. https://doi.org/10.3233/CBM-170670 (2017).
Huang, T. et al. MicroRNA-186 suppresses cell proliferation and metastasis through targeting MAP3K2 in non-small cell lung cancer. Int. J. Oncol. 49(4), 1437–1444. https://doi.org/10.3892/ijo.2016.3637 (2016) (epub 2016 Jul 29).
Morais, M. et al. Cancer cells’ metabolism dynamics in renal cell carcinoma patients’ outcome: Influence of GLUT-1-related hsa-miR-144 and hsa-miR-186. Cancers (Basel). 13(7), 1733. https://doi.org/10.3390/cancers13071733 (2021).
Lu, X. et al. miR-186-3p attenuates the tumorigenesis of cervical cancer via targeting insulin-like growth factor 1 to suppress PI3K-Akt signaling pathway. Bioengineered 12(1), 7079–7092. https://doi.org/10.1080/21655979.2021.1977053 (2021).
Liu, H., Liu, R., Hao, M., Zhao , X. & Li, C. Kinesin family member 3C (KIF3C) is a novel non-small cell lung cancer (NSCLC) oncogene whose expression is modulated by microRNA-150-5p (miR-150-5p) and microRNA-186-3p (miR-186-3p). Bioengineered 12(1), 3077–3088. https://doi.org/10.1080/21655979.2021.1942768 (2021).
Acknowledgements
This study was supported by National Natural Science Foundation of China (81860534); Natural Science Foundation of Inner Mongolia (2021MS08152); Inner Mongolia Autonomous Region science and technology planning project (2019GG039, 2019GG086, 2021GG0167); Program for Young Talents of Science and Technology in Universities of Inner Mongolia Autonomous Region (NJYT22004); Health science and technology planning project of of Inner Mongolia Autonomous Region (202201356); Zhi Yuan Talent Projects of Inner Mongolia Medical University (ZY0202011); Science and Technology achievement transformation Project of Inner Mongolia Medical University (YKD2019CGZH004); Youth Cultivation Project of Inner Mongolia Medical University (YKD2021QN029); Scientific and Technological Innovative Research Team for Inner Mongolia Medical University of Transformation application of organoid in medical and industrial interdiscipline (YKD2022TD002); Major Project of Inner Mongolia Medical University (YKD2022ZD002); Radiotherapy discipline construction project of National Cancer Regional Medical Center for for Inner Mongolia Medical University (YKD2022XK009); Radiobiology system and team construction of radiotherapy for Inner Mongolia Medical University (YKD2022XK014).
Author information
Authors and Affiliations
Contributions
J.G. and W.Z. designed the experiments; L.S., H.Y., Y.W. performed the experiments and analyzed the data; L.F. interpreted the results of the experiments; J.G. and W.Z. drafted the manuscript; H.Y. edited and revised the manuscript; all authors had a final approval of the manuscript submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Guo, J., Zhang, W., Sun, L. et al. KIF2C accelerates the development of non-small cell lung cancer and is suppressed by miR-186-3p via the AKT-GSK3β-β-catenin pathway. Sci Rep 13, 7288 (2023). https://doi.org/10.1038/s41598-023-30073-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-30073-5
This article is cited by
-
MicroRNA-203a inhibits breast cancer progression through the PI3K/Akt and Wnt pathways
Scientific Reports (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.