Abstract
The Antarctic continent is one of the most inhospitable places on earth, where living creatures, mostly represented by microorganisms, have specific physiological characteristics that allow them to adapt to the extreme environmental conditions. These physiological adaptations can result in the production of unique secondary metabolites with potential biotechnological applications. The current study presents a genetic and antibacterial characterization of four Antarctic fungi isolated from soil samples collected in Pedro Vicente Maldonado Scientific Station, at Fort William Point, Greenwich Island, Antarctica. Based on the sequences of the internal transcribed spacer (ITS) region, the fungi were identified as Antarctomyces sp., Thelebolus sp., Penicillium sp., and Cryptococcus gilvescens. The antibacterial activity was assessed against four clinical bacterial strains: Escherichia coli, Klebsiella pneumoniae, Enterococcus faecalis, and Staphylococcus aureus, by a modified bacterial growth inhibition assay on agar plates. Results showed that C. gilvescens and Penicillium sp. have potential antibiotic activity against all bacterial strains. Interestingly, Thelebolus sp. showed potential antibiotic activity only against E. coli. In contrast, Antarctomyces sp. did not show antibiotic activity against any of the bacteria tested under our experimental conditions. This study highlights the importance of conservation of Antarctica as a source of metabolites with important biomedical applications.
Similar content being viewed by others
Introduction
The Antarctic continent is the coldest desert on Earth and contains nearly 90% of Earth's ice1. The climatic characteristics of this hostile environment include: temperatures below 0 °C, freezing and melting seasons, high UV radiation, arid conditions, and scarcity of nutrients2,3,4. Despite these harsh conditions, diverse groups of organisms have colonized the continent, with the microbiota (i.e. bacteria, archaea, and fungi) contributing to the most abundant biomass5,6. Among these, the Antarctic fungi are represented by endemic, native and cosmopolitan species, adapted to the cryosphere7,8,9,10.
Initial reports of Antarctic fungi were in the early twentieth century, and more than 1000 non-lichenized fungal species had been reported in this continent11. Nowadays, metagenomics and metabarcoding provide a pivotal contribution to biodiversity surveys in this continent and its sub-Antarctic islands12,13,14,15. Recent reports have described new mycological species in the maritime Antarctica region. Rosa and collaborators12 used metabarcoding to analyze fungal diversity in soil samples from Deception Island (South Shetland Islands). However, a significant number of sequences were only grouped at the Kingdom taxonomic level12. Similarly, a recent study sequenced 184 fungal taxa from the Antarctic Peninsula and South Shetland Islands, of which 37 taxa were detected for the first time in Antarctica; among maritime sampling sites, Greenwich Island showed more mycological diversity13.
Studies related to the diversity of fungi in Antarctica are essential to characterize Antarctic microbiology, but also to discover novel fungi metabolites. However, metabolic mechanisms for Antarctic fungi adaptation and their bioprospecting potential is still considered poorly studied16,17,18. Nevertheless, numerous researches expose the potential biotechnological applications of this kingdom, particularly in biomedicine19,20,21. This is likely due to their specialized metabolic adaptation that includes high catalytic activity at low temperatures, extracellular enzyme production, synthesis of antifreeze protein and elevated unsaturated fatty acids, among others22,23. Unique metabolic properties such as these result in the production of diverse secondary metabolites that are mainly regulated by internal24,25 and external environmental factors26 (e.g., sexual stage, luminous intensity and pH). Thus, the extreme environment of the Antarctic continent is thought to contribute to the development of distinctive metabolites with potential antimicrobial properties that can lead to the discovery of new antibiotics27.
The discovery of novel compounds with bioactivity is crucial to face the increasing threat of multidrug-resistant (MDR), pandemic drug-resistant (PDR) and extensively drug-resistant bacteria (XDR)28, a major public health concern. To this end, microorganisms like Streptomyces coelicolor, Amycolatopsis orientalis and Penicillium chrysogenum have become valuable bioresources for the production of antibiotics29,30,31. Important medical compounds, such as beta-lactam penicillin, benzopyrenes, macrolides, and alkaloids have been isolated from fungi32,33. Fungi from polar regions represent a source of novel metabolites, with unique biomolecules that evolved under selective pressure34. Cold-adapted fungi showed antibacterial potential35,36,37,38, with distinctive structure and biological activity39. It is considered that new drugs derived from them may be currently understudied34, as expression of these compounds might be linked to environmental cues that are challenging to emulate under standard laboratory growth conditions40,41.
The present study shows the genetic, morphology and antibacterial characterization of four Antarctic fungi isolated from soil samples collected at Fort William Point, Greenwich Island, Antarctica. The phylogenetic analysis was based on sequences of the internal transcribed spacer (ITS) region and their potential antibacterial activity was assessed by a modified bacterial growth inhibition assay on agar plates.
Results
Fungi phylogeny
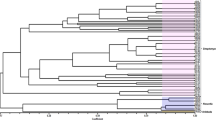
The phylogenetic analysis of the ITS sequences generated in this study (MZ958929, MZ958928, MZ958926, MZ958927) revealed that the T4-400-5E, T4-1K-1A and T4-1K-1G isolates clustered with the phylum Ascomycota, identified as Penicillium sp., Antarctomyces sp. and Thelebolus sp., respectively. The isolate T4-200-3B, clustered with the phylum Basidiomycete, was identified as Cryptococcus gilvescens (Fig. 1). All clades were strongly supported by bootstrap values higher than 70%. The tree grouped the family Thelebolaceae (Thelebolus and Antarctomyces genera) in a monophyletic group with a bootstrap value of 99% and segregated the family Trichocomaceae (Penicillium genus) in a separate group with a high 100% bootstrap value. Cryptococcus gilvescens was described as a more genetically distant species and the sequences contemplated in the tree were further divided into two additional clades with bootstraps well supported and within the family Tremellaceae.
Phylogenetic tree inferred using the Maximum Likelihood estimation based on the Kimura 2-parameter model. New sequences described in this study are preceded by the symbol ▲. Taxonomic relatedness is indicated on the right side. Scale bar shows nucleotide substitutions per site. Bootstrap values higher than 70% are shown. The name of each sequence corresponds to the species, location, accession number and date, if available.
Morphological observations
On PDA media the macro and microscopic morphological identification of the fungi isolates corroborated the topology of the analyzed genetic sequences.
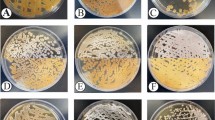
Antarctomyces sp. (T4-1K-1A) showed a smooth white colony appearance, with 3 cm of diameter after 10 days of growth. After 15 days, the colony changed to a furrowed appearance and progressively started darkening (Fig. 2a,b). At 30 days, Antarctomyces sp. had a blue coloration with an undulated margin and 4 cm of diameter. Microscopically, hyphae was septated with asci and immature ascospores (Fig. 3a).
Microscopic structures of Antarctic fungi isolates observed under a compound microscope with 1000 × magnification. (a) Cryptococcus gilvescens, showing budding division of cells (arrow); (b) Penicillium sp., showing their septate stipes and conidia at the top of phialides (arrows); (c) Thelebolus sp., showing hyaline septate hyphae and ascospores (arrow); and (d) Antarctomyces sp., showing septate hyphae and ascospores (arrow). Microscopic structures were observed at 15 days (a), and 30 days (b–d). Scale bar 10 µm.
Thelebolus sp. (T4-1K-1G) presented a circular, smooth, and cream color colony with 2 cm in size at 10 days, 3 cm and 5.5 cm of diameters after 15 and 30 days, respectively (Fig. 2c,d). Microscopically, sexual structures were scarcely developed. Immature asci and hyaline hyphae were observed (Fig. 3b).
Penicillium sp. (T4-400-5E) showed a yellow coloration on the agar. The colony had a furrowed appearance surrounded by a white margin. The center was umbonated with blue-green coloration. On the reverse, the colony had a yellow coloration. After 20 and 30 days of growth, the colony had a diameter of 2 cm and 2.5 cm, respectively (Fig. 2e,f). Microscopically, the anamorphic structure was represented with monoverticillate penicillin, the stipes were septated and the phialides measured 9 × 3 µm. The conidia were ovoid, with a dimension of 3.5 × 3 µm (Fig. 3c).
Cryptococcus gilvescens (T4-200-3B) presented a cream yeast-like colony with mucoid texture. The colony had a diameter between 1 and 1.7 cm at 15 and 30 days of growth, respectively (Fig. 2g,h). Microscopically, the cells were round to oval, with a diameter of 2.5–3 µm. Asexual reproduction by budding was observed (Fig. 3d).
Antibiotic susceptibility tests
Standard antibiotic resistant tests with the four bacteria used in this study showed that S. aureus was susceptibility to all the antibiotics tested, whereas K. pneumoniae, E. coli, and E. faecalis showed resistant to three or more antibiotics (Supplementary file 1).
The modified antibacterial susceptibility test performed on K. pneumoniae with the PDA and temperature treatment showed that the bacteria was still resistant to imipenem (imp) and meropenem (mem) regardless of the growth conditions (Supplementary file 2). Although a slight ring of growth inhibition was noticed on PDA as compared to LB agar, it was considered still a resistant phenotype.
Antibacterial potential
The antibacterial potential of the fungi was determined by the observation of bacterial growth inhibition zone around the mycelia plug (Fig. 4). Inhibition by Thelebolus sp. was only observed in the bioassay with E. coli at 15, 30 and 60 days of growth (Fig. 4a). At three different times of growth, C. gilvescens showed inhibitory effects to all Gram-positive and Gram-negative bacteria tested (Fig. 4b). Antarctomyces sp. did not show antibacterial activity against all the bacteria tested (Fig. 4c). Similar to C. gilvescens, Penicillium sp. showed inhibitory effects to all Gram-positive and Gram-negative bacteria tested at 15 and 30 days of growth (Fig. 4d).
In order to determine if growth time of the fungi (i.e. 15, 30 and 60 days) had a significant effect on the observed inhibition halo size, we conducted a Kruskal–Wallis and a Mann–Whitney statistical test to determine the difference between three and two different growth times, respectively.
Statistical analysis on the size of bacterial growth inhibition ring around mycelia plugs, previously incubated at 15, 30, and 60 days, before being exposed to the bacterial lawns, showed significant differences (p < 0.05) with E. coli and Thelebolus sp. plugs. On the other hand, C. gilvescens showed similar growth inhibition rings at all three growing periods for the four bacteria tested in the assay (p > 0.05). Similar results were obtained with bacteria exposed to 15 and 30 days grown of Penicillium sp. plugs (p > 0.05) (Supplementary file 3). Antarctomyces sp. was not included because it did not show antibacterial properties in our assays.
Discussion
The kingdom Fungi is considered a key contributor to the biotechnology industry42, with several applications in textile, food, and pharmaceutics processes43,44. Valuable compounds with antitumor, antiparasitic and antibacterial activity have been identified in fungi from Antarctica45,46,47,48,49,50. Although they have great potential as novel source of compounds, the genetic diversity of microbes from this pristine and unique polar environment is largely unexplored51. In this study, we describe the genetic and morphological characterization of four soil fungi isolated from Fort William Point, Greenwich Island, near Pedro Vicente Maldonado Ecuadorian Antarctic Research Station, some of which showed bioactivity against relevant clinical bacterial isolates.
For the genetic characterization of our selected isolates, we used a phylogenetic tree based on the sequence of the ITS region, which is considered as the barcode for fungal taxonomy52,53,54. There is some disagreement whether this region alone has enough variability as a reliable species-specific identification marker55,56,57,58,59. This has been shown to be the case with some genera in the Ascomycota60. Some authors suggest that using the ITS region along with other protein-coding genes such as RPB1 (RNA polymerase II largest subunit, regions E and F), RPB2 (RNA polymerase II second largest subunit, regions 5–7), Tsr1 (20S pre-rRNA processing protein), Cct8 (subunit of the cytosolic chaperonin Cct ring complex) and MCM7 (Minichromosome Maintenance Complex Component 7) to identify fungal species of the same genera with low intraspecific variation55. ITS region together with other genes such as calmodulin and β-tubulin have been useful in deeper taxonomical studies to discriminate between the genera Penicillium58, which has proven difficult to classify among the fungi taxa55. Recently, the ITS combined with fragments of β-tubulin and RPB2 were successfully used to identify a new species of Antarctomyces61, and to differentiate closely related fungi with low genetic variation62. However, other studies described β-tubulins as phylogenetically misleading, because they are present in the genome in multiple copies63,64. Species delimitation remains a challenging issue for closely related and cryptic fungal species65,66, and additional barcode markers, other than ITS, are being developed59.
In our study, we successfully confirmed the genus of our selected fungi with the use of a phylogenetic tree based on ITS sequencing. Isolates related to the Ascomycota group were confirmed as Penicillium sp., Thelebolus sp., and Antarctomyces sp. Identification to the species level for this group can be achieved with the implementation of additional gene sequence in upcoming studies. For a single isolate, the ITS sequencing allowed for species identification of the isolate T4-200-3B as C. gilvescens. Fungal morphological structures observed in this study were similar to previously descriptions for the same genera67. The integration of molecular data with other classification techniques such as morphology, ecology, new generation sequencing, and chemical profiling is nowadays our best set of tools to achieve a successful characterization of the fungi61,68,69,70,71.
Additionally, our phylogenetic analysis clearly separated the species of the Basidiomycota and Ascomycota phyla. Antarctomyces sp. and Thelebolus sp. segregated into sister clades that share an immediate common ancestor. These cryophilic genera have a slow generation time and thus accumulate only minor mutations, evolving slower than other species72. According to their geographic distribution, Thelebolus genus is known for its cryophilic nature and for its association with dung and guano72. Some species such as T. globosus and T. ellipsoideus are endemic to Antarctica, while others such as T. microsporus have a wider habitat, including Antarctica16,72,73,74. The genus Antarctomyces includes only two species, both native to the Antarctic continent16,61,75. Sharing the same phylum, Penicillium sp. clustered within the P. lividum and P. odoratum clade, and showed a strong bootstrap value with other species of the genus; all belonging to the section Aspergilloides60,76. The Basidiomycota phylum is represented by Cryptococcus gilvescens. This species distribution is restricted to cold environments, including the Antarctica77,78, where it is considered the most abundant genus of yeast79. C. gilvescens also showed a close relationship with C. gastricus, as previously reported78.
Bioactivity potential against pathogens is a promising application of the genetically diverse fungi of Antarctica. For instance, C. gilvescens and Penicillium sp. have shown antibiotic potential against Gram-negative bacteria, such as E. coli and K. pneumoniae, and Gram-positive bacteria, such as E. faecalis and S. aureus. This agrees in part with previous reports on the antibacterial activity of Cryptococcus species against Gram-positive bacteria22,80. In our study, C. gilvescens also showed antibacterial potential for Gram-negative bacteria. Additionally, C. gilvescens was reported to express extracellular lipolytic/esterasic activity, starch-degrading activity81, extracellular amylase, lipase, and protease activities78, anti-yeast activity82, and laccasse activity83. Various Cryptococcus isolated from Antarctic marine sediments had also exhibited lipase, esterase, and pectinase activity84.
In relation to species of Penicillium isolated from diverse polar ecosystems, such as marine sediments, deep-sea sediments, and sea-bed sediments, it is known that this genus has cytotoxic effects against cancer cell lines, anti-inflammatory effect, anti-allergic effect, antifungal and antibacterial activities84. A novel strain of Penicillium found in Antarctic soil showed production of three new indolyl diketopiperazine derivatives and seven known alkaloid compounds85. Some of these compounds had significant in vitro cytotoxic activity against cancer cell lines and one of them had antituberculosis activity85. An early study described nephrotoxicity in humans and strong antibiotic activity with P. odoratum86. This fungus produces the hazardous citrinin toxin, a mycotoxin that causes nephrotoxicity in humans87,88,89. Because the citrinin gene appears to be highly conserved within the genus Penicillium90, it is likely that citrinin is present in our Penicillium sp. isolate. P. lividum presented cytotoxic activity associated with the production of meroterpenoid compounds91. Furthermore, we report a Penicillium strain (Penicillium sp.) that produced antibacterial activity against Gram-negative and Gram-positive bacteria.
Previous studies have documented antitumoral20 and antibiotic potential in the Thelebolus genus, although the latter was less potent than Penicillium35. Thelebolus sp. from the Himalayas showed no antimicrobial activity against Gram-negative bacteria, but did exhibit antimicrobial activity against Gram-positive bacteria92. In contrast, Thelebolus sp. isolated in this study showed antibacterial activity against the Gram-negative bacteria E. coli. Several biotechnological applications have been attributed to T. microsporus due to the synthesis of linolenic acid, carotenoid pigments and extracellular α-amylase activity93. Lastly, our Antarctomyces isolate did not show any antibacterial activity against the tested bacteria in our in vitro assay conditions. Members of this genus, A. psychrotrophicus and A. pellizariae were attributed with potential biotechnological applications16. A. psychrotrophicus produced an antifreeze protein94, presented hydrocarbon biodegradation activity95 and showed antitumoral and antiprotozoal activity96. In addition, agar-block assays with A. psychrotrophicus described that this fungi has low antibacterial potential against E. coli, showing an inhibition growth zone between 7–10 mm97. On the other hand A. pellizariae produced a blue pigment with potential use in the food industry61.
To screen for bioactivity, this study used a low-cost in vitro assay adapted to the low temperature growth requirement of the fungi and the high temperature requirement for bacterial growth. This quick assay allowed us to detect bacterial growth inhibition zones around the fungi plugs as indicative of potential antibacterial activity. Without a complete knowledge of the environmental and nutrient requirements for the Antarctic fungi to produce bioactive compounds, we believe that this bioassay has its merit in detecting potential antibacterial metabolites that would have been missed otherwise. This bioassay may be extended to screen for antiviral and anticancer compounds, as well. Future studies will aim to isolate, identify, and characterize the putative bioactive compound(s). This work contributes to the preliminary description of soil fungi of Antarctica and to underscore its potential biotechnological applications and, thus, the importance of its environment conservation.
Material and methods
Soil sampling
The fungi evaluated in this study were isolated from soil samples collected in the Antarctic summer of 2008, near the “Pedro Vicente Maldonado” Antarctic Ecuadorian Scientific Station, located in Fort William Point, Greenwich Island. A total of three sites (stations GIT4-200, GIT4-400, and GIT4-1K) were sampled along a 1000 m linear transect (Fig. 5). At each sampling site, five soil sample replicates were collected with a sterile scoop in a 5 m radius from the registered GPS coordinate. The first 10 cm of soil surface from these five replicates were pooled and filtered with a 2 mm mesh. Soil samples were sealed in sterile polyethylene bags (Whirl-Pack) and transported in a cooler at 0–4 °C until their arrival to laboratory facilities at ESPOL (Guayaquil, Ecuador), where they were kept at 4 °C.
Location of Pedro Vicente Maldonado Scientific Station in Greenwich Island, Antarctica. The geographic location of the three land stations sampled are: GIT4-200 (62°26′53.9″S 59°44′07.7″W), GIT4-400 (62°26′58.7″S 59°43′58.9″W), and GIT4-1 K (62°27′16.4″S 59°43′39.8″W). The map was generated in QGIS version 3.10.14-A Coruña (https://www.qgis.org) using a geospatial vector of Antarctica's administrative boundaries obtained from http://www.diva-gis.org/.
Fungi isolation
The soil-plate method98 was used to grow the fungi on Potato Dextrose Agar, PDA (BD Difco™) with chloramphenicol (100 µg/ml) as the growth media. Under a vertical laminar flow cabinet, 0.1–0.3 g of soil were evenly dispersed on the solid media and incubated at 4 °C for a maximum of 12 days. Then, individual colonies were transferred to new PDA plates. The isolates analyzed in this report were codified as T4-200-3B, T4-400-5E, T4-1K-1G, and T4-1K-1A.
Fungal DNA extraction
DNA was extracted from 20-day old fungi cultures according to a previously described protocol, with minor modifications99. Briefly, 200–500 mg of fungi mycelium was mixed with 200 mg of 0.1 mm Zirconia/Silica beads (Biospec) and 500 µl of a bead beating solution (0.1 M NaCl, 5% sodium dodecyl sulfate and 0.5 M Tris–HCl, pH 8) into a 1.5 ml microcentrifuge tube. The tube was vortexed at maximum speed for 10 min. The mixture was cleared by centrifugation at 11,000×g for 10 min. The supernatant was transferred to a new tube containing 200 mg of clean beads and vortexed. After centrifugation, the supernatant was transferred to a clean 1.5 ml microcentrifuge tube and mixed with an equal volume of phenol (Sigma-Aldrich), and chloroform-isoamyl alcohol (Sigma-Aldrich) (25:24:1). The mixture was briefly vortexed and centrifuged at 11,000×g for 5 min. The aqueous layer was transferred to a new tube and treated with an equal volume of chloroform: isoamyl alcohol (49:1) solution (Sigma-Aldrich), vortexed and centrifuged for 5 min at 10,000×g. The aqueous layer was transferred to a new tube and mixed with 2.5 volumes of isopropanol, incubated for 1 h at 4 °C, and centrifuged 14,000×g for 10 min. The DNA pellet was washed twice with ice-cold 70% ethanol, dried and resuspended in 0.1 × TE buffer (1 mM Tris–HCl, 0.1 mM EDTA pH 8).
Amplification and cloning of the ITS region
The ITS region was amplified with Platinum Taq DNA polymerase (Invitrogen), using the universal primers ITS1 and ITS4100. PCR program consisted of 2 min at 94 °C, 35 cycles of 94 °C for 30 secs, 55 °C for 30 secs, 72 °C for 1 min, and a final extension of 3 min at 72 °C. Amplified products were resolved in a 1% agarose gel, stained with SYBR Safe (Invitrogen). The ITS fragment was cut from the gel, purified using High Pure PCR Product Purification Kit (Roche), cloned into a pGEM®-T Easy Vector (Promega), and sequenced by the Sanger method using SP6 and T7 universal primers (GENEWIZ, South Plainfield, NJ).
Sequence analysis
The four ITS sequences from this study were matched with sequences from GenBank using the BLAST software (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The final dataset included 43 sequences. They were then aligned using MAFFT101 with default settings. The final alignment was 737 bp long (Supplementary file 4). Sequences with complete information like species name, location and collection date were mainly selected for the phylogeny. The substitution model that best fit the data was selected using jModelTest 2.1.7102,103. The phylogenetic tree was constructed using the Maximum-likelihood method on MEGA Version 10.2.6104, using Kimura parameter-2 substitution model105 with uniform rate among sites, and 1000 Bootstrap replications.
Microscopic observation
Microscopic structures were observed using a compound microscope with 1000 × magnification after 15 and 30 days of growth on PDA media. A small amount of fungal culture was removed from the edge of the colony using an inoculation loop and then stained with 60% of lactophenol blue solution on a microscopic glass slide. The software Motic Image Plus 2.0 was used to measure fungal structures.
Antibiotic susceptibility tests
The clinical bacterial isolates used for the fungi antibacterial activity were previously diagnosed by classical antibiotic susceptibility tests using the Kirby Bauer method with the Agar Muller–Hinton media (Thermo Scientific™).
Because the Antarctic fungi were grown at low temperature and it is unknown the environmental conditions that may affect their potential antimicrobial activity, we performed the in vitro antibacterial assays on PDA plates at 4 °C and 37 °C. To this end, we first tested if an antibiotic resistant clinical isolate of Klebsiella pneumoniae was able to grow on PDA at 4 °C and 37 °C and still show antibiotic resistance. Bacteria streaked on PDA and Luria broth agar (LBA) were grown at 4 °C and 37 °C. The plates grown at 37 °C were incubated for 24 h, but the plates grown at 4 °C were incubated for 5 days, then the plates were transfer to 37 °C and incubated for further 24 h. On each plate antibiotic disks impregnated with 10 ug of imipenem and meropenem were deposited. Antibiotic resistance, depicted as clear rings around the antibiotic disks, were read after the 37 °C incubation in all treatments.
Assay of antibacterial potential
The antibacterial potential for the fungi was determined using the mycelia plugs method106, with fungi isolates grown at 4 °C on PDA. The fungi were analyzed at three sampling times of growth i.e. 15, 30, and 60 days. The clinical bacterial strains used in this assay were: Escherichia coli, Klebsiella pneumoniae, Enterococcus faecalis and Staphylococcus aureus. Plates with mycelia plugs and bacterial lawn were first incubated at 4 °C for 5 days on PDA to allow for fungi to grow and then transferred to 37 °C for 24 h for bacterial growth. The bioassays were performed with a minimum of three replicates, and the mean inhibition zone was calculated by measuring the border of the fungi colony to the border of the bacterial growth. This was photographed and measured in millimeters (mm) using the Motic Images Plus 2.0 software. The software SPSS 19 was used for the statistical analysis of the bacterial inhibition zone around the mycelia plug. The heterogeneity between days of growth was determined by applying Kruskal–Wallis and Mann–Whitney tests, with a statistical confidence level of 95%.
Data availability
All the new sequences will be available from the GenBank (https://www.ncbi.nlm.nih.gov/genbank/) database with the accession codes: MZ958926, MZ958927, MZ958928, MZ958929. The datasets generated or analyzed in this study are included within the article and its supplementary files.
References
Cook, D. & Zolnikov, T. R. Antarctica. In Global Adaptation and Resilience to Climate Change (ed. Zolnikov, T. R.) 31–49 (Springer International Publishing, 2019). https://doi.org/10.1007/978-3-030-01213-7_3.
Campbell, I. B. & Claridge, G. G. C. Antarctic permafrost soils. In Permafrost Soils (ed. Margesin, R.) 17–31 (Springer, 2009). https://doi.org/10.1007/978-3-540-69371-0_2.
Onofri, S. et al. Evolution and adaptation of fungi at boundaries of life. Adv. Space Res. 40, 1657–1664 (2007).
Ruisi, S., Barreca, D., Selbmann, L., Zucconi, L. & Onofri, S. Fungi in Antarctica. Rev. Environ. Sci. Biotechnol. 6, 127–141 (2007).
Wynn-Williams, D. D. Antarctic microbial diversity: The basis of polar ecosystem processes. Biodivers. Conserv. 5, 1271–1293 (1996).
Pointing, S. B. et al. Highly specialized microbial diversity in hyper-arid polar desert. Proc. Natl. Acad. Sci. U.S.A. 106, 19964–19969 (2009).
Stotz, G. C. et al. Trends in Antarctic ecological research in Latin America shown by publications in international journals. Polar Res. 32, 19993 (2013).
Godinho, V. M. et al. Diversity and bioprospecting of fungal communities associated with endemic and cold-adapted macroalgae in Antarctica. ISME J. 7, 1434–1451 (2015).
Durán, P. et al. Occurrence of soil fungi in Antarctic pristine environments. Front. Bioeng. Biotechnol. 7, 28 (2019).
Alves, I. M. S. et al. The diversity, distribution, and pathogenic potential of cultivable fungi present in rocks from the South Shetlands archipelago, Maritime Antarctica. Extremophiles 23, 327–336 (2019).
Arenz, B. E., Blanchette, R. A. & Farrell, R. L. Fungal diversity in Antarctic soils. In Antarctic Terrestrial Microbiology: Physical and Biological Properties of Antarctic Soils (ed. Cowan, D. A.) 35–53 (Springer, 2014). https://doi.org/10.1007/978-3-642-45213-0_3.
Rosa, L. H. et al. DNA metabarcoding uncovers fungal diversity in soils of protected and non-protected areas on Deception Island, Antarctica. Sci. Rep. 10, 21986 (2020).
de Menezes, G. C. A. et al. Fungi in the Antarctic cryosphere: Using DNA metabarcoding to reveal fungal diversity in glacial ice from the Antarctic peninsula region. Microb. Ecol. 83, 647–657 (2022).
de Souza, L. M. D. et al. Diversity, distribution and ecology of fungal communities present in Antarctic lake sediments uncovered by DNA metabarcoding. Sci. Rep. 12, 8407 (2022).
Doytchinov, V. V. & Dimov, S. G. Microbial community composition of the Antarctic ecosystems: Review of the bacteria, fungi, and archaea identified through an NGS-based metagenomics approach. Life 12, 916 (2022).
Rosa, L. H. et al. Fungi in Antarctica: Diversity, ecology, effects of climate change, and bioprospection for bioactive compounds. In Fungi of Antarctica: Diversity, Ecology and Biotechnological Applications (ed. Rosa, L. H.) 1–17 (Springer International Publishing, 2019). https://doi.org/10.1007/978-3-030-18367-7_1.
Newsham, K. K., Davey, M. L., Hopkins, D. W. & Dennis, P. G. Regional diversity of maritime Antarctic soil fungi and predicted responses of guilds and growth forms to climate change. Front. Microbiol. 11, 615659 (2021).
Krishnan, A. et al. Temperature and pH profiling of extracellular amylase from Antarctic and Arctic soil microfungi. Fermentation 8, 601 (2022).
Gerday, C. et al. Cold-adapted enzymes: From fundamentals to biotechnology. Trends Biotechnol. 18, 103–107 (2000).
Henríquez, M. et al. Diversity of cultivable fungi associated with Antarctic marine sponges and screening for their antimicrobial, antitumoral and antioxidant potential. World J. Microbiol. Biotechnol. 30, 65–76 (2014).
Gonçalves, V. N. et al. Antibacterial, antifungal and antiprotozoal activities of fungal communities present in different substrates from Antarctica. Polar Biol. 38, 1143–1152 (2015).
Singh, P., Tsuji, M., Singh, S. M., Roy, U. & Hoshino, T. Taxonomic characterization, adaptation strategies and biotechnological potential of cryophilic yeasts from ice cores of Midre Lovénbreen glacier, Svalbard, Arctic. Cryobiology 66, 167–175 (2013).
Maggi, O. et al. Adaptation of fungi, including yeasts, to cold environments. Plant Biosyst. 147, 247–258 (2013).
Hoff, B. et al. Two components of a velvet-like complex control hyphal morphogenesis, conidiophore development, and penicillin biosynthesis in Penicillium chrysogenum. Eukaryot. Cell 9, 1236–1250 (2010).
Bayram, O. et al. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 320, 1504–1506 (2008).
Yin, W. & Keller, N. P. Transcriptional regulatory elements in fungal secondary metabolism. J. Microbiol. 49, 329–339 (2011).
Svahn, K. S., Chryssanthou, E., Olsen, B., Bohlin, L. & Göransson, U. Penicillium nalgiovense Laxa isolated from Antarctica is a new source of the antifungal metabolite amphotericin B. Fungal Biol. Biotechnol. 2, 1 (2015).
Magiorakos, A.-P. et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281 (2012).
Bentley, S. D. et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417, 141–147 (2002).
Böhm, J. et al. Sexual reproduction and mating-type-mediated strain development in the penicillin-producing fungus Penicillium chrysogenum. Proc. Natl. Acad. Sci. 110, 1476–1481 (2013).
Xu, L. et al. Complete genome sequence and comparative genomic analyses of the vancomycin-producing Amycolatopsis orientalis. BMC Genom. 15, 363 (2014).
Silver, L. L. Challenges of antibacterial discovery. Clin. Microbiol. Rev. 24, 71–109 (2011).
Saleem, M. et al. Antimicrobial natural products: An update on future antibiotic drug candidates. Nat. Prod. Rep. 27, 238–254 (2010).
Lo Giudice, A. & Fani, R. Antimicrobial Potential of Cold-Adapted Bacteria and Fungi from Polar Regions 83–115 (Springer, 2016). https://doi.org/10.1007/978-3-319-13521-2_3.
Brunati, M. et al. Diversity and pharmaceutical screening of fungi from benthic mats of Antarctic lakes. Mar. Genom. 2, 43–50 (2009).
Li, Y. et al. Bioactive asterric acid derivatives from the Antarctic ascomycete fungus Geomyces sp. J. Nat. Prod. 71, 1643–1646 (2008).
Encheva-Malinova, M. et al. Antibacterial potential of streptomycete strains from Antarctic soils. Biotechnol. Biotechnol. Equip. 28, 721–727 (2014).
Godinho, V. M. et al. Diversity and bioprospection of fungal community present in oligotrophic soil of continental Antarctica. Extremophiles 19, 585–596 (2015).
Hemala, L., Zhang, D. & Margesin, R. Cold-active antibacterial and antifungal activities and antibiotic resistance of bacteria isolated from an alpine hydrocarbon-contaminated industrial site. Res. Microbiol. 165, 447–456 (2014).
Lim, F. Y., Sanchez, J. F., Wang, C. C. C. & Keller, N. P. Toward awakening cryptic secondary metabolite gene clusters in filamentous fungi. Methods Enzymol. 517, 303–324 (2012).
Vester, J. K., Glaring, M. A. & Stougaard, P. Improved cultivation and metagenomics as new tools for bioprospecting in cold environments. Extremophiles 19, 17–29 (2015).
Demain, A. L., Velasco, J., Adrio, J. L., Velasco, J. & Adrio, J. L. Industrial Mycology: Past, Present, and Future 20–45 (CRC Press, 2004). https://doi.org/10.1201/9780203970553-5.
Ghorai, S. et al. Fungal biotechnology in food and feed processing. Food Res. Int. 42, 577–587 (2009).
Adrio, J. L. & Demain, A. L. Fungal biotechnology. Int. Microbiol. 6, 191–199 (2003).
Gonzalez del Val, A. et al. Novel illudins from Coprinopsis episcopalis (syn. Coprinus episcopalis), and the distribution of illudin-like compounds among filamentous fungi. Mycol. Res. 107, 1201–1209 (2003).
Kelner, M. J. et al. Enhanced antitumor activity of irofulven in combination with antimitotic agents. Investig. New Drugs 20, 271–279 (2002).
Stone, E. A., Fung, H. B. & Kirschenbaum, H. L. Caspofungin: An echinocandin antifungal agent. Clin. Ther. 24, 351–377 (2002) (discussion 329).
Bianchini, L. F. et al. Microbial biotransformation to obtain new antifungals. Front. Microbiol. 6, 1433 (2015).
Pelaez, F. Biological Activities of Fungal Metabolites 68–111 (CRC Press, 2004). https://doi.org/10.1201/9780203970553-7.
Strohl, W. R. Industrial antibiotics: Today and the future. In Biotechnology of Antibiotics Vol. 82 1–47 (Marcel Dekker, 1997).
Ortiz, M. et al. Multiple energy sources and metabolic strategies sustain microbial diversity in Antarctic desert soils. PNAS 118, e202532218 (2021).
Blaalid, R. et al. ITS1 versus ITS2 as DNA metabarcodes for fungi. Mol. Ecol. Resour. 13, 218–224 (2013).
Begerow, D., Nilsson, H., Unterseher, M. & Maier, W. Current state and perspectives of fungal DNA barcoding and rapid identification procedures. Appl. Microbiol. Biotechnol. 87, 99–108 (2010).
Tekpinar, A. D. & Kalmer, A. Utility of various molecular markers in fungal identification and phylogeny. Nova Hedwigia https://doi.org/10.1127/nova_hedwigia/2019/0528 (2019).
Houbraken, J. & Samson, R. A. A. Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Stud. Mycol. 70, 1–51 (2011).
Carbone, I. & Kohn, L. M. Ribosomal DNA sequence divergence within internal transcribed spacer 1 of the Sclerotiniaceae. Mycologia 85, 415 (1993).
Skouboe, P. Phylogenetic analysis of nucleotide sequences from the ITS region of terverticilllate Penicillium species. Mycol. Res. 103, 873–881 (1999).
Houbraken, J., Frisvad, J. C. & Samson, R. A. Taxonomy of Penicillium section Citrina. Stud. Mycol. 70, 53–138 (2011).
Doilom, M. et al. Can ITS sequence data identify fungal endophytes from cultures? A case study from Rhizophora apiculata. Mycosphere 8, 1869–1892 (2017).
Visagie, C. M. M. et al. Identification and nomenclature of the genus Penicillium. Stud. Mycol. 78, 343–371 (2014).
de Menezes, G. C. A., Godinho, V. M., Porto, B. A., Gonçalves, V. N. & Rosa, L. H. Antarctomyces pellizariae sp. nov., a new, endemic, blue, snow resident psychrophilic ascomycete fungus from Antarctica. Extremophiles 21, 259–269 (2017).
Houbraken, J. et al. New penicillin-producing Penicillium species and an overview of section Chrysogena. Persoonia Mol. Phylogeny Evol. Fungi 29, 78–100 (2012).
Landvik, S., Eriksson, O. E. & Berbee, M. L. Neolecta—A fungal dinosaur? Evidence from β-tubulin amino acid sequences. Mycologia 93, 1151–1163 (2001).
Peterson, S. W. Phylogenetic analysis of Aspergillus species using DNA sequences from four loci. Mycologia 100, 205–226 (2008).
Lumbsch, H. T. & Leavitt, S. D. Goodbye morphology? A paradigm shift in the delimitation of species in lichenized fungi. Fungal Divers. 50, 59–72 (2011).
Balasundaram, S. V., Engh, I. B., Skrede, I. & Kauserud, H. How many DNA markers are needed to reveal cryptic fungal species?. Fungal Biol. 119, 940–945 (2015).
Coello Aguilar, S. Aislamiento e identificación de microhongos terrestres de la isla greenwich, antartida, y su potencial como controladores biologos (Escuela Superior Politécnica del Litoral, 2012).
Xu, M. et al. DNA barcoding and LC–MS metabolite profiling of the lichen-forming genus Melanelia: Specimen identification and discrimination focusing on Icelandic taxa. PLoS One 12, e0178012 (2017).
Wang, Z., Nilsson, R. H., James, T. Y., Dai, Y. & Townsend, J. P. Future Perspectives and Challenges of Fungal Systematics in the Age of Big Data 25–46 (Springer, 2016). https://doi.org/10.1007/978-3-319-29137-6_3.
Merényi, Z. et al. Challenges in the delimitation of morphologically similar species: A case study of Tuber brumale agg. (Ascomycota, Pezizales). Mycol. Prog. 16, 613–624 (2017).
Hibbett, D. et al. Sequence-based classification and identification of fungi. Mycologia 108, 1049–1068 (2017).
De Hoog, G. S. et al. Evolution, taxonomy and ecology of the genus Thelebolus in Antarctica. Stud. Mycol. 51, 33 (2005).
Arenz, B. E., Held, B. W., Jurgens, J. A., Farrell, R. L. & Blanchette, R. A. Fungal diversity in soils and historic wood from the Ross Sea Region of Antarctica. Soil Biol. Biochem. 38, 3057–3064 (2006).
Anupama, P., Praveen, K., Singh, R., Srivastava, A. & Arora, D. A psychrophilic and halotolerant strain of Thelebolus microsporus from Pangong Lake, Himalaya. Mycosphere 2, 601–609 (2011).
Stchigel, A. M., Cano, J., Mac Cormack, W. & Guarro, J. Antarctomyces psychrotrophicus gen. et sp. nov., a new ascomycete from Antarctica. Mycol. Res. 105, 377–382 (2001).
Samson, R. A. & Houbraken, J. Phylogenetic and taxonomic studies on the genera Penicillium and Talaromyces. Stud. Mycol. 70 (2011).
Branda, E. et al. Yeast and yeast-like diversity in the southernmost glacier of Europe (Calderone Glacier, Apennines, Italy). FEMS Microbiol. Ecol. 72, 354–369 (2010).
Carrasco, M. et al. Diversity and extracellular enzymatic activities of yeasts isolated from King George Island, the sub-Antarctic region. BMC Microbiol. 12, 251 (2012).
Shivaji, S. & Prasad, G. S. Antarctic yeasts: Biodiversity and potential applications. In Yeast Biotechnology: Diversity and Applications (Springer Netherlands, 2009). https://doi.org/10.1007/978-1-4020-8292-4_1.
Dilika, F., Bremner, P. D. & Meyer, J. J. M. Antibacterial activity of linoleic and oleic acids isolated from Helichrysum pedunculatum: A plant used during circumcision rites. Fitoterapia 71, 450–452 (2000).
Turchetti, B. et al. Psychrophilic yeasts in glacial environments of Alpine glaciers. FEMS Microbiol. Ecol. 63, 73–83 (2008).
Troncoso, E. et al. Identification and characterization of yeasts isolated from the South Shetland Islands and the Antarctic Peninsula. Polar Biol. 40, 649–658 (2017).
Rovati, J. I., Pajot, H. F., Ruberto, L., Mac Cormack, W. & Figueroa, L. I. C. Polyphenolic substrates and dyes degradation by yeasts from 25 de Mayo/King George Island (Antarctica). Yeast 30, 459–470 (2013).
Varrella, S. et al. Diversity, ecological role and biotechnological potential of Antarctic marine fungi. J. Fungi 7, 391 (2021).
Wang, J. et al. Three new indolyl diketopiperazine metabolites from the Antarctic soil-derived fungus Penicillium sp. SCSIO 05705. RSC Adv. 5, 68736–68742 (2015).
Wang, Y., Hong, F. K., Hwang, F. T. & Fan, C. S. Citrinin as an antibiotic. Science 106, 291–292 (1947).
Frisvad, J. C., Thrane, U., Samson, R. A. & Pitt, J. I. Important mycotoxins and the fungi which produce them. Adv. Exp. Med. Biol. 571, 3–31 (2006).
Perrone, G. & Susca, A. Penicillium Species and Their Associated Mycotoxins 107–119 (Humana Press, 2017). https://doi.org/10.1007/978-1-4939-6707-0_5.
Degen, G. H., Ali, N. & Gundert-Remy, U. Preliminary data on citrinin kinetics in humans and their use to estimate citrinin exposure based on biomarkers. Toxicol. Lett. 282, 43–48 (2018).
Schmidt-Heydt, M., Stoll, D. & Geisen, R. Whole-genome sequencing of the fungus Penicillium citrinum reveals the biosynthesis gene cluster for the mycotoxin citrinin. Microbiol. Resour. Announc. 8, e01419-18 (2019).
Zhuravleva, O. I. et al. Meroterpenoids from the alga-derived fungi Penicillium thomii Maire and Penicillium lividum Westling. J. Nat. Prod. 77, 1390–1395 (2014).
Hassan, N. et al. Potential of psychrotrophic fungi isolated from Siachen glacier, Pakistan, to produce antimicrobial metabolites. Appl. Ecol. Environ. Res. 15, 1157–1171 (2017).
Singh, S. K. S. M., Singh, P. N., Singh, S. K. S. M. & Sharma, P. K. Pigment, fatty acid and extracellular enzyme analysis of a fungal strain Thelebolus microsporus from Larsemann Hills, Antarctica. Polar Rec. 50, 31–36 (2013).
Xiao, N. et al. Antifreeze activities of various fungi and Stramenopila isolated from Antarctica. N. Am. Fungi 5, 215–220 (2010).
Ferrari, B. C., Zhang, C. & van Dorst, J. Recovering greater fungal diversity from pristine and diesel fuel contaminated sub-Antarctic soil through cultivation using both a high and a low nutrient media approach. Front. Microbiol. 2, 217 (2011).
Santiago, I. F. et al. Leishmanicidal and antitumoral activities of endophytic fungi associated with the Antarctic angiosperms Deschampsia antarctica Desv. and Colobanthus quitensis (Kunth) Bartl. Extremophiles 16, 95–103 (2012).
Abneuf, M. A. et al. Antimicrobial activity of microfungi from maritime Antarctic soil. Czech. Polar Rep. 6, 141–154 (2016).
Warcup, J. H. The soil-plate method for isolation of fungi from soil. Nature 166, 117–118 (1950).
Płaza, G. A., Upchurch, R., Brigmon, R. L., Whitman, W. B. & Ulfig, K. Rapid DNA extraction for screening soil filamentous fungi using PCR amplification. Pol. J. Environ. Stud. 13, 315–318 (2004).
White, T. et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications (Academic Press, 1990).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Darriba, D., Taboada, G. L., Doallo, R. & Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 9, 772 (2012).
Guindon, S. & Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704 (2003).
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549 (2018).
Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120 (1980).
Fusaro, R. M. Inoculation technique for fungus cultures. Appl. Microbiol. 23, 174–176 (1972).
Acknowledgements
We want to thank Instituto Antártico Ecuatoriano (INAE) for their extraordinary support and logistic assistance for traveling to and from Antarctica. We are in debt for their significant effort in the sample collection process and for the accommodation at Pedro Vicente Maldonado Scientific Station in Antarctica. This work was supported by the grant Seed Funding of Escuela Superior Politécnica del Litoral (ESPOL).
Author information
Authors and Affiliations
Contributions
W.B.C. designed the study. E.O.-E. and P.E. developed the protocols and conducted data recording. E.O.-E., and R.C. conducted bioinformatic analyses. S.C. isolated fungal samples. N.O. conducted soil sampling. E.O.-E. performed the statistical analysis and wrote the first draft of the manuscript. W.B.C., D.C., R.V.C., N.O. and P.E. contributed and commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ordóñez-Enireb, E., Cucalón, R.V., Cárdenas, D. et al. Antarctic fungi with antibiotic potential isolated from Fort William Point, Antarctica. Sci Rep 12, 21477 (2022). https://doi.org/10.1038/s41598-022-25911-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-25911-x
This article is cited by
-
Antarctic marine sediment as a source of filamentous fungi-derived antimicrobial and antitumor compounds of pharmaceutical interest
Extremophiles (2024)
-
Guest edited collection: fungal evolution and diversity
Scientific Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.