Abstract
So far, few studies have examined the effect of salt taste receptors genetic variation on dietary intake in the Iranian population. We aimed to evaluate associations between single nucleotide polymorphisms (SNPs) in salt taste receptors’ genes with dietary salt intake and blood pressure. A cross-sectional study was carried out among 116 randomly selected healthy adults aged ≥ 18 in Isfahan, Iran. Participants underwent sodium intake determination by 24-h urine collection, as well as dietary assessment by semi-quantitative food frequency questionnaire and blood pressure measurement. Whole blood was collected to extract DNA and genotype of SNP rs239345 in SCNN1B and rs224534, rs4790151 and rs8065080 in TRPV1 gene. Sodium consumption and diastolic blood pressure were significantly higher in carriers of the A-allele in rs239345 compared to subjects with the TT genotype (4808.4 ± 824.4 mg/day vs. 4043.5 ± 989.3 mg/day; P = 0.004) and 83.6 ± 8.5 mmHg vs. 77.3 ± 7.3 mmHg; P = 0.011), respectively. The level of sodium intake was lower in the TT genotype of TRPV1 (rs224534) than the CC genotype (3767.0 ± 713.7 mg/day vs. 4633.3 ± 793.5 mg/day; P = 0.012). We could not find any association between genotypes of all SNPs with systolic blood pressure as well as genotypes of rs224534, rs4790151 and rs8065080 with diastolic blood pressure. Genetic variations can relate with salt intake and consequently may associate with hypertension and finally cardiovascular disease risk in the Iranian population.
Similar content being viewed by others
Introduction
Approximately, 1.7 million cardiovascular disease (CVD) deaths in 2010 were attributed to high dietary sodium consumption, accounting for 10% of all CVD deaths. Furthermore, in a recent study by Messerli, it has been shown that sodium intake correlates positively with life expectancy and inversely with all-cause mortality worldwide and in high-income countries claims against dietary sodium intake are a reason of decreasing life span or a risk factor for premature deaths 1,2. Both individual- and population-based studies have shown that genetic and environmental factors significantly influence sodium consumption and consequently, the blood pressure (BP) level 3. It has been shown that excessive sodium consumption can be attributed to higher preferences for salty foods, which may be linked to genetic factors such as salt taste receptor function 4.
The amiloride-sensitive (AS) fibers using the ENaC protein moderate sodium taste preference, which typically shows a lower taste concentration threshold. However, the amiloride-insensitive (AI) fibers and hence TRPV1 can regulate aversive responses to salt concentrations 5. Therefore, AS and AI fibers structures facilitate salt taste transduction. It has been shown that SNPs in the SCNN1B gene polymorphism of AA/AT, rs239345, coded for ENaC β subunit and TRPV1 genes including rs239345, rs3785368, rs8065080 are associated with differences in salt taste perception and BP among adults and children 6,7. In addition, inter-individual variation sources such as environmental and cultural determinants of dietary intake, may have an important controversial role in salt preference, as well 8,9. Consequently, understanding the genetic variation in taste perception may lead to new personalized dietary approaches, which can reduce the risk of CVDs 10. There are many controversies on the association of various SNPs in the SLC4A5, SCNN1B and TRPV1 genes like SLC4A5 rs7571842, SLC4A5 rs10177833, SCNN1B rs239345 and TRPV1-rs8065080 with the salt intake, preference and sensitivity along with health markers like SBP and DBP 6,11,12,13,14,15,16,17. However, studies have mostly been conducted in Caucasians from Europe, US or Canada 11,12,13,14 and few studies have examined the effect of genetic variations on the sodium intake in diverse populations such as Middle East countries like Iran. Therefore, more research is needed to determine the effect of single nucleotide polymorphisms (SNPs) in different taste receptor genes on taste sensitivity. This study aimed to examine the effect of a set of SNPs on sodium intake, food contribution in sodium intake and BP among Iranian adults.
Materials and methods
Design and subjects
The present cross-sectional study consisted of 116 adults aged > 18 living in Isfahan, Iran. Exclusion criteria included diagnosis of diabetes, renal insufficiency, having special dietary regimen, fasting or menstruation (for women) on the day of sampling, using diuretics (because of 24-h urine collection to estimate salt intake) and oral contraceptives, the women who are pregnant and lactating, participants with impaired taste, excessive sweating during an unusually hot day or unusual physical activity and incomplete 24-h urine collection.
Participants were selected using multi-stage random cluster sampling method. One adult person aged > 18 was selected from each household. Considering the minor allele frequency of 26.42 for SCNN1B (rs239345), sample size calculated by 106 and after predicting 10% non-response rate, final sample size for this study was 116. They were referred to Isfahan Cardiovascular Research Institute (ICRI) for data collection. Our participation rate was 92%. We obtained written informed consents from all participants. All methods were carried out in accordance with relevant guidelines and regulations (e.g. Helsinki declaration). This study was approved by ethical committee of National Institute for medical research Development (IR.NIMAD.REC.1397.346).
Data collection
Trained health professionals conducted the detailed interviews to obtain information about participants’ socioeconomic status including education, occupation, demographic characteristics and smoking habit. Physical activities were assessed by means of International Physical Activity Questionnaire (IPAQ) 18.
Anthropometrics and blood pressure measurements
Trained health professionals measured standing height without shoes and recorded to the nearest 0.5 cm. Body weight was measured for subjects wearing light clothes, without shoes and recorded to the nearest 0.5 kg. Body mass index (BMI) was calculated as weight divided by height squared (kg/m2). While subjects were standing, waist circumference (WC) was measured midway between the lower ribs and the iliac crest in a horizontal plane. Hip circumference was also measured at the point yielding the maximum circumference over the buttocks using a non-elastic meter.
BP was measured (twice in each arm) manually with a mercury sphygmomanometer according to standard protocols 19, after resting for 5 min in a seated position. The first Korotkoff sound was recorded as the systolic BP (SBP) and the disappearance of the sounds (V phase) was considered as the diastolic BP (DBP). We used the mean BP from the arm with the highest value 16.
Dietary assessment
Dietary intake was assessed using validated 136-item semi-quantitative food frequency questionnaire (FFQ) 20. This FFQ was prepared to determine the amount of sodium intake and food contribution in sodium intake. We calculated the discretionary salt which was added at table and in cooking through questioning about the weight of salt packages, the number of households, and the period of time that each salt package is used 20. This FFQ was validated for assessment of sodium intake against two reference methods including twelve 24-h dietary recall which were completed monthly during a year among 113 heathy adults aged ≥ 19 in Isfahan, Iran. They administered two FFQ at the beginning and after 1 year to evaluate the reproducibility. The deattenuated Spearman correlation coefficient between the contribution of food sources of sodium based on the FFQ and 24-h dietary recalls varied from 0.39 for canned food to 0.53 for added salt (P < 0.001). The deattenuated Spearman correlation coefficient between the FFQ and 24-h dietary recall for total sodium intake was 0.65 (P < 0.001). Intraclass correlation coefficient (95% CI) between two FFQ ranged from 0.20 (0.005–0.37) (P = 0.031) for sauces to 0.49 (0.28–0.69) for bread. According to the Bland–Altman plots, we observed an acceptable level of agreement between the two methods for sodium intake 20.
All participants were asked to answer how often they had consumed food items over the past year in 9 options including never or rarely (less than 1 per month), 1–3 per month, 1 per week, 2–4 per week, 5–6 per weak, 1 per day, 2–3 per day, 4–5 per day and 6 or more per day. The Iranian Food Consumption Program (IFCP) 21 was used to calculate sodium intake and food group servings for all foods reported in the FFQ using the Iranian Food Composition Table 22. The FFQ contained four questions about discretionary salt consumption including the salt used at the table; the weight of the salt package used; time taken to consume each salt package and the number of family members 23.
Urine collection
The morning urine samples were collected on 2 days at 7:00 a.m. (the first sample of the first day was excluded) and was poured into a sterile plastic container labeled with the participants’ ID and a special code. The samples with low volume or lack of proper collection were excluded. The urine samples were collected at home for those that the delivery of their samples was impossible for any reason. Fasting venous blood samples were taken to measure serum biochemical parameters including, fasting blood glucose (FBG), serum albumin level, and lipid profile. Each participant provided two samples for the urinary sodium, potassium and chloride. The mean of 24-h urine sodium excretion was used to estimate daily salt intake. In order to obtain the whole 24-h urinary Na (24hUNa), we multiplied Na concentration by the volume in liters. The urinary sodium, potassium and chloride were measured by emission flame photometry and creatinine (Cr) was measured by the Jaffe method (Technical SMA 12–60) 24 in 24-h urine samples. The 24-h urine samples’ completeness was evaluated through the following criteria: Total 24-h urine volume sample ≥ 500 mL, missing no more than 1 void during collection, and collection of ≥ 20-h and 24-h urine creatinine (24hUCr) ≥ 20 mg/dL per kg of body weight in men and ≥ 15 mg/mL per kg of body weight in women aged < 50 and 24hUCr ≥ 10 mg/dL per kg of body weight in men and ≥ 7.5 mg/mL per kg of body weight for men and women aged ≥ 50 25.
Single nucleotide polymorphisms selection and genotyping
A PubMed SNP search was conducted for the following genes associated to taste detection. The selected SNPs were filtered by global minor allele frequency (MAF), and SNPs with a minor allele frequency below 5% were removed. The resulting SNPs were filtered using HaploView 4.2 software to obtain tag SNPs (tSNPs). Each tSNP was considered independent due to low linkage disequilibrium (r2 < 0.05). SCNN1B rs239345 (MAF = 0.2642) and TRPV1 gene rs4790151 (MAF = 0.2138), rs224534 (MAF = 0.378) and rs8065080 (MAF = 0.3177). Dias et al.’s study showed that TRPV1 receptor might have an important role in salt preference 6. Rs8065080, in the TRPV1 gene, affect functional activity of TRPV1 receptors and be involved in different risk factors and pain conditions 26. González-Mercado et al.’s study showed that four SNPs including rs224534 and rs8065080 are located in this gene as haplotype 24. Previous studies reported the role of TRPV1-rs8065080 polymorphism in ion channel function, rs224534 and two other variants in higher sodium intake 27,28. There is little evidence about rs3785368 variant to suggest its role in salt intake. Therefore, due to lack of budget, we selected the most important polymorphism which could have role and other variants will be studied in the future. Genotyping design of this variant is based on reported variant in dbSNP database.
Genotyping analysis
DNA was isolated from peripheral blood lymphocytes using the standard salting out method 29. Genotyping was carried out using ARMS method for allelic discrimination, and validated by capillary sequencing (AB3730, Applied Biosystems). Primers and annealing temperature used in the study are listed in Supplementary Appendix 1. The reaction details are as follows: PCR using an Eppendorf gradient type master cycler (Eppendorf, Germany) with a total volume of 16 µL (8 µL Taq PCR Master Mix, 0.4 µL each outer primer (10 mM), 0.2 µL each inner primer (10 mM), 1 µL genomic DNA and 3.2 µL H2O). After initial denaturation (95 °C 3 min) 30 cycles (95 °C 45 s, annealing temperature (Supplementary Appendix 1) 45 s, 72 °C 15 s) of amplification were performed, followed by an extension (72 °C 15 s) and a final elongation step (72 °C 1 min).
Statistical analysis
Kolmogorov–Smirnov test was used for assessing the data distribution in all datasets. Data were reported as mean ± standard deviation (SD) for quantitative variables and number (percentage) for qualitative variables. A chi-square (χ2) test was performed to determine whether distributions of the genotypes of the study polymorphisms were in Hardy–Weinberg equilibrium. One-way analysis of variance (ANOVA) and then Bonferroni as post hoc method (for correcting significance level due to multiple tests) were utilized to compare means in variables with normal distribution. Kruskal–Wallis test and then Mann–Whitney test as post hoc method were used to evaluate means of continuous variables across different genotypes of each SNP, when the assumptions of one-way analysis of variance were not met. Distribution of categorical variables across quartiles of different genotypes of each SNP were assessed using Chi-square test. We considered 2-tailed P values of less than 0.05 to be statistically significant. Analyses were conducted using SPSS statistical software version 19.0 for windows (SPSS Inc., Chicago, USA).
Ethics approval and consent to participate
This study was funded by National Institute for Medical Research Development (NIMAD, Grant Number 977549). Written informed consents were obtained from adult participants and the parents of children.
Results
Totally, 116 healthy adults (56% male and 44% female) were chosen for the study. Participants’ characteristics are summarized in Table 1. Participants had a mean age of 35.4 ± 7.7 years, BMI of 27.3 ± 4.9 kg/m2, total physical activity of 774.1 ± 667.3 METs minute per week, SBP of 122.6 ± 16.3 mmHg, DBP of 80.3 ± 14.7 mmHg and 11.2% of subjects were currently smokers. The 24UNa mean as a surrogate of sodium intake was 4651.0 ± 1647.7 mg/day. Table 2 shows the dietary intake in participants based on sex. The mean energy intake was 2080.5 ± 1656.5 kcal/day and the percentage of energy from carbohydrate, protein, total fat, saturated fatty acid (SFA), monounsaturated fatty acid (MUFA) and polyunsaturated fatty acid (PUFA) were 54.8 ± 8.7, 15.4 ± 2.5, 30.6 ± 7.4, 12.4 ± 5.9, 8.9 ± 2.5 and 8.2 ± 3.1, respectively and the mean fiber intake was 22.2 ± 10.7. The genotypic frequencies of the polymorphisms of rs239345 in the SCNN1B and rs4790151, rs224534 and rs8065080 in theTRPV1 in the participants of our study were consistent with the Hardy–Weinberg equilibrium (Table 3). Table 4 shows genotypes and alleles distributions of the SNP rs239345 in the SCNN1B and SNPs rs4790151, rs224534, rs8065080 in theTRPV1 genes.
Single nucleotide polymorphisms and their association with sodium sources intake
The mean sodium intake from its major sources according to genotypes of studied SNPs are shown in Table 5. The intake of added salt in subjects with AA genotype (SNP rs239345, P = 0.010) was significantly higher than those with TT genotype. However, there was no significant association between various genotypes and mean intake of dietary sources of sodium in other SNPs.
Single nucleotide polymorphisms and their association with salt intake and blood pressure
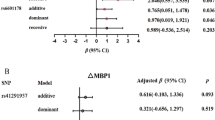
The comparison of mean sodium intake, SBP and DBP between related genotypes of different SNPs are shown in Table 6. There were significant differences between AA and TT genotypes of SNP rs239345 for sodium intake (4808.4 ± 824.4 mg/day vs. 4043.5 ± 989.3 mg/day; P = 0.004) and DBP (83.6 ± 8.5 mmHg vs. 77.3 ± 7.3 mmHg; P = 0.011). In addition, the mean sodium intake and DBP was significantly higher in A vs. T allele in SNP rs239345 (P = 0.035). The sodium intake was higher in the CC genotype of rs224534 in the TRPV1 than the TT genotype (4633.3 ± 793.5 mg vs. 3767.0 ± 713.7 mg/day/day; P = 0.012). It was also higher in C than T allel in SNP rs224534 (P = 0.029). There was no relationship between all SNPs and SBP, also SNPs including rs224534, rs4790151 and rs8065080 with DBP. Furthermore, non-significant differences were found in dietary sodium intake, with all genotypes of rs4790151 and rs8065080 SNPs in the TRPV1 gene.
Discussion
The current study examined the association between variations in taste detection genes and sodium intake as well as BP for the first time in Iran and Eastern Mediterranean region. The findings of this study showed that among four SNPs which were studied, only two SNPs including rs239345 and rs224534 were significantly related to sodium intake and only rs239345 was involved in DBP level.
Regulation of salt or sodium intake is partly due to genes variation related to homeostatic sodium regulation and to hedonic responses to the salt taste 6,30. In this study, it was shown that individuals with SCNN1B gene polymorphism of AA/AT, rs239345, coded for SCNN1B and the TRPV1 gene polymorphism of CC, rs224534 had higher sodium intake. Moreover, individuals with AA genotype of SNP rs239345 had higher consumption of added salt, as one of main source of salt intake 31. In addition, individuals with AA genotype of SNP rs239345 had higher DBP level than those with TT genotype. Cheilat’s cross-sectional study on 70 families including children and parents in Canada showed that these SNPs were involved in sodium intake, BP and CVD, as A allele carriers had higher sodium intake and DBP. However, as Cheilat’s study read the reverse strand, it found this association with T allele 32. Moreover, Chamoun’s study on Canadian young adults and preschool children illustrated associations between rs4790522 and rs222745 SNPs in the TRPV1 salt taste receptor gene and salt taste sensitivity in young adults and salt taste preference in children 33. Pilic’s study among young Caucasians in the UK revealed that the subjects with AA genotype in SNP SLC4A5 rs7571842 had the highest increase in SBP and DBP; however, SNP rs10177833 (SLC4A5), rs239345 (SCNN1B) and rs8065080 (TRPV1) had no statistically significant effects on the BP response to dietary Na manipulation and with the increasing number of A alleles in SNP SLC4A5 rs10177833, sodium intake increased 12. In addition, Barragan’ study among Caucasians aged 18–80 in Spain indicated that those with AA genotype had the highest salty taste intensity rate 13. Conversely, the SNP rs239345 (SCNN1B) was not significantly associated with salt sensitivity or salt taste thresholds in Hungarian Roma and young Caucasian subjects 11,12,17.
It is well established that sodium consumption is associated with elevated BP in multiple populations 34. In the present study, in line with Cheilat’s study, it has been shown that high DBP belongs to the carriers of A allele. Genetic polymorphisms or acquired over-activity of the ENaC is accompanied with arterial hypertension 35, despite the fact that the link between sodium intake CVD events was controversial in subjects without hypertension 36. Research on the alpha subunit of ENaC suggests a potential implication on BP regulation in mice 37. Furthermore, the rs239345 polymorphism on the beta subunit of ENaC) may be associated with another SNP, perhaps, the same one that related to BP in the alpha subunit of the ENaC in rats 38.
The taste responses to salt can be obstructed by the ENaC blocker amiloride without similar effect on other taste manners in mice 39. However, this mechanism can inhibit about 20% of salt taste perception in humans. Thus, to some extent, salt taste responses are regulated by ENaC in humans 40. However, other markers related to sodium intake were studied including salt taste thresholds and learned responses 17,41.
Similar to our findings, Ferraris et al. implied that there were no significant associations between the rs8065080 SNP in TRPV1 gene with an individual’s salt intake, SBP and DBP 14. However, the studies by Dias et al., Dioszegi et al. and Pilic et al., showed that T allele carriers perceived salt solutions significantly stronger than those homozygous for the C allele 6,11,17. An isoleucine (C) to valine (T) amino acid replacement (585 position of the TRPV1 protein, missense mutation) leads to rs8065080 (C > T) polymorphism. Contrary to our results, Pilic et al. 17 reported that TRPV1 rs8065080 T allele carriers had higher sodium intake than C allele carriers in a small sample of young predominantly Caucasian participants. The potential reason of these contradiction might be due to TRPV1 rs8065080 missense mutation and hence altering one amino acid 42. Moreover, age difference between the studies could be another reason, since age is a factor in phenotypical variance in genetic expression 43.
However, owing to the restricted research in the salt taste genotypes, the comparison of findings are limited, and therefore, highlighting the necessity of performing further well-designed studies.
Strengths and limitations
To the best of our knowledge, it was the first genetic study on salt intake in the Eastern Mediterranean region. In addition, we examined sodium intake by the precise method of 24-h urine sodium measurement and food contribution with validated FFQ in our population. However, we also had some limitations including limited coverage of polymorphisms within the most important SCNN1B -associated gene and TRPV1, low sample size and lack of sequencing possibility for all samples and the fact that collecting a single 24-h urine was not enough to reflect a true customary intake. Although it has been proposed that some other SNPs might be associated with sodium intake, because of financial limitation, we did not examine them. Finally, not adjusting age, sex and BMI was another limitation. It might alter the association of salt intake and blood pressure with the SNPs.
Conclusion
This study demonstrated that SNPs in the SCNN1B and TRPV1 genes associated with sodium intake and BP level. Therefore, genetic variations can relate with salt intake and consequently may associate with hypertension and finally CVD risk in the Iranian population. Further studies with larger sample size are warranted to replicate these results in order to better understand the genetic basis for salt taste and hypertension risk and also examine the potential effect of interactions between the diverse SNPs with the valid urine collection method can be effective in accurate estimation of sodium intake.
Data availability
The authors confirm that the data supporting the findings of this study are available.
Abbreviations
- CVD:
-
Cardiovascular disease
- AS:
-
Amiloride
- AI:
-
Amiloride-insensitive
- SNP:
-
Single nucleotide polymorphisms
- BP:
-
Blood pressure
- ICRI:
-
Isfahan Cardiovascular Research Institute
- IPAQ:
-
International Physical Activity Questionnaire
- BMI:
-
Body mass index
- WC:
-
Waist circumference
- DBP:
-
Diastolic blood pressure
- SBP:
-
Systolic blood pressure
- FFQ:
-
Food frequency questionnaire
- IFCP:
-
Iranian Food Consumption Program
- FBG:
-
Fasting blood glucose
- 24hUNa:
-
24-h urinary sodium
- CR:
-
Creatinine
- 24hUCr:
-
24-h urine creatinine
References
Mozaffarian, D. et al. Global sodium consumption and death from cardiovascular causes. N. Engl. J. Med. 371, 624–634 (2014).
Messerli, F. H. et al. Sodium intake, life expectancy, and all-cause mortality. Eur. Heart J. 42(21), 2103–2112 (2021).
Grillo, A., Salvi, L., Coruzzi, P., Salvi, P. & Parati, G. Sodium intake and hypertension. Nutrients 11, 1970 (2019).
Bigiani, A. Salt taste, nutrition, and health. Nutrients 12, 1537 (2020).
Yoshida, R. et al. NaCl responsive taste cells in the mouse fungiform taste buds. Neuroscience 159, 795–803 (2009).
Dias, A. G. et al. Genetic variation in putative salt taste receptors and salt taste perception in humans. Chem. Senses 38, 137–145 (2013).
Tapanee, P., Tidwell, D. K., Schilling, M., Peterson, D. G. & Tolar-Peterson, T. Genetic variation in taste receptor genes (SCNN1B, TRPV1) and its correlation with the perception of saltiness in normotensive and hypertensive adults. Int. J. Hypertens 2021, 5559831 (2021).
Bertino, M., Beauchamp, G. K. & Engelman, K. Long-term reduction in dietary sodium alters the taste of salt. Am. J. Clin. Nutr. 36, 1134–1144 (1982).
Wise, P. M., Hansen, J. L., Reed, D. R. & Breslin, P. A. Twin study of the heritability of recognition thresholds for sour and salty taste. Chem. Senses 32, 749–754 (2007).
Mozaffarian, D. Dietary and policy priorities for cardiovascular disease, diabetes, and obesity: A comprehensive review. Circulation 133, 187–225 (2016).
Diószegi, J. et al. Association of single nucleotide polymorphisms with taste and food preferences of the Hungarian general and Roma populations. Appetite 164, 105270 (2021).
Pilic, L. et al. The associations between genetics, salt taste perception and salt intake in young adults. Food Qual. Prefer. 84, 103954 (2020).
Barragán, R. et al. Bitter, sweet, salty, sour and umami taste perception decreases with age: Sex-specific analysis, modulation by genetic variants and taste-preference associations in 18- to 80-year-old subjects. Nutrients 10(10), 1539 (2018).
Ferraris, C. et al. Salt taste genotype, dietary habits and biomarkers of health: No associations in an elderly cohort. Nutrients 12(4), 1056 (2020).
DeSimone, J. A. & Lyall, V. Taste receptors in the gastrointestinal tract III. Salty and sour taste: Sensing of sodium and protons by the tongue. Am. J. Physiol. Gastrointest. Liver Physiol. 291(6), G1005–G1010 (2006).
Gu, X. et al. resequencing epithelial sodium channel genes identifies rare variants associated with blood pressure salt-sensitivity: The GenSalt study. Am. J. Hypertens. 31(2), 205–211 (2018).
Pilic, L. & Mavrommatis, Y. Genetic predisposition to salt-sensitive normotension and its effects on salt taste perception and intake. Br. J. Nutr. 120, 721–731 (2018).
Craig, C. L. et al. International physical activity questionnaire: 12-country reliability and validity. Med. Sci. Sports Exerc. 35(8), 1381–1395 (2003).
Chobanian, A. V. et al. Seventh report of the Joint National Committee on prevention, detection, evaluation, and treatment of high blood pressure. Hypertension 42(6), 1206–1252 (2003).
Mohammadifard, N. et al. validation of simplified tools for assessment of sodium intake in Iranian population: Rationale, design and initial findings. Arch. Iran Med. 19, 652–658 (2016).
Rafiei, M., Boshtam, M. & Marandi, A. The Iranian food consumption program (IFCP): A unique nutritional sotware in Iran. Iran. J. Public Health 31, 105–107 (2002).
Dorosti Motlagh, A. R. & Tabatabaei, M. Iranain Food Composition Table 1 (Iran Donyaye Taghzieh Press, 2007).
Mohammadifard, N. et al. Dietary sodium and potassium intake and their association with blood pressure in a non-hypertensive Iranian adult population: Isfahan salt study. Nutr. Diet 74, 275–282 (2017).
Hedayati, S. S. et al. Association of urinary sodium/potassium ratio with blood pressure: Sex and racial differences. Clin. J. Am. Soc. Nephrol. 7, 315–322 (2012).
Wang, C. Y. et al. Urinary excretion of sodium, potassium, and chloride, but not iodine, varies by timing of collection in a 24-hour calibration study. J. Nutr. 143(8), 1276–1282 (2013).
Yakubova, A. et al. Searching for predictors of migraine chronification: A pilot study of 1911A>G polymorphism of TRPV1 gene in episodic versus chronic migraine. J. Mol. Neurosci. 71, 618–624 (2021).
González-Mercado, A. et al. The relationship of single nucleotide polymorphisms in the TRPV1 gene with lipid profile, glucose, and blood pressure in Mexican population. Genet. Test Mol. Biomark. 24, 42042–42044 (2020).
Cantero-Recasens, G. et al. Loss of function of transient receptor potential vanilloid 1 (TRPV1) genetic variant is associated with lower risk of active childhood asthma. J. Biol. Chem. 285, 27532–27535 (2010).
Koshy, L. et al. Evaluating genomic DNA extraction methods from human whole blood using endpoint and real-time PCR assays. Mol. Biol. Rep. 44, 97–108 (2017).
Zhao, Q. et al. Common variants in epithelial sodium channel genes contribute to salt sensitivity of blood pressure: The GenSalt study. Circ. Cardiovasc. Genet. 4, 375–80 (2011).
Mohammadifard, N. et al. Salt intake and its sources in children, adolescents and adults in Isfahan, Islamic Republic of Iran. East Mediterr. Health J. 27, 279–286 (2021).
Chleilat, F. Genetic Variation in Salt Taste Receptors Impact Salt Intake, Blood Pressure and Cardiovascular Disease Risk Factors in the Guelph Family Health Study. http://hdl.handle.net/10214/10069 (2016). (Accessed 12 February 2020).
Chamoun, E. et al. The relationship between single nucleotide polymorphisms in taste receptor genes, taste function and dietary intake in preschool-aged children and adults in the Guelph Family Health Study. Nutrients 10, 990 (2018).
Mente, A. et al. Association of urinary sodium and potassium excretion with blood pressure. New Engl. J. Med. 371, 601–611 (2014).
Bubien, J. K. Epithelial Na+ channel (ENaC), hormones, and hypertension. J. Biol. Chem. 285, 23527–23531 (2010).
Mente, A. et al. Associations of urinary sodium excretion with cardiovascular events in individuals with and without hypertension: A pooled analysis of data from four studies. Lancet 388, 465–475 (2016).
Shigemura, N. et al. Amiloride-sensitive NaCl taste responses are associated with genetic variation of ENaC alpha-subunit in mice. Am. J. Physiol. Regul. Integr. Comp. Physiol. 294, R66–R75 (2008).
Shehata, M. F. The alternatively spliced form “b” of the epithelial sodium channel α subunit (α ENaC): Any prior evidence of its existence? Clin. Med Insights Cardiol. 4, 73–75 (2010).
Stewart, R. E., DeSimone, J. A. & Hill, D. L. New perspectives in a gustatory physiology: Transduction, development, and plasticity. Am. J. Physiol. 272, C1–C26 (1997).
Bigiani, A. Does ENaC work as sodium taste receptor in humans? Nutrients 12, 1195 (2020).
Gilman, T. L. et al. High salt intake lowers behavioral inhibition. Front. Behav. Neurosci. 13, 271 (2019).
National Center for Biotechnology Information. Data Base of Single Nucleotide Polymorphisms (dbSNP). https://www.ncbi.nlm.nih.gov/snp/ (Accessed 20 January 2022).
Viñuela, A. et al. Age-dependent changes in mean and variance of gene expression across tissues in a twin cohort. Hum. Mol. Gen. 27(4), 732–741 (2018).
Acknowledgements
The authors are indebted to the participants for their close cooperation. The efforts of all staff of Cardiovascular Research Institute and laboratories who helped us to run this study is highly appreciated. Also we are thankful Institute for Medical Research Development (NIMAD, Grant Number 977549) which granted this study.
Funding
This study was funded by National Institute for Medical Research Development (NIMAD, Grant Number 977549).
Author information
Authors and Affiliations
Contributions
N.M., S.S. & N.S. designed the study; F.M. collected the data; L.S. & E.K. did lab measurement; A.M. analyzed the data; N.M., M.G. & L.S. interpreted the data; N.M. & F.A. wrote the first draft; All authors read and confirmed the manuscript. All authors consent to the publication of the manuscript in Jenes & Nutrtiom Journal.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mohammadifard, N., Moazeni, F., Azizian-Farsani, F. et al. Genetic variation in salt taste receptors impact salt intake and blood pressure. Sci Rep 13, 4037 (2023). https://doi.org/10.1038/s41598-022-23827-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-23827-0
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.