Abstract
The erythrocyte sedimentation rate (ESR) is a non-specific blood test for determining inflammatory conditions. However, the long measurement time (60 min) to obtain ESR is an obstacle for a prompt evaluation. In this study, to reduce the measurement time of ESR, deep neural networks (DNNs) were applied to the sedimentation tendency of blood samples. DNNs using multilayer perceptron (MLP), long short-term memory (LSTM), and gated recurrent unit (GRU) were assessed and compared to determine a suitable length of time for the input sequence. To avoid overfitting, a stacking ensemble learning was adopted, which combines multiple models by using a meta model. Four meta models were compared: mean, median, least absolute shrinkage and selection operator, and partial least squares regression (PLSR) schemes. From the empirical results, LSTM and GRU models have better prediction than MLP over sequence lengths of 5 to 20 min. The decrease in \(\overline{\mathrm{MAPE} }\) and \(\overline{\mathrm{RMSE} }\) of GRU and LSTM was attenuated after a sequence length of 15 min, so the input sequence length is determined as 15 min. In terms of the meta model, the statistical comparison suggests that GRU combined with PLSR (GRU–PLSR) is the best case. Then, the GRU–PLSR was tested for prediction of ESR data obtained from periodontitis patients to check its applicability to a specific disease. The Bland–Altman plot shows acceptable agreement between measured and predicted ESR values. Based on the results, the GRU–PLSR can predict ESR with improved performance within 15 min and has potential applicability to ESR data with inflammatory and non-inflammatory conditions.
Similar content being viewed by others
Introduction
The erythrocyte sedimentation rate (ESR) is a simple and inexpensive blood test for providing general information about an inflammatory or acute response. As the gold standard of ESR measurements, the Westergren method measures the distance from the meniscus to the top of the column of sedimented erythrocytes separated from plasma in a vertical Westergren tube (inner diameter = 2.5 mm, length = 200 mm, and whole blood volume = 5 mL) after 1 h1,2. The ESR value represents the sedimentation speed in units of millimeters per hour.
Under low shear conditions, erythrocytes tend to form one-dimensional coin stacks (rouleaux) or three-dimensional aggregates in a reversible process. Erythrocyte aggregation (EA) is affected by two major factors: aggregating force induced by the macromolecules, and disaggregating forces including negative surface charge and shear force3. Although negative charges of erythrocytes repel each other, the presence of certain plasma proteins (fibrinogen, immunoglobulins, lipoproteins, and α-2 macroglobulin) and inflammation-related proteins (cytokines and chemokines) in plasma enhance the degree of EA. Elevated EA is considered to augment blood viscosity4, which increases flow resistance and wall shear stress. In addition, the high extent of EA is somewhat correlated with cardiovascular diseases (CVDs)4 and diabetes mellitus5. Considering Stokes’ law, the sedimentation speed of a spherical particle in a medium is dependent on its size6. Therefore, the ESR value is closely associated with the degree of EA7.
Although ESR with minor modifications is still considered to be a useful index of nonspecific disease, the Westergren method has some disadvantages, including a time-consuming procedure (60 min) and a repetitive cleaning process. In addition, the Westergren method provides only the final sedimentation speed (the ESR value) after 1 h. It is insufficient to describe the temporal variation of the sedimentation tendency. Hong et al. proposed the micro-vibrational erythrocyte sedimentation rate (MV-ESR) and demonstrated the possibility of shortening the time of ESR measurement8. Shteinshnaider et al. demonstrated that ESR values at 60 min can be predicted by ESR values at 30 min by using simple linear regression9. Nishisako et al. applied a formula for calculating a normal maximum range of ESR with only age information of patients10. However, this formula was only applicable to Asian patients aged more than 65 years. Lapić et al. compared automated ESR methods (TEST1, Ves-Matic Cube 200) with the Westergren method11. Ves-Matic Cube 200 showed larger dispersion, so they concluded that automated ESR methods should be used carefully. To gain information about the temporal sedimentation tendency, a characteristic time representing the time required for sedimentation by a specific length was proposed12.
Although ESR elevation typically results from inflammatory factor such as fibrinogen, some noninflammatory factors such as age, sex, and pregnancy can also cause elevation of the ESR, which might make it difficult to identify the patterns of ESR13. For analysis of complex biological systems, machine learning algorithms have been proposed to recognize, classify, and predict the patterns14. Thus, deep learning methods were used in this study considering their ability to learn the various patterns in ESR.
The sedimentation tendency of erythrocytes is time series data. For forecasting a time series, several deep neural networks (DNNs) have been proposed in several fields15,16,17,18,19. Multilayer perceptron (MLP) is a type of feed-forward neural network that does not have cycles in the same layer. Unlike the MLP, long short-term memory (LSTM) and gated recurrent unit (GRU) have cycles within a hidden layer and filter the information through gate structure15,16. Kong et al. compared various benchmarks with LSTM, including the k-Nearest Neighbors’ algorithm and extreme learning machine17. They found that LSTM outperformed the other rival algorithms in short-term forecasting for loads in individual residential households. AlDahoul et al. investigated the usage of ElasticNet linear regression, MLP, extreme gradient boosting, and LSTM for the prediction of suspended sediment in the Johor river in Malaysia18. The results showed that the LSTM is better than the other models. Che et al. developed a GRU-based model (GRU-D) that captures the informative missing patterns in time series analysis19. It was shown that GRU-D performed better than deep learning models built on GRU as well as other machine learning methods. Yilmaz and Buyrukoğlu proposed the hybrid model combining Back Propagation-Based Artificial Neural Network (BP-ANN), Correlated Additive Model (CAM) and Auto-Regressive Integrated Moving Average (ARIMA) to forecast Coronavirus disease (Covid-19) case20. They showed that the proposed hybrid model provides the best results, which can be used as the prediction model for Covid-19 to obtain better prediction results and to take immediate measures. Buyrukoğlu analyzed five promising cryptocurrencies with the ensembles of LSTM and single-based LSTM networks21. This study revealed both ensembles of LSTM and single-based LSTM can be used separately, considering that ensemble of LSTM does not always outperform the single-based LSTM.
When a small amount of data is available, a single neural network algorithm tends to exhibit generalization error for unseen instances22. Generalization means the capacity of the model to predict the correct output for previously unseen data23. Because small datasets are more prone to overfitting than large datasets, an ensemble approach was proposed to reduce the generalization error by avoiding overfitting24. Specifically, ensemble learning refers to the combination of multiple algorithms with different opinions and then weighting and combining them to make a complex decision. Ensemble learning is mainly used to improve prediction performance, or to avoid selection of a poor predictions by combining multiple models. In several studies, ensemble learning could improve the forecasting performance25,26,27.
There are commonly used ensemble methods like bagging, boosting, and stacking. Bagging, also known as bootstrap aggregation, constructs multiple classifiers trained on bootstrap samples and averages the outputs of the predictors to obtain the final prediction with the appropriate size28. Since the bootstrap samples are randomly drawn with replacement from the entire training data, it causes individual bootstrap samples to overlap significantly, with many of the same instances in the training data appearing in most samples, and some instances appearing multiple times in a given sample. Unlike bagging, boosting creates different classifiers by sequentially reweighting the instances in the training data29. Each instance misclassified by the previous base classifier get larger weights, and the next classifier is boosted on the reweighted training data. In this way, a sequence of training data and classifiers is obtained, which can be combined by a simple majority voting or by weight majority voting in the final decision. In stacking, which stands for stacked generalization, a set of base models are constructed from the training data, then their outputs are used to train a meta model30. In most cases, the performance of an ensemble is better than a single model21,31,32,33,34. Ensemble learning has been widely used in various fields such as: health science31, sport science32, agriculture33,34, finance21.
In this study, the MLP, LSTM, and GRU models were compared to reduce the measurement time of ESR. Specifically, the sequence length (input data) is determined by monitoring predictive performance with evaluation metrics for each model. After determining the sequence length, stacking ensemble models with different ensemble sizes were investigated to select an adequate ensemble structure. Furthermore, all models including the individual DNNs were compared statistically. From the statistical comparison, the best performing meta model was determined. The proposed model was then applied to ESR prediction for patients with a particular disease. The aim of this study can be summarized as:
-
To reduce measurement time of ESR, using ensemble deep learning models.
-
To find the best performing ensemble model based on the statistical scores.
-
To validate the applicability of the selected ensemble model for predicting ESR in nontrained samples (Patients with a particular disease).
Material and methods
Blood sample preparation
A total of 310 subjects from the Pusan National University Dental Hospital (Busan, Korea) participated in this study. The study population included 304 normal subjects and 6 patients who have periodontal disease. The age of the subjects ranged from 5 to 95 years (48.369 ± 18.823). The percentages of males and females were 46% and 54%, respectively. Blood samples were collected via 3-mL BD Vacutainer® K2 EDTA tubes (Becton, Dickinson and Co., Franklin Lakes, NJ) for anticoagulation. A centrifuge (DSC-102SD, Nasco Korea) was used for separating erythrocytes from plasma. Separated erythrocytes were mixed with autologous plasma with the hematocrit (packed volume percentage of erythrocytes) fixed at 35% by mixing. Ethical permission for this study was granted by the institutional review board (IRB) of Pusan National University Hospital (H- 1911–028–085), and the study followed the rules of the Declaration of Helsinki.
Experimental setup
Figure 1a shows the experimental setup for recording the images for erythrocyte sedimentation. A disposable syringe (1 mL syringe, BD, USA) filled with a 1 mL blood sample was mounted vertically. Consecutive images of the blood in the syringe were acquired by a digital camera (D5600, Nikon Imaging Korea Co., Ltd., Korea) with a micro-lens (AF Micro-Nikkor 105 mm f/2.8D, Nikon Co., Japan). Using software (Camera control pro 2, Nikon Co., Japan), images were taken automatically at intervals of 0.5 min for 60 min. The pixel size of the CMOS camera was 6000 × 4000 pixels. All experiments were conducted at room temperature (25 °C) with 50% humidity.
(a) Measurement system composed of a disposable syringe (1 mL) and digital camera. (b) Typical images of sedimented erythrocytes in a blood sample with respect to time (t). Height of the sedimented erythrocytes (HE) is gradually decreased during 60 min. (c) Data processing procedure. The original image is transformed into a binary image using Otsu’s method. The binary image is averaged in the horizontal direction to detect an interface (marked with a black circle) between sedimented erythrocytes and plasma. This interface, denoted by HE/HT, was extracted during 60 min (marked with a gray circle), and forms time series data.
Image processing procedure
Figure 1b shows the process of erythrocyte sedimentation in a blood sample at various times (t = 0, 20, 40, and 60 min). The total height of the blood (HT) was about 53 mm, corresponding to 1 mL. The height of the sedimented erythrocytes (HE) decreased continuously due to gravity sedimentation with the lapse of time. Figure 1c shows the image processing procedure using commercial software (MATLAB, Mathworks, USA). The captured images were cropped to the region of interest in the syringe (120 × 2900 pixels) and then converted into gray images. The images were transformed into binary images using Otsu’s method. By averaging the binary images in the horizontal direction, variation of the intensity along the gravity direction was obtained. The interface between sedimented erythrocytes and separated plasma was detected by utilizing the ‘findchangepts’ function in MATLAB, which detects a drastic change in signal. Based on the detected interface, HE/HT was determined at a given time. During ESR measurement (60 min), temporal variation of HE/HT for one sample was obtained by repeating the procedure, as illustrated in the right inset of Fig. 1c. For learning and training for the prediction of ESR, normalized time series data (HE/HT) were used as model input.
Deep learning methodology for ESR prediction
MLP model
The MLP is the most widely used algorithm for forecasting. It consists of multiple layers of interconnected elements called neurons with a nonlinear activation function (φ)35. Each neuron is only forward connected to every neuron in the subsequent layers. This structure is combined with the hidden layer activation function and allows the MLP to approximate any continuous function36. The mathematical model of the MLP is represented by:
where yi is the output variable, H represents the number of neurons in the hidden layer, wij is the weight connecting the jth neuron and the ith neuron, xi is an input variable, and bj is the bias of the jth neuron.
LSTM
A recurrent neural network (RNN) has a recurrent hidden state with an activation at each time that depends on the one from the previous time. RNN based algorithms have been developed in time series forecasting applications37,38,39. Due to infinite lookback windows, RNNs are vulnerable to long-range dependencies in the data, and the gradients tend to either vanish or explode40. LSTM is a variant of RNN that was developed to deal with the limitation of RNNs by using memory cells. A memory cell in each LSTM unit stores long-term information and these cells are modulated through three gates: an input gate (it), output gate (ot), and forget gate (ft). These gates adjust the hidden state (ht) and cell states (ct) of the LSTM as follows:
where W and b are the weights and biases of the LSTM layer, respectively. σ is a sigmoid function, and tanh is a hyperbolic tangent function. ⊙ is the element-wise (Hadamard) product.
GRU
GRU was introduced by Cho et al.16 and makes each recurrent unit with an adaptive selection of dependencies of different time scales. Unlike LSTM, GRU modulates the flow of information inside the unit without memory cells. GRU has two gates: an update gate (gt) and reset gate (rt).
where \({\widetilde{\mathrm{h}}}_{\mathrm{t}}\) denotes candidate hidden state.
Selection of sequence length
To reduce ESR measurement time, the length of the input sequence of the prediction should be decreased. The starting time is fixed as 0 min. To select the optimal length of time, different time spans were tested for MLP, LSTM, and GRU. The input time ranged from 5 to 20 min with intervals of 2.5 min. When the input time is shorter than 5 min, the predicted results are extremely poor.
Ensemble model
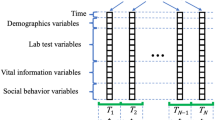
In the present work, stacking ensemble using bootstrapped samples of the training data was proposed. As shown in Fig. 2, stacking is a two-stage approach. In the first stage, multiple base models were built on bootstrap samples, and corresponding predictions were obtained. Bootstrap samples were randomly drawn with replacement from the entire training data. In the second stage, meta model combines the predictions of the base models and produces the final prediction. The final prediction should be a weighted average of the individual predictions since some base models are more accurate than others. However, given that all base models attempt to solve the same task, their predictions are expected to be highly correlated, which can lead to performance degradation of the ensemble24,28,41. Feature selection technique is necessary to remove the redundant or irrelevant features or features correlated in the data. Some studies proposed deep autoencoder or deep belief network to extract the nonlinear features42,43. However, the predictions of the base models didn’t show any complex patterns but high correlation in this study. Thus, LASSO and PLSR were used to cope with the multicollinearity due to the high degree of correlation between the individual predictions. The meta models are briefly introduced.
Mean ensemble
The mean is the simplest weight combination approach and assigns equal weights to all the individual forecasts44. The mean operator is intuitive, but it has a limitation that is widely known: the mean is susceptible to outliers and skewed distribution. The mean ensemble forecast (za) can be calculated as:
where M is the number of base models, and zi (i = 1, …, M) denotes the forecasted output vector of the ith base model.
Median ensemble
The median simply provides the middle value of zi in a sorted list if M is odd or the mean of the two middle values otherwise44. The median is less affected by the influence of outliers. The median ensemble forecast (zm) can be calculated as:
Least absolute shrinkage and selection operator (LASSO)
The LASSO is a regularized regression to deal with multicollinearity in data45. The LASSO penalty can force certain coefficients to be zero by imposing a penalty on their size, and it yields interpretable results with improved accuracy. The LASSO estimator (\({\widehat{\upbeta }}_{\mathrm{L}})\) is the solution to the following optimization problem:
where y represents the vector of observed ESR values, Z = [z1, z2, …, zM] is the individual forecasts matrix, and β denotes the regression coefficient. λ is a positive regularization (penalty/tuning) parameter that controls the amount of shrinkage of β. As the value of λ depends on the data, it can be determined using data-driven methods such as cross-validation (CV). The intercept is ignored for convenience. The LASSO ensemble forecast (zL) can be calculated as:
Partial least squares regression (PLSR)
The basic idea of PLSR is to project the original variables onto a set of orthogonal latent variables. The latent variables, called components, are linear combinations of the original variables. The number of the latent components (J) is determined by CV, similar to the LASSO method. PLSR extracts these components by modeling both input and output data46. PLSR builds a relationship between the input matrix Z and the output vector y. The PLSR technique attempts to find a linear decomposition of Z and y by the outer relations47:
where T and U are score matrices, P and Q are loading matrices, and Ex and Ey are residual errors for input and output. pj and qj are loading vectors that can be considered as weights. The superscript “T” means transposition. Latent vectors tj and uj are obtained to maximize the covariance between Z and y while minimizing Ex and Ey. The inner relation between U and T is given by:
where B denotes the coefficients, and EU is an error term between U and T. If the error terms are minimized, the PLSR ensemble forecast (zP) can be calculated as:
Deep neural network implementation
DNN models ware built using TensorFlow 2.8.0 and Keras packages in Python 3.7.12 in the Google Colaboratory environment. For training and evaluation of the model, time series data of 304 normal subjects were randomly split into training, validation, and test sets (194, 50 and 60 subjects, respectively). Models were trained for 300 epochs with an early stopping method. Early stopping was adopted that specifies an arbitrarily large number of epochs and stops training the model when there is no improvement in the validation loss for 15 epochs. Moreover, an exponential decay schedule was utilized, which decays the learning rate using an exponential function. The loss function was the mean squared error. The optimizer used in this study was the adaptive moment estimation (ADAM), which showed the highest performance among other optimizers including Adadelta, Adagrad, RMSprop, and SGD in a preliminary test.
The neural network architecture was determined empirically by monitoring the performance with different architectures. MLP consisted of two hidden layers: one with 15 neurons and one with 10 neurons. The architecture of the GRU consisted of one hidden layer with 15 neurons. The architecture of LSTM was identical to that of GRU. A dense layer with a single neuron was chosen for the output layer in all DNNs.
Performance metrics
The forecasting performance of each model was evaluated through some error metrics. The mean absolute percentage error (MAPE) and root mean squared error (RMSE) were adopted48. The formulas for MAPE and RMSE are as follows:
where ns and ŷ denote the number of samples and the forecasted ESR, respectively.
Statistical test for all pairwise comparison
The Friedman test is a non-parametric test that detects significant differences among models49. It produces ranks of models’ performance averaged over multiple datasets. The null hypothesis of the Friedman test states that all the models are equivalent and their mean ranks are nearly equal. If the null hypothesis is rejected with very small p-values, the Nemenyi post-hoc test can be done50. Although the Nemenyi test is known to be conservative, the procedure is the simplest among post-hoc tests for pairwise multiple comparisons51. The Nemenyi test was used to compare all models with each other. The performance between any two models is significantly different if their corresponding mean ranks differ by at least the critical distance (CD). The CD is given by:
where k and D are the numbers of models and datasets, respectively, and \({\mathrm{q}}_{\mathrm{\alpha }}\) is the critical value based on studentized range statistics divided by \(\sqrt{2}\).
Results and discussion
Tendency of the erythrocyte sedimentation
Figure 3a shows the temporal variations of HE/HT for all subjects (n = 304) and mean trend of all samples (black dotted line). Since the hematocrit of blood samples was fixed at 35%, ESR values (HE/HT at 60 min) ranged from 0.350 to 0.859. Although some samples show similar ESR values, the variation tendency of HE/HT is somewhat different. To further investigate the variation tendency during the erythrocyte sedimentation process, the instantaneous time rate of sedimentation (VE = −dHE/dt) for 304 subjects is shown in Fig. 3b as the mean and standard deviation (SD). The extent of VE can be approximately divided into two stages according to time. Within 13 min, VE shows an acceleration stage. After that, it tends to decrease with the lapse of time. Since VE can describe the extent of erythrocyte sedimentation for a specific time, it is closely related to the ESR sedimentation process.
The sedimentation process can be approximately described by 3 phases: aggregation, sedimentation, and packing52,53. During the aggregation phase, erythrocytes aggregate and form the rouleaux or aggregates. Since the sedimentation speed is proportional to the particle size6, large rouleaux and aggregates produced in the aggregation stage promote sedimentation. As a result, erythrocytes rapidly fall in the sedimentation phase, whereas erythrocytes slowly fall and pile up at the bottom of the tube in the packing phase. It can be inferred that times with high VE (10–20 min) may be included in the sedimentation phase (Fig. 3b). Thus, temporal variations of HE/HT at 0–20 min are important to predict the ESR value because they provide information on the erythrocyte sedimentation process.
Performance comparison of DNNs depending on input sequence lengths
Figure 4a,b show the variations of \(\overline{\mathrm{MAPE} }\) and \(\overline{\mathrm{RMSE} }\) according to the sequence length (input data at 0-t min). \(\overline{\mathrm{MAPE} }\) and \(\overline{\mathrm{RMSE} }\) are mean values of MAPE and RMSE obtained by repeating the prediction over 30 times. Due to the random setting of initial weights and biases, the 30 results are different. In general, prediction errors decrease with increasing sequence length for all DNNs. MLP shows relatively poor performance for all sequence lengths. This might result from the inherent structure of MLP, which cannot learn the sequential order of the time series input. As shown in Supplementary Table 1, SD values of MLP are usually higher than those of LSTM and GRU. This indicates that the prediction results of LSTM and GRU are less dependent on the initial setting than those of MLP. These results are well matched with a previous study that compared the prediction uncertainty of LSTM, GRU, and ANN54. Since similar accuracy was observed between LSTM and GRU, the optimal sequence length was determined based on LSTM and GRU results. \(\overline{\mathrm{MAPE} }\) and \(\overline{\mathrm{RMSE} }\) of both GRU and LSTM gradually decrease until the sequence length reaches 15 min and show a marginal decrease from 15 min onward.
(a) \(\overline{\mathrm{MAPE} }\) (%) of the individual models according to sequence length (min). \(\overline{\mathrm{MAPE} }\) is the mean MAPE over 30 repeated experiments. (b) \(\overline{\mathrm{RMSE} }\) of the individual models according to sequence length (min). \(\overline{\mathrm{RMSE} }\) is the mean RMSE over 30 repeated experiments.
Determination of the exact time input for the ESR is challenging since many factors also affect ESR, including sex, age, and even vertical installation angle of the specimen tubes. Previous studies reported that the ESR can be estimated by the ESR recorded at 30 min9,55. The inconsistent optimal time can be explained by the method to monitor erythrocyte sedimentation. In this experiment, relatively dense temporal variations of erythrocyte sedimentation (time interval of 0.5 min) were recorded. In contrast, previous reports measured the ESR values at 15, 20, 30, and 40 min. Considering the marginal decrease from 15 min with small values of \(\overline{\mathrm{MAPE} }\) and \(\overline{\mathrm{RMSE} }\), the optimal sequence length was selected as 15 min.
Performance of ensemble model according to ensemble size
As the number of base models (NB) is an important parameter in ensemble performance, its impact was investigated in terms of \(\overline{\mathrm{MAPE} }\) and \(\overline{\mathrm{RMSE} }\). Both LSTM and GRU were selected as base models for the ensemble. All meta models were implemented in MATLAB, where the parameters of LASSO and PLSR were determined by fivefold CV. All base models were trained on bootstrap samples and run 30 times for the fixed sequence length (15 min) of the test set. To select an adequate ensemble size, NB was varied from 3 to 10. In a preliminary test, \(\overline{\mathrm{MAPE} }\) and \(\overline{\mathrm{RMSE} }\) of each ensemble model were almost saturated when NB exceeded 10 DNNs.
Figures 5,6 show the variations of \(\overline{\mathrm{MAPE} }\) and \(\overline{\mathrm{RMSE} }\) for LSTM and GRU coupled with four meta models. It can clearly be observed that the results of the mean are similar to those of the median. This marginal difference can be explained by the strong correlation between the individual forecasts. As the mean and median are measures of the central tendency, multicollinearity in the individual forecasts results in similar performance between them. However, LASSO and PLSR methods are more accurate than the mean and median over all NB, which show superior ability to handle the multicollinearity.
Combinations of LSTM with PLSR and LASSO show similar performance, as illustrated in Fig. 5. However, compared to LASSO, PLSR shows a different trend for NB ranging from 3 to 7. Particularly, PLSR exhibits a significantly low error when NB increases from 4 to 5. Both methods show marginal improvement with the increase in NB (NB > 8). In the case of the GRU base model, PLSR achieves better results, followed by LASSO (Fig. 6). The variations of PLSR fluctuate with increasing NB, whereas the variations of LASSO gradually decrease. PLSR shows relatively high reduction of error when NB varies from 4 to 5, which was similarly observed in LSTM results (Fig. 5).
Although PLSR shows the minimum error at NB of 10, there is no significant difference at NB ranging from 8 to 10 in terms of PLSR performance (Figs. 5,6). The performance degradation with the increase in NB indicates that usage of all base models may not contribute to the improvement of the ensemble forecast56,57. The pattern learned by some base models has already been represented by other models due to serious correlation between the models when NB is sufficiently high. Thus, the candidates for the optimal NB are 8 and 10. In this study, the optimal NB was determined as 8 because there was no significant difference between 8 and 10.
Statistical comparison of model performance
To examine significant differences among all models, statistical tests were carried out with the selected NB of 8. Extremely low p-values were obtained by the Friedman test for both MAPE and RMSE at a significance level of 0.05, so the Nemenyi test was performed as illustrated in Fig. 7. P-values and CD are shown in the title, and the mean rank is marked next to each model. The ensemble models are named based on their base models and the meta models, such as LSTM-Mean or GRU-Median. As the base models coupled with meta models show better performance (low mean ranks), the models are positioned at the top in ascending order. The methods are not significantly different if their mean ranks share the same color bar that is less than or equal to the CD.
According to the Nemenyi test rank, GRU–PLSR achieves the lowest mean rank in both of \(\overline{\mathrm{MAPE} }\) and \(\overline{\mathrm{RMSE} }\). Furthermore, the ensemble models coupled with PLSR and LASSO outperform the other meta models and the individual models with 95% confidence. The findings suggest that both PLSR and LASSO are useful meta models and that GRU–PLSR is the best-performing ensemble model for the ESR prediction.
Application of GRU–PLSR for ESR prediction of periodontitis patients
Periodontal disease is a bacterial infection that damages the supporting tissues of the teeth. It may result in tooth loss and elevation of the body’s inflammation. Some studies reported relatively high ESR values in patients with periodontitis disease58,59. The ESR difference between normal subjects and periodontitis patients was monitored to examine the effect of periodontitis disease. For comparison, 6 control subjects were randomly sampled from 304 normal subjects. Temporal variation of HE/HT for the control group and periodontitis patients is illustrated in Fig. 8a.
The ESR tendency shows differences between the two groups because ESR is increased by infectious and inflammatory diseases. The mean values of ESR for the control and periodontitis groups are 0.537 ± 0.141 and 0.414 ± 0.083, respectively. This indicates that a fast sedimentation trend was observed in periodontitis patients. The different tendency from periodontitis disease is similar to a previous result of the faster sedimentation for a patient group in a comparison of characteristic time8. To further examine the effect of periodontitis disease, variations of VE for the control and periodontitis groups are illustrated in Fig. 8b. Based on VE, the erythrocytes for periodontitis patients rapidly decreased in 15 min in comparison to the control subjects, which indicates that the periodontitis disease accelerates the aggregation and sedimentation phases.
GRU–PLSR trained using normal subjects was applied to the ESR prediction for periodontitis patients to demonstrate the practical applicability of the ensemble model. The Bland–Altman method was used to compare the similarity between methods60. Figure 9 displays the difference between the actual ESR values and predicted ESR values according to their mean values. The bold line and dashed lines denote the mean value of bias and 95% limits of agreement, respectively. As the presence of data in the 95% limits indicates that the two methods are well matched, ESR values estimated by the GRU–PLSR are in good agreement with the actual ESR values. However, the limited sample size of patients with periodontitis may lead to biased results, and further study with more data is needed.
Bland–Altman plot demonstrating difference between the actual ESR values and predicted ESR values obtained by the GRU–PLSR model for periodontitis subjects. The horizontal axis represents the mean ESR values estimated by the two values, and the vertical axis shows the difference between them. The bold line and dashed lines denote the mean value of bias and 95% limits of agreement, respectively (n = 6).
Contributions of the study
To the best of our knowledge, this is the first paper that analyzes and predicts the sedimentation tendency of erythrocytes using deep learning algorithms. The key contributions of this study are as follows:
-
The use of deep learning approaches to predict ESR, in an attempt to find faster means of obtaining the ESR
-
The use of ensemble models to improve the generalization ability on unseen data.
-
The performance evaluation of various deep learning and ensemble models to find a best predictive model for ESR based on post-hoc analysis.
Conclusion
In this study, the optimal sequence length was determined based on LSTM and GRU to reasonably predict the ESR value before 60 min. For improvement of ESR prediction, ensemble models with four meta models were suggested and compared. Empirical and statistical results demonstrated that PLSR and LASSO were superior meta models, and GRU–PLSR achieved the lowest mean ranks among all methods. Finally, the GRU–PLSR model was applied to the prediction of ESR values for patients with periodontitis. Based on our results, it can be concluded that GRU–PLSR is able to predict ESR values from inflammatory and non-inflammatory conditions with high accuracy in only 15 min. Therefore, GRU–PLSR could be useful for clinical applications in ESR measurement, where prompt decision-making is needed.
Data availability
The datasets used and analysed during the current study available from the corresponding authors on reasonable request.
References
Westergren, A. Studies of the suspension stability of the blood in pulmonary tuberculosis 1. Acta Med. Scand. 54, 247–282 (1921).
Jou, J. et al. ICSH review of the measurement of the erythocyte sedimentation rate. Int. J. Lab. Hematol. 33, 125–132 (2011).
Yeom, E., Nam, K. H., Paeng, D. G. & Lee, S. J. Effects of red blood cell aggregates dissociation on the estimation of ultrasound speckle image velocimetry. Ultrasonics 54, 1480–1487 (2014).
Yeom, E., Jun Kang, Y. & Joon Lee, S. Hybrid system for ex vivo hemorheological and hemodynamic analysis: A feasibility study. Sci. Rep. 5, 1–15 (2005).
Yeom, E., Byeon, H. & Lee, S. J. Effect of diabetic duration on hemorheological properties and platelet aggregation in streptozotocin-induced diabetic rats. Sci. Rep. 6, 1–11 (2016).
Stokes, G. G. On the effect of the internal friction of fluids on the motion of pendulums. Trans. Cambridge Philos. Soc. 9, 8 (1851).
Olshaker, J. S. & Jerrard, D. A. The erythrocyte sedimentation rate. J. Emerg. Med. 15, 869–874 (1997).
Hong, H., Song, J. M. & Yeom, E. Micro-vibrational erythrocyte sedimentation rate (ESR) for sensitive measurement of erythrocyte aggregation. J. Vis. 24, 749–760 (2021).
Shteinshnaider, M. et al. Shortened erythrocyte sedimentation rate evaluation is applicable to hospitalised patients. Eur. J. Intern. Med. 21, 226–229 (2010).
Nishisako, H. et al. Investigation on prediction formulae for calculating erythrocyte sedimentation rate. J. Gen. Fam. Med. 18, 146 (2017).
Lapić, I. et al. Automated measurement of the erythrocyte sedimentation rate: method validation and comparison. Clin. Chem. Lab. Med. 57, 1364–1373 (2019).
Yeom, E. & Lee, S. J. Microfluidic-based speckle analysis for sensitive measurement of erythrocyte aggregation: A comparison of four methods for detection of elevated erythrocyte aggregation in diabetic rat blood. Biomicrofluidics 9, 024110 (2015).
Ormseth, M. J. et al. Utility of a novel inflammatory marker, GlycA, for assessment of rheumatoid arthritis disease activity and coronary atherosclerosis. Arthritis Res. Ther. 17, 1–8 (2015).
Tarca, A. L., Carey, V. J., Chen, X. W., Romero, R. & Drăghici, S. Machine learning and its applications to biology. PLoS Comput. Biol. 3, e116 (2007).
Hochreiter, S. & Schmidhuber, J. Long short-term memory. Neural Comput. 9, 1735–1780 (1997).
Cho, K. et al. Learning phrase representations using RNN encoder-decoder for statistical machine translation. arXiv preprint arXiv:1406.1078 (2014).
Kong, W. et al. Short-term residential load forecasting based on LSTM recurrent neural network. IEEE Trans. Smart Grid 10, 841–851 (2017).
AlDahoul, N. et al. Suspended sediment load prediction using long short-term memory neural network. Sci. Rep. 11, 1–22 (2021).
Che, Z., Purushotham, S., Cho, K., Sontag, D. & Liu, Y. Recurrent neural networks for multivariate time series with missing values. Sci. Rep. 8, 1–12 (2018).
Yilmaz, Y. & Buyrukoğlu, S. Hybrid machine learning model coupled with school closure for forecasting COVID-19 cases in the most affected countries. Hittite J. Sci. Eng. 8, 123–131 (2021).
Buyrukoğlu, S. Promising cryptocurrency analysis using deep learning. In 2021 5th International symposium on multidisciplinary studies and innovative technologies (ISMSIT). 372–376 (IEEE, 2021).
Sagi, O. & Rokach, L. Ensemble learning: A survey. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 8, 1249 (2018).
Mc Loone, S. & Irwin, G. Improving neural network training solutions using regularisation. Neurocomputing 37, 71–90 (2001).
Hansen, L. K. & Salamon, P. Neural network ensembles. IEEE Trans. Pattern Anal. Mach. Intell. 12, 993–1001 (1990).
Moretti, F., Pizzuti, S., Panzieri, S. & Annunziato, M. Urban traffic flow forecasting through statistical and neural network bagging ensemble hybrid modeling. Neurocomputing 167, 3–7 (2015).
Li, C., Tang, G., Xue, X., Saeed, A. & Hu, X. Short-term wind speed interval prediction based on ensemble GRU model. IEEE Trans. Sustain. Energy 11, 1370–1380 (2019).
Puleio, A. Recurrent neural network ensemble, a new instrument for the prediction of infectious diseases. Eur. Phys. J. Plus 136, 1–15 (2021).
Breiman, L. Bagging predictors. Mach. Learn. 24, 123–140 (1996).
Schapire, R. E. The strength of weak learnability. Mach. Learn. 5, 197–227 (1990).
Wolpert, D. H. Stacked generalization. Neural Netw. 5, 241–259 (1992).
Buyrukoğlu, S. Improvement of machine learning models’ performances based on ensemble learning for the detection of Alzheimer disease. In 2021 6th International Conference on Computer Science and Engineering (UBMK). 102–106 (IEEE, 2021).
Buyrukoğlu, S. & Savaş, S. Stacked-based ensemble machine learning model for positioning footballer. Arab. J. Sci. Eng.1–13 (2022).
Buyrukoğlu, S. New hybrid data mining model for prediction of Salmonella presence in agricultural waters based on ensemble feature selection and machine learning algorithms. J. Food Saf. 41, e12903 (2021).
Buyrukoğlu, S., Yılmaz, Y. & Topalcengiz, Z. Correlation value determined to increase Salmonella prediction success of deep neural network for agricultural waters. Environ. Monit. Assess. 194, 1–12 (2022).
Rosenblatt, F. Principles of neurodynamics. perceptrons and the theory of brain mechanisms. (Cornell Aeronautical Lab Inc Buffalo NY, 1961).
Hornik, K., Stinchcombe, M. & White, H. Universal approximation of an unknown mapping and its derivatives using multilayer feedforward networks. Neural Netw. 3, 551–560 (1990).
Salinas, D., Flunkert, V., Gasthaus, J. & Januschowski, T. DeepAR: Probabilistic forecasting with autoregressive recurrent networks. Int. J. Forecast. 36, 1181–1191 (2020).
Rangapuram, S. S. et al. Deep state space models for time series forecasting. Adv. Neural Inf. Process. Syst. 31, 7785–7794 (2018).
Geravanchizadeh, M. & Roushan, H. Dynamic selective auditory attention detection using RNN and reinforcement learning. Sci. Rep. 11, 1–11 (2021).
Bengio, Y., Simard, P. & Frasconi, P. Learning long-term dependencies with gradient descent is difficult. IEEE Trans. Neural Netw. 5, 157–166 (1994).
Kang, D. K. & Kim, M. J. Performance enhancement of SVM ensembles using genetic algorithms in bankruptcy prediction. In 2010 3rd International Conference on Advanced Computer Theory and Engineering (ICACTE). V2–154 (IEEE, 2010).
Liu, Y., Fan, Y. & Chen, J. Flame images for oxygen content prediction of combustion systems using DBN. Energy Fuels 31, 8776–8783 (2017).
Liu, K., Zheng, M., Liu, Y., Yang, J., & Yao, Y. Deep Autoencoder Thermography for Defect Detection of Carbon Fiber Composites. IEEE Trans. Industr. Inform. (2022).
Kourentzes, N., Barrow, D. K. & Crone, S. F. Neural network ensemble operators for time series forecasting. Expert Syst. Appl. 41, 4235–4244 (2014).
Tibshirani, R. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. Ser. B Methodol. 58, 267–288 (1996).
Rosipal, R. & Krämer, N. Overview and recent advances in partial least squares. in International Statistical and Optimization Perspectives Workshop" Subspace, Latent Structure and Feature Selection" (ed. Saunders, C., Grobelnik, M., Gunn, S., & Shawe-Taylor, J) 34–51 (Springer, 2005).
Geladi, P. & Kowalski, B. R. Partial least-squares regression: a tutorial. Anal. Chim. Acta 185, 1–17 (1986).
Botchkarev, A. Performance metrics (error measures) in machine learning regression, forecasting and prognostics: Properties and typology. arXiv preprint arXiv:1809.03006 (2018).
Friedman, M. A comparison of alternative tests of significance for the problem of m rankings. Ann. Math. Stat. 11, 86–92 (1940).
Nemenyi, P. B. Distribution-free multiple comparisons. (Princeton University, 1963).
Garcia, S. & Herrera, F. An extension on “statistical comparisons of classifiers over multiple data sets" for all pairwise comparisons. J. Mach. Learn. Res. 9, 1 (2008).
Rovel, A., L’Huillier, J. & Vigneron, C. Mechanisms of erythrocyte sedimentation. Biomedicine 28, 248–255 (1978).
Bull, B. Clinical and laboratory implications of present ESR methodology. Clin. Lab. Haematol. 3, 283–298 (1981).
Gao, S. et al. Short-term runoff prediction with GRU and LSTM networks without requiring time step optimization during sample generation. J. Hydrol. 589, 125188 (2020).
Harrow, C., Singer, A. J. & Thode, H. C. Jr. Facilitating the use of the erythrocyte sedimentation rate in the emergency department. Acad. Emerg. Med. 6, 658–660 (1999).
Zhou, Z. H., Wu, J. & Tang, W. Ensembling neural networks: many could be better than all. Artif. Intell. 137, 239–263 (2002).
Ahmad, Z. & Zhang, J. Selective combination of multiple neural networks for improving model prediction in nonlinear systems modelling through forward selection and backward elimination. Neurocomputing 72, 1198–1204 (2009).
Hutter, J. et al. Lower numbers of erythrocytes and lower levels of hemoglobin in periodontitis patients compared to control subjects. J. Clin. Periodontol. 28, 930–936 (2001).
Prakash, S., Dhingra, K. & Priya, S. Similar hematological and biochemical parameters among periodontitis and control group subjects. Eur. J. Dent. 6, 287–294 (2012).
Bland, J. M. & Altman, D. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 327, 307–310 (1986).
Acknowledgements
This work was supported by the National Research Foundation (NRF) of Korea grant, which is funded by the Korean government (MSIP) (NRF-2018R1A5A2023879 and 2021R1I1A3047664).
Author information
Authors and Affiliations
Contributions
E.Y., and J.L. proposed the study. J.L. and H.H. performed the experiment and analyzed the experimental data. J.L., H.H., and E.Y. wrote the paper. All authors participated in completing the manuscript. All authors contributed to revising the manuscript, and approved its final version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lee, J., Hong, H., Song, J.M. et al. Neural network ensemble model for prediction of erythrocyte sedimentation rate (ESR) using partial least squares regression. Sci Rep 12, 19618 (2022). https://doi.org/10.1038/s41598-022-23174-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-23174-0
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.