Abstract
Non-coding RNAs (ncRNAs) have long been considered the "white elephant" on the genome because they lack the ability to encode proteins. However, in recent years, more and more biological experiments and clinical reports have proved that ncRNAs account for a large proportion in organisms. At the same time, they play a decisive role in the biological processes such as gene expression and cell growth and development. Recently, it has been found that short sequence non-coding RNA(miRNA) and long sequence non-coding RNA(lncRNA) can regulate each other, which plays an important role in various complex human diseases. In this paper, we used a new method (JSCSNCP-LMA) to predict lncRNA–miRNA with unknown associations. This method combined Jaccard similarity algorithm, self-tuning spectral clustering similarity algorithm, cosine similarity algorithm and known lncRNA–miRNA association networks, and used the consistency projection to complete the final prediction. The results showed that the AUC values of JSCSNCP-LMA in fivefold cross validation (fivefold CV) and leave-one-out cross validation (LOOCV) were 0.9145 and 0.9268, respectively. Compared with other models, we have successfully proved its superiority and good extensibility. Meanwhile, the model also used three different lncRNA–miRNA datasets in the fivefold CV experiment and obtained good results with AUC values of 0.9145, 0.9662 and 0.9505, respectively. Therefore, JSCSNCP-LMA will help to predict the associations between lncRNA and miRNA.
Similar content being viewed by others
Introduction
NcRNAs are a class of RNAs that don’t have the function of translating proteins in organisms1,2,3. Therefore, ncRNAs have always been neglected by biological researchers. With the progress of science and technology, gene detection technology is also developing. Researchers have found that RNAs don’t participate in protein coding account for about 98% of RNA in organisms4. As a result, researchers are increasingly interested in ncRNAs. Studies have found that ncRNAs can be divided into many types, including lncRNA, miRNA, circRNA, and snRNA5,6. MiRNAs are a class of ncRNAs with a short sequence of 18–25 nucleotides in length, while lncRNAs are a class of ncRNAs with a length of more than 200 nucleotides7,8,9,10,11. Experiments have found that lncRNAs play an important role in various biological processes such as transcription, translation and differentiation12,13,14,15,16. Meanwhile, mutations and dysregulations of these lncRNAs have also been shown to have complex relationships with many human complex diseases17, such as lung cancer18, AIDS19, cardiovascular disease20, Alzheimer's disease (AD)21, and diabetes22. For another example, lncRNA HOTAIR, PCA3 and H19 have been treated as potential biomarkers of hepatocellular carcinoma recurrence23, prostate cancer aggressiveness24 and breast cancer detection, respectively25. Similarly, miRNAs also play a key role in the differentiation, proliferation and apoptosis of biological cells26,27,28,29,30. Correspondingly, more and more miRNAs have been proved to have an impact on the occurrence of certain diseases. For example, in the midbrain of patients with Parkinson’s disease, miRNAs were regarded as a regulator to the maturation and function of midbrain dopaminergic neurons31. Overexpression of mir-128 in glioma cells was proved to inhibit cell proliferation31,32. Furthermore, mir-375 could regulate insulin secretion33; the miR-1 was involved in heart development; deletion of miRNA-1–2 interrupted the regulation of carcinogenesis34,35.
Recently, studies have shown that some specific lncRNAs and miRNAs can be detected in serum and blood of some cancer patients36,37. At the same time, lncRNAs and miRNAs can interact with each other in some diseases, which jointly affects the occurrence of human diseases38,39. For example, in the COVID-19 study, after comparing the transcriptomic data from different patient groups, researchers observed that COVID-19 patients had abnormally expressed mRNA and lncRNA when they were admitted to the ICU. Shaath and Alajez suggested that further studies on the identification and role of these mRNA and lncRNA-based biomarkers, as well as their impact on the onset and severity of COVID-19, which could play a crucial role in patient stratification and help select appropriate treatment options40. Therefore, the research on the associations between lncRNAs and miRNAs has become a research boom. However, it is very complicated to verify the associations between lncRNAs and miRNAs only through biological experiments. In order to improve the research efficiency of biological researchers, researchers in the computer field use existing biological data to analyze and predict the unknown associations between lncRNAs and miRNAs.
Before that, some prediction models of ncRNA-disease were well-established and had good results. For example, Chen et al. developed a reliable computational tool of LRLSLDA to predict novel human lncRNA-disease associations based on the assumption that similar diseases could tend to be related with functionally similar lncRNAs. This model was mainly based on a semi-supervised learning framework of Laplacian Regularized Least Squares41, which integrated known disease-lncRNA associations and lncRNA expression profile. In 2015, based on the assumption that similar diseases could tend to be associated with lncRNAs with similar functions, Chen et al. further developed two novel lncRNA functional similarity calculation models (LNCSIM)42. In the model of LNCSIM, disease semantic similarity was first calculated based on the directed acyclic graph (DAG) which represented the relationships among different diseases. Then, lncRNA functional similarity was further obtained by calculating the semantic similarity between their associated disease groups. Yang et al. implemented a propagation algorithm on the coding-non-coding gene-disease bipartite network to infer potential lncRNA-disease associations43. The coding-non-coding gene-disease bipartite network was constructed by integrating known lncRNA-disease associations and gene-disease associations. Chen developed a computational model of KATZLDA to identify potential lncRNA-disease associations by known lncRNA-disease associations and various similarity measures of diseases and lncRNAs44. Considering the limitations of traditional Random Walk with Restart (RWR), the model of Improved Random Walk with Restart for LncRNA-Disease Association prediction (IRWRLDA45) was developed by Chen et al. to predict novel lncRNA-disease associations by integrating known lncRNA-disease associations, disease semantic similarity, and various lncRNA similarity measures.
Meanwhile, Chen et al.46 proposed a novel computational model of Within and Between Score for MiRNA-Disease Association prediction (WBSMDA) by incorporating miRNA functional similarity, disease semantic similarity, miRNA-disease associations and Gaussian interaction profile kernel similarity for diseases and miRNAs. Li et al.47 developed a matrix completion for MiRNA-disease association prediction model (MCMDA). This model used the Lagrange multiplier method to update the adjacency matrix of known miRNA-disease associations and further predict potential associations. Chen et al.48 further proposed a model called graph regression for MiRNA-disease association prediction. The model carried out graph regression in three spaces, including association space, miRNA similarity space and disease similarity space. Then, Chen et al.49 put forward Inductive Matrix Completion for MiRNA-Disease Association prediction (IMCMDA), which could apply to new diseases without known miRNAs. Furthermore, Chen et al.50 developed another prediction miRNA-disease association prediction model of Bipartite Network Projection for MiRNA-Disease Association prediction (BNPMDA). This model first constructed the bias ratings for miRNAs and diseases based on three networks, including the known miRNA-disease association network, the disease similarity network and the miRNA similarity network. Then bipartite network recommendation algorithm was implemented to reveal potential miRNA-disease associations. Chen et al.51, proposed a novel computational method named Ensemble of Decision Tree based MiRNA-Disease Association prediction (EDTMDA), which innovatively built a computational framework integrating ensemble learning and dimensionality reduction. The model adopted ensemble learning strategy that integrated multiple classifiers (base learners) to get final prediction results. Then, Chen et al.52, developed the model of deep-belief network for miRNA-disease association prediction (DBNMDA). DBNMDA innovatively utilized the information of all miRNA-disease pairs during the pre-training process. This step could reduce the impact of too few known associations on prediction accuracy to some extent. In addition, Chen et al.53, proposed a new computational model named Neighborhood Constraint Matrix Completion for MiRNA-Disease Association prediction (NCMCMDA) to predict potential miRNA-disease associations. The model innovatively integrated neighborhood constraint with matrix completion, which provided a novel idea of utilizing similarity information to assist the prediction. After the recovery task was transformed into an optimization problem, this model solved it with a fast iterative shrinkage-thresholding algorithm.
However, the current lncRNA–miRNA association prediction models mainly use machine learning algorithms. Huang et al.54 proposed a method named EPLMI, which relied on the assumption that lncRNAs having similar expression profiles were prone to associate with a cluster of miRNAs that had similar expression profiles. However, a new question had arisen as to how to use the expression profile of ncRNAs to define the similarity between them. The EPLMI model calculated the similarity using the Person correlation coefficient, which was basically consistent with the hypothetical ncRNAs feature similarity score of each element pair. Nevertheless, the method still had some problems due to the nature of its mechanism. Liu et al.55 proposed the LMFNRLMI model, which utilized the strongest neighborhood relationship and established a neighborhood matrix to predict the lncRNA–miRNA association by using the K nearest neighbor method. However, there was still a lack of high-performance and high-precision models to predict potential lncRNA–miRNA associations. At the same time, Huang et al.56 developed a novel group preference Bayesian collaborative filtering model (GBCF), which picked up a top-k probability ranking list for an individual miRNA or lncRNA based on known lncRNA–miRNA interaction network. However, the Bayesian classifier needed to have negative samples to improve its performance. There were no negative samples in the lncRNA–miRNA association studies, and a random selection of positional association as negative samples would affect the prediction performance. A sequence-derived linear neighborhood propagation method (SLNPM) to predict lncRNA–miRNA associations was proposed by Zhang et al.57. Firstly, miRNA–miRNA similarity and lncRNA–lncRNA similarity were calculated by using miRNA sequence and lncRNA sequence and the known lncRNA–miRNA associations. Secondly, the integrated lncRNA similarity-based graph and the integrated miRNA similarity-based graph were respectively constructed, and the label propagation processes were respectively implemented on two graphs to score lncRNA–miRNA pairs. Finally, the averages of their outputs were adopted as final predictions. However, these methods still have some limitations, which will inspire us to develop better models. In the association prediction of between lncRNA and miRNA, the focus and difficulty of the next step is to further reduce the dependence of the model on the quality of the lncRNA and miRNA similarity matrix, pay more attention to the difference of correlation strength, reduce the complexity of model calculation, and avoid the prediction model to bias towards some well-studied lncRNAs or miRNAs. In the future development of lncRNA–miRNA association prediction, cloud computing can further make it possible to mine complex large-scale information. It can further explore the deep correlation between lncRNAs and miRNAs related indicators, so as to find out that the joint action of lncRNAs and miRNAs leads to the occurrence of diseases. Therefore, the diseases can be accurately predicted before they are formed, so as to carry out manual intervention as early as possible, make a more accurate description of the degree of disease, and find a series of changes in the body to form the root (lncRNAs and miRNAs) of the disease, so as to achieve accurate and efficient treatment.
In this paper, in order to more effectively predict potential associations between lncRNA and miRNA, we proposed a new computational method called Network Consistency projection for the Human LncRNA–miRNA Association (JSCSNCP-LMA). JSCSNCP-LMA achieved excellent prediction performance by using Jaccard similarity algorithm, self-correcting spectral clustering similarity algorithm, cosine similarity algorithm and known lncRNA–miRNA association network to predict lncRNA–miRNA of unknown associations. There are three advantages to this method. First of all, the algorithm in our prediction model is relatively simple and has no complex parameters. And the algorithm can also get good prediction results. Furthermore, our method could be also used for other association prediction, which has good expansibility. Last but not least, we can use lncRNA–miRNA association prediction to further study lncRNA-disease association prediction or miRNA-disease association prediction, so as to improve the accuracy of lncRNA-disease association prediction or miRNA-disease association prediction. To demonstrate the prediction performance of the JSCSNCP-LMA, LOOCV and fivefold CV were used to test the model. The results showed that the AUC values of the proposed JSCSNCP-LMA were 0.9268 and 0.9145, respectively.
Datasets and methods
Datasets
For lncRNA, miRNA, and lncRNA–miRNA interactions data, there are many open-source datasets available for online download. For example, miRBase58, miRmine59, NONCODE60, and lncRNASNP61. We obtained three different datasets from different databases in order to verify the accuracy of our experiment. The specific operations are as follows. Firstly, we downloaded data from lncRNASNP, and obtained 8091 experimentally verified lncRNA–miRNA interactions. After removing duplicated associations, we obtained 275 miRNAs and 780 lncRNAs. Then, we collected lncRNAs’ sequences from NONCODE and miRNAs’ sequences from miRbase. We finally obtained 417 lncRNAs and 265 miRNAs, which could be used as our Data 1. Secondly, we downloaded and cleaned the starBasev2.062 database on the ENCORI (open source platform). After processing, we obtained 1089 lncRNAs and 246 miRNAs, which could be used as our Data 2. Thirdly, we obtained the lncRNA–miRNA interactions from the known lncRNASNP2 database. After processing, we finally obtained 8634 lncRNA–miRNA interactions, including 468 lncRNAs and 262 miRNAs, which could be used as our Data 3. Three datasets were finally obtained, as shown in Table 1 below.
In this paper, we let \(L=\left\{{l}_{1},{l}_{2},{l}_{3},\dots ,{l}_{r}\right\}\) and \(M=\left\{{m}_{1},{m}_{2},{m}_{3},\dots ,{m}_{n}\right\}\), which represented the set of r lncRNAs and n miRNAs. We defined adjacency matrix \({\varvec{Y}}\) to represent the relationship between lncRNA and miRNA interactions. If \(lncRNA {l}_{i}\) was verified to interact with \(miRNA {m}_{j}\), then \(Y (i, j)\) was assigned 1, otherwise 0. We let \(Y\left({l}_{i}\right)=\left\{{l}_{1},{l}_{2},{l}_{3},\dots ,{l}_{r}\right\}\) and \(Y\left({m}_{j}\right)=\left\{{m}_{1},{m}_{2},{m}_{3},\dots ,{m}_{n}\right\}\), which represented the row i-vector of matrix \({\varvec{Y}}\) and the column j-vector of matrix \({\varvec{Y}}\). \(Y({l}_{i})\) and \(Y({m}_{j})\) represented the interactions of \(lncRNA {l}_{i}\) and \(miRNA {m}_{j}\), respectively. Matrix \({\varvec{Y}}\) is defined as follows:
Methods
Cosine similarity for lncRNA and miRNA
Previously, cosine similarity algorithm has been widely used by researchers in the collaborative filtering recommendation algorithm63,64. Recently, Gaussian distribution kernel similarity algorithm has been widely used to calculate the similarity of individual biomolecules in human body. However, its performance is lower than the cosine similarity algorithm. Therefore, in this paper we decided to use cosine similarity as a complementary dimension of lncRNA and miRNA similarity. The principle of lncRNA cosine similarity was based on the assumption that if \(lncRNA {l}_{i}\) and \(lncRNA {l}_{j}\) were similar to each other, then in the lncRNA–miRNA association matrix, binary vector \(Y({l}_{i})\) and binary vector \(Y({l}_{j})\) should be similar to each other. The same assumption should also be true for miRNA. Based on known lncRNA–miRNA associations data, the cosine similarity matrix CL of lncRNA is calculated as follows:
The binary vector \(Y({l}_{i})\) indicates whether there is an association between \(lncRNA {l}_{i}\) and each miRNA (the row i of the adjacency matrix \({\varvec{Y}}\), 1 if \({l}_{i}\) is related to miRNA, otherwise 0). Meanwhile, \(CL({l}_{i},{l}_{j})\) is the cosine similarity of between \(lncRNA {l}_{i}\) and \(lncRNA {l}_{j}\). The \({\varvec{C}}{\varvec{L}}\) is the lncRNA cosine similarity matrix.
Similarly, the cosine similarity of \(miRNA {m}_{i}\) and \(miRNA {m}_{j}\) is calculated as follows:
The binary vector \(Y({m}_{i})\) indicates whether there is an association between \(miRNA {m}_{i}\) and each lncRNA (the column j of adjacency matrix \({\varvec{Y}}\), 1 if \({m}_{j}\) is related to lncRNA, otherwise 0). Meanwhile, \(CM({m}_{i},{m}_{j})\) is the cosine similarity of between \(miRNA {m}_{i}\) and \(miRNA {m}_{j}\). The \({\varvec{C}}{\varvec{M}}\) is the miRNA cosine similarity matrix.
Jaccard similarity for lncRNA and miRNA
Recently, Jaccard similarity coefficient has been widely used in recommendation algorithms65. Meanwhile, it has been used by researchers to predict the associations of biological factors, because it is mainly used to compare the similarity between limited sample sets, and it does not consider the potential value size in the vector. In this paper, we used Jaccard similarity to calculate the similarity of lncRNA and miRNA respectively. The Jaccard similarity matrix \({\varvec{J}}{\varvec{L}}\) of lncRNA is calculated as follows:
where \(Y({l}_{i})\) and \(Y({l}_{j})\) represent the number of miRNAs sets associated with \(lncRNA {l}_{i}\) and \(lncRNA {l}_{j}\), respectively. The \({\varvec{J}}{\varvec{L}}\) is the lncRNA Jaccard similarity matrix. Similar to lncRNA, the Jaccard similarity between \(miRNA {m}_{i}\) and \(miRNA {m}_{j}\) can be calculated as follows:
where \(Y({m}_{i})\) and \(Y({m}_{j})\) represent the number of miRNAs sets associated with \(miRNA {m}_{i}\) and \(miRNA {m}_{j}\), respectively. The \({\varvec{J}}{\varvec{M}}\) is the miRNA Jaccard similarity matrix.
Self-tuning spectral clustering similarity for lncRNA and miRNA
After Jaccard similarity calculation, the similarity matrix we obtained was still relatively sparse. To improve the accuracy of the experiment, we used self-tuning spectral clustering similarity algorithm to fill the sparse matrix66. Spectral clustering is one of the methods of clustering, which can deal with complex and diverse structured data. It does not require an explicit model for estimating the data distribution, but rather performs a spectral analysis of the point-to-point similarity matrix. In this paper, we concluded that similar lncRNAs were related to similar function miRNAs, so we used self-tuning spectral clustering similarity algorithm to calculate the similarity between \(lncRNA {l}_{i}\) and \(lncRNA {l}_{j}\). We represented the rows i and j vectors of the matrix \({\varvec{Y}}\) as \(VL({l}_{i})\) and \(VL({l}_{j})\), respectively. Then, the self-tuning spectral clustering similarity of lncRNAs can be calculated as follows:
where, \(VL({l}_{iK})\) is the K th adjacent point of the \(VL({l}_{i})\) sample point, and here we default K value is 5.
where, \({\varvec{S}}{\varvec{L}}\) is the self-tuning spectral cluster similarity matrix of lncRNAs.
Similarly, we represented the column i and j vectors of the matrix \({\varvec{Y}}\) as \(VM({m}_{i})\) and \(VM({m}_{j})\), respectively. The self-tuning spectral clustering similarity of miRNAs can be calculated as follows:
where, \(VM({m}_{iK})\) is the K th adjacent point of the \(VM({m}_{i})\) sample point, and here we default K value is 5.
where, \({\varvec{S}}{\varvec{M}}\) is the self-tuning spectral cluster similarity matrix of miRNAs.
Integrated lncRNA similarity and miRNA similarity
After the above steps, we obtained the cosine similarity matrix, Jaccard similarity matrix, and self-tuning spectral cluster similarity matrix of lncRNAs and miRNAs. Then, we integrated various similarity matrices to obtain a more complete similarity matrix. First, we combined the Jaccard similarity matrix of lncRNAs with the self-tuning spectral cluster similarity matrix of lncRNAs. The integrated similarity matrix JSL of the lncRNAs can be calculated, as follows:
where, we used the self-tuning spectral cluster similarity to supplement the Jaccard similarity. If \(JL({l}_{i},\,{l}_{j})\) was 0, it was filled directly by \(SL({l}_{i},{l}_{j})\), otherwise the mean was taken, so that the matrix was more complete to make the sparse matrix dense and improve the accuracy of the experimental results. Second, we combined the Jaccard similarity matrix of miRNAs with the self-tuning spectral cluster similarity matrix of miRNAs. The integrated similarity matrix JSM of the miRNAs can be calculated, as follows:
To better supplement the dimension of the similarity matrix, we would re-integrate the cosine similarity with the new integrated similarity matrix. Specifically, if \(lncRNA {l}_{i}\) and \(lncRNA {l}_{j}\) had no common associated miRNA in the adjacency matrix \({\varvec{Y}}\), then the value between them was 0 in the cosine similarity matrix \({\varvec{C}}{\varvec{L}}\). Therefore, when the \(lncRNA {l}_{i}\) and \(lncRNA {l}_{j}\) had no similarity scores in the \({\varvec{C}}{\varvec{L}}\), we directly used the \({\varvec{J}}{\varvec{S}}{\varvec{L}}\) similarity score between them as the integrated similarity score. If \(lncRNA {l}_{i}\) and \(lncRNA {l}_{j}\) had similar scores in \({\varvec{C}}{\varvec{L}}\), then we integrated the similarity scores in \({\varvec{C}}{\varvec{L}}\) and \({\varvec{J}}{\varvec{S}}{\varvec{L}}\) as the final similarity scores. We conducted experiments on the weight parameters of the integration process and found that 0.5 was the best integration. The integrated similarity matrix JSCL of the lncRNAs can be calculated, as follows:
Similarly, if the \(miRNA {m}_{i}\) and \(miRNA {m}_{j}\) had no common associated lncRNA in the adjacency matrix \({\varvec{Y}}\), then the value between them was 0 in the cosine similarity matrix \({\varvec{C}}{\varvec{M}}\). Therefore, when the \(miRNA {m}_{i}\) and \(miRNA {m}_{j}\) had no similarity scores in the \({\varvec{C}}{\varvec{M}}\), we directly used the \({\varvec{J}}{\varvec{S}}{\varvec{M}}\) similarity score between them as the integrated similarity score. If \(miRNA {m}_{i}\) and \(miRNA {m}_{j}\) had similar scores in \({\varvec{C}}{\varvec{M}}\), then we integrated the similarity scores in \({\varvec{C}}{\varvec{M}}\) and \({\varvec{J}}{\varvec{S}}{\varvec{M}}\) as the final similarity scores. The integrated similarity matrix JSCM of the miRNAs can be calculated, as follows:
Network consistent projection of the human
LncRNA–miRNA association
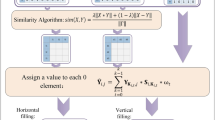
In our study, network consistency projection predicted potential lncRNA–miRNA associations through heterogeneous networks. The heterogeneous networks included the known lncRNA–miRNA association networks, the integrated lncRNAs similarity network (\({\varvec{J}}{\varvec{S}}{\varvec{C}}{\varvec{L}}\)), and the integrated miRNAs similarity network (\({\varvec{J}}{\varvec{S}}{\varvec{C}}{\varvec{M}}\)). JSCSNCP-LMA was divided into two parts: lncRNAs spatial consistency projection score and miRNAs spatial consistency projection score. The flow chart of the JSCSNCP-LMA method is shown in Fig. 1.
Flow chart of JSCSNCP-LMA applied to lncRNAs–miRNAs association prediction. The JSCSNCP-LMA including four steps: First, construct the lncRNA–miRNA association matrix. Second, calculate the cosine similarity matrix of lncRNA and miRNA, Jaccard similarity matrix of lncRNA and miRNA and self-tuning spectral cluster similarity matrix of lncRNA and miRNA. Third, integrate the above matrix to obtain the comprehensive similarity matrix of lncRNA and miRNA. Finally, the final prediction score matrix is calculated by the network consistency projection.
LncRNA space consistency projection score
Network consistency projection refers to the higher similarity score between \(lncRNA {l}_{i}\) and other lncRNA (including \(lncRNA {l}_{i}\) itself) in the integrated lncRNA similarity matrix (\({\varvec{J}}{\varvec{S}}{\varvec{C}}{\varvec{L}}\)), more lncRNAs related to \(miRNA {m}_{j}\), and the high spatial similarity of \(lncRNA {l}_{i}\) with \(miRNA {m}_{j}\). In the adjacency matrix \({\varvec{Y}}\), the values of the unknown associations are all 0; but in fact, these unknown associations are uncertain. Therefore, we replaced each of them with one small positive integer \(\delta\), and the value of \(\delta\) was set as 10–30. This approach prevents the denominator from being 0 and will not affect the result of the calculation. The lncRNA space consistency projection scores are calculated as follows:
where, \({JSCL}_{i}\) is the i th row of the integrated lncRNAs similarity matrix \({\varvec{J}}{\varvec{S}}{\varvec{C}}{\varvec{L}}\), which represents the similarity of \(lncRNA {l}_{i}\) with all lncRNAs. \({Y}_{j}\) is the jth column of the adjacency matrix \({\varvec{Y}}\), which represents the association of \(miRNA {m}_{j}\) with all lncRNAs. \(\left|{Y}_{j}\right|\) is the length of the vector \({Y}_{j}\), which is also the modules of the vector. \(JSCLSNCP(i,j)\) represents the network consistency projection score of \({JSCL}_{i}\) on \({Y}_{j}\). Specifically, if the angle of the network projection of \({JSCL}_{i}\) on \({Y}_{j}\) is smaller, then the value of \(JSCLSNCP(i,j)\) is larger.
MiRNA space consistency projection score
Similarly, the miRNA space consistency projection scores are calculated as follows:
where, \({JSCM}_{j}\) is the j th column of the integrated miRNAs similarity matrix JSCM, which represents the similarity of \(miRNA {m}_{j}\) with all miRNAs. \({Y}_{i}\) is the ith row of the adjacency matrix \({\varvec{Y}}\), which represents the association of \(lncRNA {l}_{i}\) with all miRNAs. \(\left|{Y}_{i}\right|\) is the length of the vector \({Y}_{i}\), which is also the modules of the vector. \(JSCMSNCP(i,j)\) represents the network consistency projection score of \({JSCM}_{j}\) on \({Y}_{i}\). Specifically, if the angle of the network projection of \({JSCM}_{j}\) on \({Y}_{i}\) is smaller, then the value of \(JSCMSNCP(i,j)\) is larger.
With the integration of \(JSCLSNCP(i,j)\) and \(JSCMSNCP(i,j)\) calculated above, the final similarity score matrix can be integrated and normalized, as follows:
where, \(JSCLSNCP(i,j)\) and \(JSCMSNCP(i,j)\) represent the network consistency projection scores for \(lncRNA {l}_{i}\) and \(miRNA {m}_{j}\) in the lncRNA space and miRNA space, respectively. The \(\left|\Delta \right|\) is a normalized operation to standardize the final prediction scores. Therefore, the value of \(JSCSNCP(i,j)\) ranges between 0 and 1. Matrix \({\varvec{J}}{\varvec{S}}{\varvec{C}}{\varvec{S}}{\varvec{N}}{\varvec{C}}{\varvec{P}}\) is the final projection score matrix in lncRNA space and miRNA space, and each value in the matrix represents the final score of each lncRNA–miRNA pair. The final score is used to predict lncRNA with miRNA association. The higher the score, the higher the association.
Results and discussion
Self-performance evaluation of the JSCSNCP-LMA model
The performance evaluation of the JSCSNCP-LMA model was divided into two parts: the self-performance evaluation and the performance evaluation with other methods. For the self-performance evaluation, we verified the performance of JSCSNCP-LMA by using k-fold CV. We set the k values to 2,3,4,5, respectively, to perform the comparison test. In the k-fold CV scheme, 2272 known lncRNA–miRNA associations were divided into k equal subsets randomly. For each cross-validation experiment, k − 1 of them were used as the training set and the remaining one subset was used as the test sample. The predicted scores were calculated and sorted by JSCSNCP-LMA, the special ranking position was selected as the threshold value, and the offline area (AUC value) of the receiver operating characteristic (ROC) curve was used as a performance index to evaluate the prediction performance67,68. The ROC curve can plot the relationship between true positive rate (TPR) and false positive rate (FPR) at different thresholds. If the AUC is closer to 1, then the predicted performance is better. The TPR and FPR can be calculated as follows:
where, TP, FN, FP, and TN, each represent True Positive, False Negative, False Positive and True Negative.
As shown in Fig. 2, it represented the ROC curves and AUC values under different k values in k-fold CV in Data 1, respectively.
To verify the generality of the method, we selected the data obtained from three different datasets by fivefold CV experiments to evaluate and compare its predictive analysis power. In the fivefold CV, 2272 lncRNAs–miRNAs associations in Data 1, 9086 lncRNAs–miRNAs associations in Data 2 and 8634 lncRNAs–miRNAs associations in Data 3 were included. For each cross-validation experiment, 4 of them were used as the training set and the remaining one subset was used as the test sample. The results of experiment are shown in Fig. 3.
To further verify the results of experiment, in this study, we used LOOCV and fivefold CV to compare their prediction performance on Data 1. The specific results are shown in Fig. 4.
After comparing the results of different validation methods, we also considered the following conditions of the self-performance evaluation of JSCSNCP-LMA: (1) predictive performance with all information (JSCSNCP-LMA); (2) considering only the prediction performance of lncRNA space projection; (3) considering only the prediction performance of miRNA space projection. According to the above situation, the ROC curves and the AUC values in LOOCV on Data 1 are shown in Fig. 5.
Influence of self-model’ s change on prediction accuracy. The figure shows that JSCSNCP-LMA has a reliable predictive performance with an AUC value of 0.9268. In the lncRNA projection space, the AUC value of JSCSNCP-LMA reach 0.8991. In the miRNA projection space, the AUC value is 0.9078. If the projection of two spaces is integrated, then the prediction performance can be greatly improved. Therefore, the JSCSNCP-LMA is reliable and has achieved good performance.
Meanwhile, we also compared influence of self-model’ s change by using AUPR in Data 1, as shown in Table 2.
Where, SSNCP-LMA is method JSCSNCP-LMA only including self-tuning spectral clustering similarity algorithm. JSSNCP-LMA is method JSCSNCP-LMA only including self-tuning spectral clustering similarity algorithm and Jaccard similarity algorithm. JSCLSNCP-LMA is considering only the prediction performance of lncRNA space projection. JSCMSNCP-LMA is considering only the prediction performance of miRNA space projection.
Comparison with the other methods
To further verify the advantages of JSCSNCP-LMA, we used the known lncRNAs–miRNAs associations to compare JSCSNCP-LMA with other five methods. To the best of our knowledge, there were only a few machine-learning based methods for lncRNA–miRNA associations prediction. Here, we adopted EPLMI and INLMI69 as the benchmark methods. EPLMI is a two-way diffusion model which uses the known lncRNA–miRNA interaction-based bipartite graph and expression profiles to predict lncRNA–miRNA associations. We implemented EPLMI using its publicly available source code. INLMI integrates the expression similarity networks and the sequence similarity networks to predict lncRNA–miRNA associations. And we implemented this model according to descriptions in Ref.69. Since predicting lncRNA–miRNA associations could be considered as a link prediction task, we adopted several network link inference methods as baseline methods, i.e. the collaborative filtering method (CF)63 and the resource allocation algorithm (RA)70. In collaborative filtering method takes known lncRNA–miRNA interactions as a bipartite graph and exploits external information, such as expression profiles to calculate the lncRNA–lncRNA similarity and miRNA–miRNA similarity. Then, the CF method finds neighbors for each lncRNA and each miRNA, then uses the weighted average of its neighbor-interacting miRNA (lncRNA) to predict unknown associations, and then combines the lncRNA-based neighbor prediction and the miRNA-based neighbor prediction with a trade-off parameter. The resource allocation algorithm also formulates lncRNAs (miRNAs) as nodes and lncRNA–miRNA interactions as links in a bipartite graph. Interaction information is iteratively propagated from miRNAs to their linked lncRNAs, and then the information is allocated from lncRNAs back to miRNAs. After finite iteration, final resources for miRNAs are probabilities that the lncRNA interacts with these miRNAs. We used open source code to implement the method of sequence-derived linear neighborhood propagation (SLNPM) to predict the lncRNA–miRNA associations. Finally, we adjusted the parameters to achieve the best performance of each method. In this study, the fivefold CV was used to compare their prediction performance in Data 1. The results of the JSCSNCP-LMA comparison with the other methods are shown in Fig. 6. In Fig. 6, we can see that the AUC of the JSCSNCP-LMA model has a score of 0.9145. The proposed model performs much better than EPLMI (AUC score 0.8494), INLMI (AUC score 0.8477), RA (AUC score 0.8637), CF (AUC score 0.8610) and SLNPM-SC (AUC score 0.9115).
In addition, we also compared the model with the recent and more popular algorithms(LMI-INGI71 NDALMA72) by using data in Refs.71,72. The comparison results (AUC values) are shown in Table 3.
Case studies
To further investigate JSCSNCP-LMA model proposed by us for predicting lncRNA–miRNA associations, we constructed JSCSNCP-LMA based on the Data1 to predict unkown lncRNA–miRNA associations that didn’t include in the Data 1. After predicting, our prediction results had been verified by other databases or relevant literature. Therefore, we selected the top ten lncRNAs–miRNAs data pairs of the prediction scores. As shown in Table 4.
Overall, 9 of the 10 data pairs of lncRNA–miRNA obtained by ranking prediction are confirmed by the corresponding database.
In addition, we also investigated the proportions of correctly predicted lncRNA–miRNA pairs among the top 100 highly scored predictions based on the bench marking dataset and compared their results with other methods. The results are shown in Table 5.
Therefore, we can argue that JSCSNCP-LMA can predict lncRNA–miRNA associations with high accuracy.
In addition, a number of studies have shown that lncRNA and miRNA play a key role in various diseases, especially cancer. To further evaluate the ability of JSCSNCP-LMA to predict potential lncRNA–miRNA associations, we conducted case studies on two common lncRNAs: H1973 and HOTAIR74. Among all the predicted results, we found and analyzed the top 15 miRNAs in H19 and HOTAIR prediction score. In H19, 14 miRNAs related to H19 have been verified by known databases or relevant literature. In HOTAIR, 14 miRNAs related to HOTAIR have been also verified by known databases or relevant literature, as shown in Tables 6 and 7.
In recent years, H19 has become a research hotspot due to its ectopic expression in human diseases, especially malignant tumors, and plays an important role as an oncogene in human malignant tumors. Meanwhile, H19 has been shown to be involved in the development and malignant progression of many tumors, and promote cell growth, invasion, migration, epithelial-mesenchymal transition, metastasis and apoptosis. In addition, H19 can isolate some miRNAs, promote multi-layer molecular regulatory mechanisms, or co-affect the occurrence of some diseases with some miRNAs. For example, in Table 3, Qin et al.75 verified that both H19 and hsa-mir-301b were prognostic factors of cervical cancer. Luo et al.76 verified that H19 played an important role in regulating inflammatory processes in retinal endothelial cells by regulating hsa-mir-93 under high-glucose condition. He et al.77 verified that H19 and hsa-mir-17/hsa-mir-106b could affect the treatment efficacy of patients with chronic hepatitis B. Zhao et al.78 verified that H19 could regulate the expression of ID2 through competitive binding to hsa-mir-19a/b, which played a role in the proliferation of acute myelocytic leukemia (AML) cells. Moreover, H19 also plays an important role in the generation or treatment of other cancers, such as colon cancer, breast cancer, lung cancer and prostate cancer79,80,81,82. Similarly, there are also many miRNAs that play an important role in the generation or treatment of cancer. For example, Rafael Sebastian Ford et al.83 verified the oncogenic role of hsa-mir-130b in prostate cancer. However, no biological researchers have directly verified the association between H19 and hsa-mir-130b at present. Therefore, the prediction results provided by the JSCSNCP-LMA are mainly used to provide biological researchers with research directions, so as to improve the efficiency of biological research.
Like H19, HOTAIR is one of the most widely studied abnormally regulated lncRNAs in human cancers. Studies have shown that in preclinical cancer research84, HOTAIR can control basic biochemical and cellular processes and promote proliferation, invasion, survival, drug resistance and metastasis through interactions with a variety of other biological factors. And HOTAIR has been also shown to promote tumor progression by regulating miRNAs expression and function. For example, in Table 4, Pan et al.85 verified that the regulatory mechanism between HOTAIR and hsa-mir-17 played an important role in ruptured intracranial aneurysm disease. Cao et al.86 verified that the regulatory mechanism between HOTAIR and hsa-mir-20a played an important role in liver cancer cells. Bao et al.87 verified that HOTAIR could affect human chondrosarcoma disease by controlling mir-454-3p. Moreover, HOTAIR also plays an important role in the generation or treatment of other cancers, such as lung cancer, rectal cancer, prostate cancer and cervical cancer88,89. Similarly, there are also many miRNAs that play an important role in the generation or treatment of cancer. For example, Liu et al.90 verified that hsa-mir-106b also played a crucial role in rectal cancer. Therefore, it is very important to study the unknown lncRNA–miRNA associations.
Conclusions
LncRNAs–miRNAs associations are critical to many biological activities and are closely related to the development of various diseases. Therefore, exploring and identifying these associations can help to understand the function of lncRNAs/miRNAs and complex disease mechanisms. In this paper, we proposed a prediction method named the JSCSNCP-LMA that was different from other traditional methods. JSCSNCP-LMA achieved excellent prediction performance by using Jaccard similarity algorithm, self-tuning spectral clustering similarity algorithm, cosine similarity algorithm and known lncRNA–miRNA association networks. JSCSNCP-LMA did not require redundant parameters and showed a obvious advantage when the known experimentally validated lncRNA–miRNA associations were insufficient. To validate the predictive performance of the JSCSNCP-LMA, LOOCV and fivefold CV were used. Results showed that the proposed method outperformed other methods and effectively identified potential lncRNAs–miRNAs associations. Meanwhile, the prediction capabilities of proposed models were also tested by case studies. We validated the results of case studies by using known literature and datasets. In conclusion, JSCSNCP-LMA is promising for lncRNA–miRNA association prediction. It not only has a good performance, but also has good expansibility. For example, metabolite-disease association prediction91, miRNA-disease association prediction92, lncRNA-disease association prediction93, and lncRNA-protein association prediction94.
Although this model has achieved good results, it still has some limitations. First of all, the known association data between lncRNAs and miRNAs is relatively small, which will affect the final prediction results. What’s more, this model only uses a single lncRNAs–miRNAs association data and does not combine other association data, such as lncRNAs-diseases, miRNAs-diseases and lncRNAs-proteins. This may lead to inaccurate prediction results due to the failure to consider the influence of other factors. Finally, in the algorithm, we only simply compare the influence of the default parameters on the algorithm and do not use the optimization algorithm to find the optimal solution automatically. This will reduce the accuracy of the prediction results. In order to better reduce the prediction bias and improve the prediction performance, our future work mainly focuses on the optimization of similarity calculation and method fusion. At the same time, we will also build an algorithm to automatically seek the optimal parameters, so as to improve the accuracy of the prediction results. Finally, we will try to combine other association data to consider the predicted results from multiple perspectives. We believe that when more biological knowledge is applied to a refined fusion method, the accuracy of model prediction can be improved. And our method can be helpful for relevant biomedical research.
Data availability
miRBase: http://www.mirbase.org/index.shtml. miRmine: http://guanlab.ccmb.med.umich.edu/mirmine. NONCODE: http://www.noncode.org. lncRNASNP: http://bioinfo.life.hust.edu.cn/lncRNASNP. ENCORI: http://starbase.sysu.edu.cn/.
References
Mercer, T. R., Dinger, M. E. & Mattick, J. S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 10(3), 155–159 (2009).
Kapranov, P. et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316(5830), 1484–1488 (2007).
Kapranov, P., Willingham, A. T. & Gingeras, T. R. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 8(6), 413–423 (2007).
Yamamura, S. et al. Interaction and cross-talk between non-coding RNAs. Cell Mol. Life Sci. 75(3), 467–484 (2018).
Mattick, J. S. & Makunin, I. V. Non-coding RNA. Hum. Mol. Genet. 15(1), R17-29 (2006).
Yong, H. et al. Molecular functions of small regulatory noncoding RNA. Biochem. Mosc. 78(3), 221–230 (2013).
Trzybulska, D., Vergadi, E. & Tsatsanis, C. miRNA and other non-coding RNAs as promising diagnostic markers. EJIFCC. 29(3), 221–226 (2018).
Hung, T. & Chang, H. Y. Long noncoding RNA in genome regulation: prospects and mechanisms. RNA Biol. 7(5), 582–585 (2010).
Guttman, M. et al. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell Camb. MA. 154(1), 240–251 (2013).
Spizzo, R. et al. Long non-coding RNAs and cancer: A new frontier of translational research?. Oncogene 31(43), 4577–4587 (2012).
Derrien, T. et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 22(9), 1775–1789 (2012).
Dechao, B. et al. NONCODE v3.0: Integrative annotation of long noncoding RNAs. Nucleic Acids Res. 40, D210–D215 (2012).
Mattick, J. S. & Lee, J. T. The genetic signatures of noncoding RNAs. PLoS Genet. 5(4), e1000459 (2009).
Qureshi, I. A., Mattick, J. S. & Mehler, M. F. Long non-coding RNAs in nervous system function and disease. Brain Res. 1338, 20–35 (2010).
Wapinski, O. & Chang, H. Y. Long noncoding RNAs and human disease. Trends Cell Biol. 21(6), 354–361 (2011).
Wang, K. C. & Chang, H. Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 43(6), 904–914 (2011).
Chen, X. et al. Long non-coding RNAs and complex diseases: from experimental results to computational models. Br. Bioinform. 18(4), 558–576 (2017).
Zhang, J. et al. Overexpression of FAM83H-AS1 indicates poor patient survival and knockdown impairs cell proliferation and invasion via MET/EGFR signaling in lung cancer. Sci. Rep. 7, 42819 (2017).
Zhang, Q. et al. NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. MBio 4(1), e00596-e612 (2013).
Congrains, A. et al. Genetic variants at the 9p21 locus contribute to atherosclerosis through modulation of ANRIL and CDKN2A/B. Atherosclerosis 220(2), 449–455 (2012).
Faghihi, M. A. et al. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat. Med. 14(7), 723–730 (2008).
Alvarez, M. L. & DiStefano, J. K. Functional characterization of the plasmacytoma variant translocation 1 gene (PVT1) in diabetic nephropathy. PLoS One 6(4), e18671 (2011).
Yang, Z. et al. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann. Surg. Oncol. 18(5), 1243–1250 (2011).
Poppel, H. V. et al. The relationship between Prostate CAncer gene 3 (PCA3) and prostate cancer significance. BJU Int. 109(3), 360–366 (2012).
Wang, J., Sun, J. & Yang, F. The role of long non-coding RNA H19 in breast cancer. Oncol. Lett. 19(1), 7–16 (2020).
Gebert, L. & Macrae, I. J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 20(1), 21–37 (2019).
Stefani, G. & Slack, F. J. Small non-coding RNAs in animal development. Nat. Rev. Mol. Cell Biol. 9(3), 219–230 (2008).
Ruan, K., Fang, X. & Ouyang, G. MicroRNAs: Novel regulators in the hallmarks of human cancer. Cancer Lett. 285(2), 116–126 (2009).
Schickel, R., Boyerinas, B., Park, S. M. & Peter, M. E. MicroRNAs: Key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene 27(45), 5959–5974 (2008).
Chen, X. et al. MicroRNAs and complex diseases: From experimental results to computational models. Br. Bioinform. 20(2), 515–539 (2019).
Huang, Y. et al. Biological functions of microRNAs: A review. J. Physiol. Biochem. 67(1), 129–139 (2011).
Zhang, Y. et al. MicroRNA-128 inhibits glioma cells proliferation by targeting transcription factor E2F3a. J. Mol. Med. (Berl). 87(1), 43–51 (2009).
Poy, M. N. et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature 432(7014), 226–230 (2004).
Yang, B. et al. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat. Med. 13(4), 486–491 (2007).
Zhao, Y. et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell 129(2), 303–317 (2007).
Pradhan, A. K. et al. The enigma of miRNA regulation in cancer. Adv. Cancer Res. 135, 25–52 (2017).
Nappi, L. & Nichols, C. MicroRNAs as biomarkers for germ cell tumors. Urol. Clin. N. Am. 46(3), 449–457 (2019).
Yang, G., Lu, X. & Yuan, L. LncRNA: A link between RNA and cancer. Biochem. Biophys. Acta. 1839(11), 1097–1109 (2014).
You, Z. et al. PBMDA: A novel and effective path-based computational model for miRNA-disease association prediction. PLoS Comput. Biol. 13(3), e1005455 (2017).
Shaath, H. & Alajez, N. M. Identification of PBMC-based molecular signature associational with COVID-19 disease severity. Heliyon. 7(5), e06866 (2021).
Chen, X. & Yan, G. Y. Novel human lncRNA-disease association inference based on lncRNA expression profiles. Bioinformatics 29(20), 2617–2624 (2013).
Chen, X. et al. Constructing lncRNA functional similarity network based on lncRNA-disease associations and disease semantic similarity. Sci. Rep. 5, 11338 (2015).
Yang, X. et al. A network based method for analysis of lncRNA-disease associations and prediction of lncRNAs implicated in diseases. PLoS One 9, e87797 (2014).
Chen, X. KATZLDA: KATZ measure for the lncRNA-disease association prediction. Sci. Rep. 5, 16840 (2015).
Chen, X. et al. IRWRLDA: Improved random walk with restart for lncRNA-disease association prediction. Oncotarget 7(36), 57919–57931 (2016).
Chen, X. et al. WBSMDA: Within and between score for MiRNA-disease association prediction. Sci. Rep. 6, 21106 (2016).
Li, J. Q. et al. MCMDA: Matrix completion for MiRNA-disease association prediction. Oncotarget 8, 21187–21199 (2017).
Chen, X. et al. GRMDA: Graph regression for MiRNA-disease association prediction. Front. Physiol. 9, 92 (2018).
Chen, X. et al. Predicting miRNA-disease association based on inductive matrix completion. Bioinformatics 34, 4256–4265 (2018).
Chen, X. et al. BNPMDA: Bipartite network projection for MiRNA-disease association prediction. Bioinformatics 34(18), 3178–3186 (2018).
Chen, X., Zhu, C. C. & Yin, J. Ensemble of decision tree reveals potential miRNA-disease associations. PLoS Comput. Biol. 15(7), e1007209 (2019).
Chen, X. et al. Deep-belief network for predicting potential miRNA-disease associations. Br. Bioinform. 22(3), 186 (2021).
Chen, X., Sun, L. G. & Zhao, Y. NCMCMDA: miRNA-disease association prediction through neighborhood constraint matrix completion. Br. Bioinform. 22(1), 485–496 (2021).
Huang, Y. A., Chan, K. C. C. & You, Z. H. Constructing prediction models from expression profiles for large scale lncRNA–miRNA interaction profiling. Bioinformatics 34(5), 812–819 (2018).
Liu, H. et al. Predicting lncRNA–miRNA interactions based on logistic matrix factorization with neighborhood regularized. Knowl.-Based Syst. 191, 10526 (2020).
Huang, Z. A. et al. Novel link prediction for large-scale miRNA–lncRNA interaction network in a bipartite graph. BMC Med. Genom. 11(Suppl 6), 113 (2018).
Zhang, W. et al. LncRNA–miRNA interaction prediction through sequence-derived linear neighborhood propagation method with information combination. BMC Genom. 20(Suppl 11), 946 (2019).
Kozomara, A. & Griffiths-Jones, S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42(Database issue), D68-73 (2014).
Panwar, B., Omenn, G. S. & Guan, Y. miRmine: A database of human miRNA expression profiles. Bioinformatics 33(10), 1554–1560 (2017).
Fang, S. et al. NONCODEV5: A comprehensive annotation database for long non-coding RNAs. Nucleic Acids Res. 46(D1), D308–D314 (2018).
Gong, J., Liu, W., Zhang, J., Miao, X. & Guo, A. Y. lncRNASNP: A database of SNPs in lncRNAs and their potential functions in human and mouse. Nucleic Acids Res. 43(Database issue), D181–D186 (2015).
Li, J. H., Liu, S., Zhou, H., Qu, L. H. & Yang, J. H. starBase v2.0: Decoding miRNA–ceRNA, miRNA–ncRNA and protein–RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 42(Database issue), D92–D97 (2014).
Herlocker, J. L. et al. Evaluating collaborative filtering recommender systems. ACM Trans. Inf. Syst. 22(1), 5–53 (2004).
Duan, R., Jiang, C. & Jain, H. K. Combining review-based collaborative filtering and matrix factorization: A solution to rating’s sparsity problem. Decis. Support Syst. 156, 113748 (2022).
Wu, M. et al. IMPMD: An integrated method for predicting potential associations between miRNAs and diseases. Curr. Genom. 20(8), 581–591 (2019).
Zelnik-Manor, L., Perona, P. Self-tuning spectral clustering. In Advances in Neural Information Processing Systems (NIPS). (2004).
Hanley, J. A. & McNeil, B. J. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 143(1), 29–36 (1982).
Fawcett, T. An introduction to ROC analysis. Pattern Recogn. Lett. 27(8), 861–874 (2005).
Hu, P., Huang, Y.-A., Chan, K. C. C. & You, Z.-H. Discovering an Integrated Network in Heterogeneous Data for Predicting lncRNA–miRNA Interactions 539–545 (Springer, 2018).
Tao, Z. et al. Solving the apparent diversity-accuracy dilemma of recommender systems. Proc. Natl. Acad. Sci. U.S.A. 107(10), 4511–4515 (2010).
Zhang, L. et al. Predicting lncRNA–miRNA interactions based on interactome network and graphlet interaction. Genomics 113(3), 874–880 (2021).
Zhang, L. et al. Using network distance analysis to predict lncRNA–miRNA interactions. Interdiscip. Sci. 13(3), 535–545 (2021).
Ghafouri-Fard, S., Esmaeili, M. & Taheri, M. H19 lncRNA: Roles in tumorigenesis. Biomed. Pharmacother. 123, 109774 (2020).
Rajagopal, T. et al. HOTAIR LncRNA: A novel oncogenic propellant in human cancer. Clin. Chim. Acta. 503, 1–18 (2020).
Qin, S. et al. Identifying molecular markers of cervical cancer based on competing endogenous RNA network analysis. Gynecol. Obstet. Investig. 84(4), 350–359 (2019).
Luo, R., Xiao, F., Wang, P. & Hu, Y. X. lncRNA H19 sponging miR-93 to regulate inflammation in retinal epithelial cells under hyperglycemia via XBP1s. Inflamm. Res. 69(3), 255–265 (2020).
He, Y. et al. Identifying potential biomarkers in hepatitis B virus infection and its response to the antiviral therapy by integrated bioinformatic analysis. J. Cell Mol. Med. 25(14), 6558–6572 (2021).
Zhao, T. F. et al. LncRNA H19 regulates ID2 expression through competitive binding to hsa-miR-19a/b in acute myelocytic leukemia. Mol. Med. Rep. 16(3), 3687–3693 (2017).
Jing, R. et al. Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics. 8(14), 3932–3948 (2018).
Wang, J. et al. The long noncoding RNA H19 promotes tamoxifen resistance in breast cancer via autophagy. J. Hematol. Oncol. 12(1), 81 (2019).
Zhao, Y. et al. LncRNA H19 promotes lung cancer proliferation and metastasis by inhibiting miR-200a function. Mol. Cell Biochem. 460(1–2), 1–8 (2019).
Hu, J. C. et al. Impact of H19 polymorphisms on prostate cancer clinicopathologic characteristics. Diagnostics. 10(9), 656 (2020).
Fort, R. S. et al. An integrated view of the role of miR-130b/301b miRNA cluster in prostate cancer. Exp. Hematol. Oncol. 7, 10 (2018).
Tr, A. et al. HOTAIR LncRNA: A novel oncogenic propellant in human cancer—ScienceDirect. Clin. Chim. Acta 503, 1–18 (2020).
Pan, Y. B. et al. Construction of competitive endogenous RNA network reveals regulatory role of long non-coding RNAs in intracranial aneurysm. BMC Neurosci. 22(1), 1–14 (2021).
Cao, M. R. et al. Bioinformatic analysis and prediction of the function and regulatory network of long non-coding RNAs in hepatocellular carcinoma. Oncol. Lett. 15(5), 7783–7793 (2018).
Bao, X. et al. Knockdown of long non-coding RNA HOTAIR increases miR-454-3p by targeting Stat3 and Atg12 to inhibit chondrosarcoma growth. Cell Death Dis. 8(2), e2605 (2017).
Liu, X. H. et al. LncRNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol. Cancer 13(1), 92 (2014).
Zhou, Y. H. et al. Long non-coding RNA HOTAIR in cervical cancer: Molecular marker, mechanistic insight, and therapeutic target. Adv. Clin. Chem. 97, 117–140 (2020).
Liu, S. et al. The miR-106b/NR2F2-AS1/PLEKHO2 axis regulates migration and invasion of colorectal cancer through the MAPK pathway. Int. J. Mol. Sci. 22(11), 5877 (2021).
Sun, F., Sun, J. & Zhao, Q. A deep learning method for predicting metabolite-disease associations via graph neural network. Br. Bioinform. 23(4), bbac266 (2022).
Liu, W. et al. Identification of miRNA-disease associations via deep forest ensemble learning based on autoencoder. Br. Bioinform. 23(3), 104 (2022).
Xie, G. et al. HBRWRLDA: Predicting potential lncRNA-disease associations based on hypergraph bi-random walk with restart. Mol. Genet. Genom. 297(5), 1215–1228 (2022).
Jia, L. & Luan, Y. Multi-feature fusion method based on linear neighborhood propagation predict plant LncRNA–protein interactions. Interdiscip. Sci. Comput. Life Sci. 14(2), 545–554 (2022).
Acknowledgements
These authors contributed equally to this work.
Funding
This work was supported in part by the Undergraduate Universities Fundamental Research Funding Project of Heilongjiang Province in 2020, No.135509112.
Author information
Authors and Affiliations
Contributions
B.W.: Conceptualization, methodology, software, formal analysis, writing-original draft, funding acquisition; X.W.: Methodology, software, investigation, formal analysis, writing-original draft, funding acquisition; X.Z.: Investigation, software, validation, supervision; Y.H.: Software, validation; X.D.: Writing—review & editing, funding acquisition; all authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, B., Wang, X., Zheng, X. et al. JSCSNCP-LMA: a method for predicting the association of lncRNA–miRNA. Sci Rep 12, 17030 (2022). https://doi.org/10.1038/s41598-022-21243-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-21243-y
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.