Abstract
In this study we evaluated eleven candidate reference genes in Avena sativa during compatible and incompatible interactions with two different pathotypes of Puccinia coronata f. sp. avenae in six time points post-inoculation. The identification of genes with high expression stability was performed by four algorithms (geNorm, NormFinder, BestKeeper and ΔCt method). The results obtained confirmed that the combination of two genes would be sufficient for reliable normalization of the expression data. In general, the most stable in the tested plant-pathogen system were HNR (heterogeneous nuclear ribonucleoprotein 27C) and EF1A (elongation factor 1-alpha). ARF (ADP-ribosylation factor) and EIF4A (eukaryotic initiation factor 4A-3) could also be considered as exhibiting high expression stability. CYP (cyclophilin) was shown by all assessment methods to be the worst candidate for normalization in this dataset. To date, this is the first report of reference genes selection in A. sativa–P. coronata interaction system. Identified reference genes enable reliable and comprehensive RT-qPCR analysis of oat gene expression in response to crown rust infection. Understanding the molecular mechanisms involved in the host–pathogen interactions may expand knowledge of durable resistance strategies beneficial to modern oat breeding.
Similar content being viewed by others
Introduction
Avena sativa L. is a species cultivated on a global scale with a high range of applications from the food industry to pharmaceuticals and animal feed1,2. One of the greatest threats to cultivated oat are fungal diseases, of which the most common is crown rust caused by Puccicnia coronata f. sp avenae P. Syd. & Syd3,4. Research on the Puccinia population biology confirms the high virulence dynamics of this pathogen5,6. Despite the identification of over 100 crown rust resistance genes (Pc), due to the high adaptive potential of the fungus, most of them have already been overcome. Hence, the need to conduct the research on obtaining durable plant resistance, effective under conditions favourable to the disease development7,8. Genetically determined disease resistance mechanisms, although universal in nature, are very complex and closely dependent on the plant-pathogen system. Plant immune studies are often based on the analysis of gene expression profiling with the use of modern molecular biology methods, of which qPCR (quantitative PCR, real-time PCR) has become very popular due to its high sensitivity and specificity. In order to obtain highly reliable and repeatable results, the MIQE (minimum information for publication of quantitative real-time PCR experiments) guideline was developed recommending the standardization of each step of the qPCR experiment9. In accordance with this laboratory practice, reliable qPCR experiment requires the selection of at least two reference genes with a stable level of expression in a given experimental system. The housekeeping genes of basic cellular metabolism are widely used for this purpose, however numerous studies have shown that none of these genes are universal10. The stability of the reference gene expression may vary depending on the species studied, tissue type, developmental stage or experimental conditions11,12,13,14. For this reason, it is crucial to analyze the expression stability of potential reference genes individually for each experimental system. This type of research was carried out mainly under abiotic stress conditions. Jaiswal et al.15 analyzed the expression of ten candidate reference genes during seed development in Cyamopsis tetragonoloba (L.) Taub. Similar studies were conducted in Rhizophora apiculata16, Hordeum brevisubulatum17 Hypericum perforatum18 and Hordeum vulgare19. In the genus Avena, studies involving the selection of reference genes to analyse the gene expression under various types of stress have so far been conducted mainly on Avena fatua L. Wrzesińska et al.20 assessed the expression of six genes (18S ribosomal RNA (18S rRNA), actin (ACT), TATA-binding protein (TBP), α-tubulin (TUB), elongation factor 1-alpha (EF1A), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in A. fatua biotypes resistant to various types of herbicides. Liu et al.21 attempted to evaluate eight candidate reference genes (18S rRNA, 28S rRNA, ACT, GAPDH, EF1A, ribosomal protein L7 (RPL7), α-TUB, and TBP) in A. fatua differend types of tissues at various growth stages under the influence of the herbicide. Similar studies verifying the four reference genes (ACT, EF1A, GAPDH, TBP) in herbicide treated stem and leaf tissues were carried out on A. ludoviciana22. The stability of reference genes was also tested for the molecular analysis of the A. fatua caryopses dormancy23. However, the large size (12.5 Gb) of the allohexaploid genome (AACCDD) and the basic chromosome number of 2n = 6x = 42 make A. sativa a very difficult research object24. Oat genome structure may indicate the presence of mainly duplicated genes. This hinders the finding of the proper RGs or design of optimal primers, as each copy of the duplicated gene may not be uniformly expressed in different samples25,26. Preferably “single-copy” genes should be used as a reference, however, the study of Yang et al.27 provides a proof of concept that using duplicated RG is also feasible and valid in polyploid oat.
The study attempts to determine the reference genes for the A. sativa–P. coronata experimental setup. The obtained results will provide valuable data necessary for a comprehensive analysis of gene expression in oat. The study will also constitute a reference point for further analyses of the new Pc genes identified in our previous research28,29,30,31.
Materials and methods
Plant material and crown rust inoculation
The study material consisted of oat line Pc39 (Pendek × Pc39) with major gene resistance to crown rust caused by P. coronata f. sp. avnae and the Polish cultivar Kasztan (Dawid × CHD 1685/84) susceptible to rust infection32. The differential near-isogenic line Pc39 was developed at the Cereal Research Centre AAFC Winnipeg, Canada33,34,35. The line carries a major race-specific crown rust resistance gene Pc39 incorporated into common oat from the wild hexaploid oat Avena sterilis F-366 collected in Israel36. Seeds of the studied genotypes were grown in plug trays filled with a universal substrate containing peat for 10 days in a phytotron at 18 °C with a 16-h photoperiod.
Two P. coronata f. sp. avenae pathotypes used in the study (13.1, 230, Table 1) with a virulence profile defined based on the standard differentials set37 and supplemental differentials (Pc14, Pc35, Pc36, Pc55, Pc57, Pc60, Pc61, Pc63, Pc67, Pc70, Pc71, Pc91, Pc94, Pc96, Pc97, Pc98, Pc101, Pc103-1 and Pc104) were selected from a collection of single-pustule isolates derived from populations collected in Poland in the years 2010–20196,38,39. Both pathotypes are virulent to ‘Kasztan’. Pc39 line is resistant to 13.1 and susceptible to 230. Dried pathotypes were stored in 1.5-ml microfuge tubes at − 70 °C. Before inoculation, urediniospores were heat-shocked for 4 min at 42 °C and multiplied on leaf fragments of the susceptible oat cultivar Kasztan32 using the host–pathogen method of Hsam et al.40, originally used for Blumeria graminis f. sp. avenae and modified by Paczos-Grzęda and Sowa6. Study was conducted on the first leaves of 10-day-old seedlings. Inoculation was performed in a settling towers by spreading uredinospores on plant material at a density of ≈ 200 spores/cm2. Pc39 line was inoculated using both P. coronata pathotypes and ‘Kasztan’ was inoculated with 230 P. coronata pathotype. Glass slides were placed between the inoculated leaves to monitor the inoculation density. The settling towers were thoroughly cleaned with ethanol between inoculations with different isolates and the room sprayed with water. Seedlings were incubated in a spore-proof growth chamber at 18 °C with 70% humidity, light intensity of approximately 4 kLx under a 16-h photoperiod.
Plant experiments were performed in accordance with relevant guidelines and regulations.
RNA extraction and reverse transcription
RNA extraction was performed after 0 (uninoculated ‘Kasztan’ and Pc39), 8, 16, 24, 48 and 72 h post inoculation (hpi). Three biological replicates, each composed of five leaves of different seedling from the study group were sampled. After harvesting, plant material was immediately frozen in liquid nitrogen and grind to a fine powder with sterile mortar and pestle. Total RNA was extracted with TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. The integrity and quality of RNA samples were evaluated by electrophoresis on 1.5% agarose gel stained with ethidium bromide. The RNA quantity was determined with NanoDrop2000 spectrophotometer (Thermo Fisher Scientific Inc., USA). Prior to cDNA synthesis, potential genomic DNA contamination was eliminated by DNase I (EURx Ltd., Poland) treatment of all RNA samples at 37 °C for 30 min and heat inactivated at 65 °C for 10 min with 50 mM EDTA. The cDNA synthesis was performed in 20 µl reaction volume using the NG dART RT kit (EURx Ltd., Poland), 2 µg RNA sample and oligo (dT)18 primer, as per manufacturer’s instructions. cDNA samples were diluted with nuclease-free water and used for RT-qPCR.
Selection of reference genes and primer design
Eleven candidate reference genes frequently used in other plants: ARF (ADP-ribosylation factor), CYP (cyclophilin), EF1A (elongation factor 1-alpha), EIF4A (eukaryotic initiation factor 4A-3), GAPDH (glyceraldehyde-3-phosphate dehydrogenase), HNR (heterogeneous nuclear ribonucleoprotein 27C), HSP70 (heat shock protein 70), TUA (alpha tubulin), UBC (ubiquitin conjugating enzyme), ACT (actin), EP (expressed protein) were chosen for expression stability assessment41,42. Specific primers for qPCR (Table S1) were obtained based on existing literature20,27,43 as well as designed with PrimerBLAST tool (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). Homologous oat sequences were retrieved from the A. sativa v2.0 genome detabase (https://wheat.pw.usda.gov/jb?data=/ggds/oat-ot3098v2-pepsico) and oat seed transcriptome44 via BLASTn search using highly conserved regionsof the corresponding Triticum aestivum, Avena fatua, and Brachypodium distachyon cDNA sequences with an expected value (E) < 10−10 and minimum base identity > 95% as blast criteria with CLC Genomics Workbench (CLCbio, Seoul, Korea).
Quantitative realtime PCR analysis of candidate reference genes
For qRT-PCR, primer specificity was determined using melting curve analysis and the PCR products were evaluated on 2% agarose gel. The amplification efficiency (E) and regression coefficients (R2) of candidate reference genes were calculated based on the standard curve generated by a fivefold serial dilution points of cDNA combined with a mix containing all the studied samples. The calculation of E values is as follows: E (%) = (10−1/slope − 1) × 100. Only specific primer pairs with efficiency ranging from 90 to 110%, the slope between − 3.6 to − 3.1 and correlation coefficient (R2) over 0.99 were left for further analysis.
Diluted aliquots of the reverse-transcribed cDNAs were used as templates in qPCR assays. Quantitative PCR was performed in three biological replicates with three technical replicates, no template control (NTC) and no reverse transcription control (NRT) on QuantStudio™ 3 Real-Time PCR System (Applied Biosystems, USA) with Power Track SYBR Green Master Mix (Thermo Fisher Scientific Inc., USA). Each 20 μL reaction mixture contained 20 ng of cDNA 1 × qPCR Mix and 400 nM of each primer. The qPCR program was as follows: 95 °C for 2 m, 40 cycles of 95 °C for 15 s, 60 °C for 1 m. To confirm the amplification specificity and lack of primer dimer formation, each run was performed with a melting curve analysis.
Analysis and validation of gene expression stability
The raw data of qPCR was processed by means of Thermo Fisher Scientific online data analysis app. The expression stability of the selected housekeeping genes was analysed using either untransformed Cq values for BestKeeper45 and ΔCt method or relative quantities for NormFinder46 and geNorm47 software packages. The input data included reaction efficiency corrections. The expression stability analysis was conducted on five datasets—separately for A. sativa–P. coronata compatible and incompatible interactions, independently for Kasztan cultivar and Pc39 oat line and additionally as full dataset consisting of all experimental samples48.
For validation of identified RGs the relative expression of the target gene encoding Phenylalanine ammonia lyase (PAL) (Table S1) was analysed. qPCR reaction was conducted as described earlier. Data normalization was performed with two most stable RGs identified for given dataset (individually and in combination) and with the least stable gene. The relative mRNA level was calculated according to the 2−∆∆Ct method48 with uninoculated samples being used as calibrator.
Results
Primer specificity and efficiency check
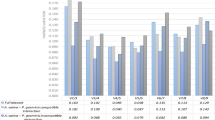
To identify the most stable reference genes combination, cDNA of tested oat genotypes was used in the qPCR. The primers specificity was estimated by qPCR melting curve analysis and further evaluated on 2% agarose gel. Single peaks were generated on the dissociation curves obtained for each primer pair thus confirming the specificity of the amplification. Moreover, single desired size bands were observed on agarose gel (Fig. S1–S2). No signal was detected in the no template control (NTC) and no reverse transcription control (NRT). PCR amplification efficiency ranged from 90.26% (EIF4A) to 128.7% (EP) and the R2 based on linear regression varied from 0.875 to 0.997. Primer pairs for ACT an EP did not meet the adopted effectiveness criteria (90% < E < 110%; R2 > 0.99), so they were excluded from the analysis (Table 2). The Tm values of the remining primer pairs varied from 83.8 °C (EF1A) to 89.5 °C (CYP), and the amplicon sizes were between 88(EIF4A) and 170 bp (EF1A) (Table S1). The raw quantification cycle (Cq) values were estimated for determination of the gene expression levels. The Cq values for analysed samples ranged between 21.37 and 32.29 (Fig. 1).
Cq values for nine candidate reference genes (ARF (ADP-ribosylation factor),CYP (cyclophilin), EF1A (elongation factor 1-alpha), EIF4A (eukaryotic initiation factor 4A-3), GAPDH (glyceraldehyde-3-phosphate dehydrogenase), HNR (heterogeneous nuclear ribonucleoprotein 27C), HSP70 (heat shock protein), TUA (alpha tubulin), and UBC (ubiquitin conjugating enzyme (E2)) across experimental samples. A line across the box is depicted as the median. The box indicates the 25th and 75th percentiles, the dots represent outlier values.
geNorm analysis
Candidate RGs were ranked by geNorm algorithm according to the stability value M. The cut-off threshold established by Vandesompele et al.47 for M value was set at 1.5, with lowest M values being exhibited by genes showing the highest expression stability. All tested genes in all analyzed datasets showed high expression stability (M < 0.7). For all samples analyzed in A. sativa–P. coronata interaction system ARF and HNR displayed the most stable expression, while CYP showed the least stable expression. Identical results were obtained for compatible interaction dataset. Incompatible interaction subgroup yielded contrary results with CYP being one of the most stable among the RGs tested. The best-performing RGs in Kasztan cultivar were EIF4A and HSP70, whereas most stable RGs found in Pc39 oat line were correspondingly ARF and HNR (Fig. 2).
NormFinder analysis
Stability value (SV) of each candidate RGs was estimated using NormFinder algorithm. RGs with low SV are considered best candidates for qPCR data normalization. Overall, HNR and EIF4A were two most stable RGs in analyzed pathosystem. When compatible and incompatible interactions were evaluated separately, ARF and EIF4A or ARF and HNR were best-scoring RGs pairs, respectively. The highest SVs representing high variation in expression were reported for GAPDH and CYP in the total dataset as well as in compatible interaction subgroup. Within incompatible interaction subgroup maximum variation was displayed by HSP70 and TUA (Fig. 3).
BestKeeper analysis
According to BestKeeper’s correlation coefficients (r) the most stably expressed gene in all experimental datasets (regardless of the samples subgroups being analyzed separately or together) was EF1A. High correlations were also obtained for HNR (total dataset and Kasztan samples set), EIF4A (incompatible subgroup and Pc39 samples set) and ARF (compatible subgroup). Out of all genes tested CYP was found to be the worst or second-worst performing RG (Fig. 4) in each analyzed samples set.
Correlation coefficients (r) of tested RGs determined by BestKeeper algorithm for full dataset, compatible interaction dataset, incompatible interaction dataset, Pc39 oat line dataset and Kasztan cultivar dataset. High coefficient of correlation indicates high expression stability across samples set.
ΔCt method
Based on average standard deviation (mean SD) generated by ΔCt method HNR and ARF were identified as the most stable RGs for the entire dataset. Similar results were obtained for incompatible interaction samples as well as for Pc39 oat line samples. For compatible interaction subgroup ARF and EF1A were found to be the least variable RGs, whereas across Kasztan cultivar samples the lowest mean SD was reported for HNR and EIF4A (Fig. 5).
Determination of the optimal number of RGs for data normalization
Pairwise variation (Vn/Vn+1) calculated by geNorm algorithm allows the prediction of optimal number of RGs that should be used for accurate qPCR data normalization. The Vn/Vn+1 value below 0.15 indicates that the inclusion of additional RG will not significantly improve reliability of data analysis. In terms of pairwise variation the V2/3 values calculated within all analyzed datasets were below 0.12 (Fig. 6). Consequently, regardless of the samples being analyzed together or in the various subgroups, only two best-performing RGs are required for sufficient data normalization.
Candidate RGs rankings generated by all tested algorithms are shown in Table 3. In the present study HNR and EF1A could be considered as most stable in tested plant-pathogen system. Their ranking positions, however, slightly differ depending on calculation method. High level of expression stability was also displayed by ARF. All algorithms conclusively pointed out to GAPDH and CYP as the least stable pair of RGs in combined samples dataset.
In general, ARF and EF1A were among top best ranked RGs in compatible interaction subgroup, whereas in incompatible interaction subgroup it was ARF in combination with EIF4A. The latter dataset produced most incongruent rankings with some RGs (such as EF1A or CYP) being ranked by various algorithms as either showing best, medium or even poor expression stability. Irrespective of this, HSP70 was shown by all assessment methods to be the worst candidate for normalization in this dataset.
Both HNR and ARF were indicated as most stable in Pc39 oat line samples. Nonetheless, in Kasztan cultivar samples HNR and EIF4A were found to be best candidates for internal controls. In this material, as opposed to other datasets, HSP70 was shown to exhibit relatively high expression stability.
Expression analysis of target gene for reference genes validation
Aiming to validate the reliability of the selected RGs, the expression analysis of PAL gene was performed in experimental samples harvested in various time points post inoculation. For each dataset, both best and worst performing RGs were used in order to demonstrate how incorrect data normalization may affect obtained results.
When normalization was carried out with most stable RGs (used either individually or in pair) consistent expression patterns were observed in every time point in all datasets (Fig. 7). However, when RGs showing poor stability were used, a clear overestimation of transcript level was noticed. This was especially true for samples collected 24 hpi. Results from full dataset normalized against best RGs (either HNR, EF1A or HNR + EF1A) show approximately twofold increase in transcript abundance. These results are distorted when CYP is used as RG, suggesting strong transcription upregulation (sevenfold increase). In consequence it shows how inaccurate and altered obtained expression profiles might be if RGs selection is not carried out properly.
Discussion
Studying complex plant resistance mechanisms is based on RNA sequencing (RNAseq) and transcriptome analysis using high-throughput next-generation sequencing approaches (NGS). Accurate quantification and validation of gene expression derived from in silico NGS data is performed by reverse transcription quantitative real-time polymerase chain reaction. This research aimed to identify reference genes with stable expression level in order to ensure obtaining repeatable and reliable RT-qPCR data.
Eleven frequently used candidate reference genes were evaluated in A. sativa during interactions with two different pathotypes of P. coronata in six time points post inoculation. The use of crown rust races with defined virulence profiles allowed to stimulate susceptibility as well as resistance response and observe compatible and incompatible interactions respectively. Previous research involving the selection of reference genes in the genus Avena enabled the use of specific primers for qPCR developed in these studies20,27. The design of the remaining primers was facilitated by the use of recently released A. sativa v2.0 genome (“Avena sativa—OT3098 v2, PepsiCo,” n.d.) and available, although significantly limited Avena transcriptomic data44.
In this study nine of eleven candidate reference genes were subjected to further analysis. Primer pairs for actin (ACT) and expressed protein (EP) were excluded for not reaching the efficiency criterion of 90 to 110%. Actin is one of the typically used housekeeping reference genes, however many studies testing larger panels of potential RGs eliminate this gene due to its low expression stability49. Yet, in the genus Avena ACT was chosen as suitable for qPCR data normalisationin A. fatuaand A. ludoviciana under herbicide treatment21,22. Nevertheless, under the biotic stress of Trichoderma polysporum infection in A. fatua, ACT did not perform very well50 in contrast toTBP (TATA-binding protein), 18S rRNA (18S ribosomal RNA) and UBC, that were the most stable internal reference genes in this pathosystem. Moreover, according to Wrzesińska et al.20 in the study of herbicide-resistant A. fatua biotypes TBP and GAPDH were chosen as the most stably expressed reference genes instead of ACT.
Remaining RGs expression stability was assessed with four algorithms, geNorm, NormFinder, BestKeeper and ΔCt method. For each algorithm, RGs were ranked from the most to least stable. Nonetheless, due to the different calculation methods, our data indicated a slight variation depending on the used algorithm.
GeNorm and NormFinder require relative quantities of Cq values (RQ). GeNorm calculates a stability measure (M-value) by the stepwise exclusion of the least stable gene and assesses the pairwise variation (Vn/Vn+1) between two sequential normalization factors that contain increasing numbers of genes47. The use of this algorithm allowed to determine the optimal number of reference gene combinations. Because the pairwise variation of V2/3 values for all of the experimental sets was lower than the cut-off threshold of 0.15, this number, in the case of the A. sativa–P. coronata pathosystem may be limited to two best-performing RGs. Similar results were obtained by Ruduś et al.23 in the study examining the best reference genes for molecular studies of dormancy in wild oat (A. fatua L.) caryopses. Based on the pairwise variation analysis, further confirmed by the validation experiment, two RGs may be sufficient for reliable normalization of the expression data. Moreover, the authors concluded that the use of two or three reference genes of high expression stability, performed better than a single gene, which is also in line with the MIQE guideline9.
In NormFinder analysis, intra- and intergroup variation within subgroups of a full dataset is calculated with ANOVA-based model. In this study experimental design allowed to divide the dataset into four subgroups: compatible reaction, incompatible reaction, Pc39 oat line and Kasztan cultivar dataset. However, in order to use ANOVA model, assumptions concerning homogeneity of variance and normality of data must be made51. In this analysis at the subgroup level, SV of all tested RGs was below the default limit of 0.5 indicating relatively high expression stability52. In the total dataset, the highest variation in expression was reported for GAPDH, CYP and UBC, however the value of SV above 0.5 was reported only for GAPDH. GAPDH was commonly used as an endogenous control in expression analysis in response to biotic53 and abiotic stress54,55 while many studies of potential RGs evaluation confirm its expression variability49. In the Triticum aestivum–Puccinia triticina pathosystem analyzed by Prasad et al.56,GAPDH was also the least stable of the five tested housekeeping genes along with 18S rRNA. The study was conducted on the wheat leaves during pre-haustorial stages of plant-pathogen compatible and incompatible interaction.
In contrast, for BestKeeper and ΔCt method, Cq values with efficiency corrections were used. BestKeeper calculates the geometric mean of Cq values and the most stable genes are indicated by high correlation coefficients and low standard deviations45. Simple ΔCt is based on comparisons between each RG and the other RGs within each sample and calculates the average standard deviation against the other RGs57. In this study, both methods indicate identical order for the five genes with the lowest expression stability (CYP, GAPDH, HSP70, TUA, UBC), whereas differences appear when a comparison of the most stable genes is made. BestKeeper ranks EF1A as one with the highest correlation coefficient, however the large standard deviation of the results for this gene should also be taken into account. The results of the ΔCt method place EF1A fourth out of nine in the stability ranking. According to this method, HNR and ARF were identified as the most stable RGs for the entire dataset.
In general, the most stable in the tested plant-pathogen system were HNR (heterogeneous nuclear ribonucleoprotein 27C) and EF1A (elongation factor 1-alpha). ARF (ADP-ribosylation factor) and EIF4A (eukaryotic initiation factor 4A-3) could also be considered as exhibiting high expression stability. Similar results were obtained by Yang et al.27, who evaluated the expression of eleven genes in A. sativa developing seeds and corresponding endosperm, as well as shoots and roots of seedlings. They have chosen EIF4A + HNR as the best performing candidate RG set across all tested samples and in developing endosperms. EF1A was among the best RGs for developing seeds. They have also pointed out GAPDH as the least stable RG which confirms this study results.
In this study, the worst candidate for normalization regardless of the assessment method was CYP (cyclophilin). CYP was tested as a candidate reference gene in the study of Tajti et al.43 validating RGs for studying different abiotic stresses in oat by RT-qPCR and was ranked as one of the least stable genes under drought stress, however had high stability in leaves under salt stress. CYP expression was also analyzed in the research of Wang et al.58 on Nitraria tangutorum seedlings under a series of experimental conditions and was ranked as one with the average stability. The least suitable as reference gene in the abovementioned experiment was HSP70, which in our study also had very low expression stability. This indicates the possible involvement of HSP70 in plant biotic interaction59.
The valdation of RGs carried out in this research through PAL expression analysis confirmed the need of using carefully chosen RGs for data normalization. The use of stable internal controls produced consistent expression patterns. It was also shown that resorting to unstable RGs could result in data misinterpretation.
To our best knowledge, this is the first report regarding reference genes selection in A. sativa–P. coronata interaction system. The obtained results will provide valuable data necessary for a comprehensive analysis of oat gene expression in response to crown rust infection. This may significantly contribute to the understanding of the complex plant resistance mechanisms involved in the host–pathogen interactions and expand knowledge of durable resistance strategies beneficial to modern oat breeding.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Stewart, D. & McDougall, G. Oat agriculture, cultivation and breeding targets: Implications for human nutrition and health. Br. J. Nutr. 112(Suppl), S50–S57 (2014).
Boczkowska, M., Podyma, W. & Łapiński, B. Oat. in Genetic and Genomic Resources for Grain Cereals Improvement (eds. Singh, M. & Upadhyaya, H. D.) 159–225 (Elsevier, 2016).. https://doi.org/10.1016/B978-0-12-802000-5.00004-6.
Chaves, M. S., Martinelli, J. A., Wesp, C. de L. & Graichen, F. A. S. The cereal rusts: An overview. Pest Technol. 2, 38–55 (2008).
Fetch, T. G., McCallum, B., Menzies, J., Rashid, K. & Tenuta, A. Rust diseases in Canada. PS&C 4, 86–96 (2011).
Sowa, S. & Paczos-Grzȩda, E. Virulence structure of Puccinia coronata f. sp. avenae and effectiveness of Pc resistance genes in Poland during 2017–2019. Phytopathology 111(7), 1158–1165 (2021).
Paczos-Grzȩda, E. & Sowa, S. Virulence structure and diversity of Puccinia coronata f. sp. avenae P. syd. & syd. in Poland during 2013 to 2015. Plant Dis. 103, 1559–1564 (2019).
Cabral, A. L. et al. Oat fungal diseases and the application of molecular marker technology for their control. in Future Challenges in Crop Protection Against Fungal Pathogens (eds. Goyal, A. & Manoharachary, C.) 343–358 (Springer Science+Business Media, 2014). https://doi.org/10.1007/978-1-4939-1188-2.
Nazareno, E. S. et al. Puccinia coronata f. sp. avenae: A threat to global oat production. Mol. Plant Pathol. 19, 1047–1060 (2018).
Bustin, S. A. et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622 (2009).
Kozera, B. & Rapacz, M. Reference genes in real-time PCR. J. Appl. Genet. 54, 391–406 (2013).
Thellin, O. et al. Housekeeping genes as internal standards: Use and limits. J. Biotechnol. 75, 291–295 (1999).
Schmittgen, T. D. & Livak, K. J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108 (2008).
Gutierrez, L. et al. The lack of a systematic validation of reference genes: A serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol. J. 6, 609–618 (2008).
Guenin, S. et al. Normalization of qRT-PCR data: The necessity of adopting a systematic, experimental conditions-specific, validation of references. J. Exp. Bot. 60, 487–493 (2009).
Jaiswal, P. S., Kaur, N. & Randhawa, G. S. Identification of reference genes for real-time PCR gene expression studies during seed development and under abiotic stresses in Cyamopsis tetragonoloba (L.) Taub. bioRxiv preprint (2018).
Saddhe, A. A., Malvankar, M. R. & Kumar, K. Selection of reference genes for quantitative real-time PCR analysis in halophytic plant Rhizophora apiculata. PeerJ 6, e5226 (2018).
Zhang, L. et al. Reference genes identification for normalization of qPCR under multiple stresses in Hordeum brevisubulatum. Plant Methods 14, 110 (2018).
Zhou, W. et al. Reference genes for qRT-PCR normalisation in different tissues, developmental stages, and stress conditions of Hypericum perforatum. PeerJ 7, e7133 (2019).
Cai, J. et al. Selection of appropriate reference genes for the detection of rhythmic gene expression via quantitative real-time PCR in Tibetan hulless barley. PLoS ONE 13, 1–19 (2018).
Wrzesińska, B., Kierzek, R. & Obrępalska-Stęplowska, A. Evaluation of six commonly used reference genes for gene expression studies in herbicide-resistant Avena fatua biotypes. Weed Res. 56, 284–292 (2016).
Liu, J. et al. Selection and evaluation of potential reference genes for gene expression analysis in Avena fatua. Plant Prot. Sci. 55, 61–71 (2019).
Akbarabadi, A., Ismaili, A., Kahrizi, D. & Firouzabadi, F. N. Validation of expression stability of reference genes in response to herbicide stress in wild oat (Avena ludoviciana). Cell. Mol. Biol. 64, 113–118 (2018).
Ruduś, I. & Kępczyński, J. Reference gene selection for molecular studies of dormancy in wild oat (Avena fatua L.) caryopses by RT-qPCR method. PLoS ONE 13, e0192343 (2018).
Bekele, W. A., Wight, C. P., Chao, S., Howarth, C. J. & Tinker, N. A. Haplotype based genotyping-by-sequencing in oat genome research. Plant Biotechnol. J. 16, 1452–1463 (2018).
Chaffin, A. S. et al. A consensus map in cultivated hexaploid oat reveals conserved grass synteny with substantial sub-genome rearrangement. Plant Genome 9(2), 1–35 (2016).
Zeng, L., Deng, R., Guo, Z., Yang, S. & Deng, X. Genome-wide identification and characterization of Glyceraldehyde-3-phosphate dehydrogenase genes family in wheat (Triticum aestivum). BMC Genomics 17, 1–10 (2016).
Yang, Z., Wang, K., Aziz, U., Zhao, C. & Zhang, M. Evaluation of duplicated reference genes for quantitative real-time PCR analysis in genome unknown hexaploid oat (Avena sativa L.). Plant Methods 16, 1–14 (2020).
Sowa, S. et al. Resistance to Puccinia coronata f. sp. avenae in Avena magna, A. murphyi, and A. insularis. Plant Dis. 100, 1184–1191 (2016).
Paczos-Grzęda, E., Sowa, S., Boczkowska, M. & Langdon, T. Detached leaf assays for resistance to crown rust reveal diversity within populations of Avena sterilis L. Plant Dis. 103, 832–840 (2018).
Paczos-Grzęda, E., Sowa, S., Koroluk, A. & Langdon, T. Characteristics of resistance to Puccinia coronata f. sp. avenae in Avena fatua. Plant Dis. 102, 1–9 (2018).
Paczos-Grzęda, E., Boczkowska, M., Sowa, S., Koroluk, A. & Toporowska, J. Hidden diversity of crown rust resistance within genebank resources of Avena sterilis L. Agronomy 11, 1–14 (2021).
Sowa, S. & Paczos-Grzęda, E. A study of crown rust resistance in historical and modern oat cultivars representing 120 years of Polish oat breeding. Euphytica 216, 1–10 (2020).
Carson, M. L. Virulence in oat crown rust (Puccinia coronata f. sp. avenae) in the United States from 2006 through 2009. Plant Dis. 95, 1528–1534 (2011).
Chong, J. et al. Virulence of Puccinia coronata f. sp. avenae in the eastern prairie region of Canada during 2007–2009. Can. J. Plant Pathol. 33, 77–87 (2011).
Menzies, J. G., Xue, A., Dueck, R. & Greunke, J. Virulence of Puccinia coronata f. sp. avenae in Canada; 2010 to 2014. In 14th International Cereal Rust and Powdery Mildew Conference 5–8 July 2015 95 (2015).
Fleischmann, G. & McKenzie, R. I. H. Inheritance of crown rust resistance in Avena sterilis L. Crop Sci. 8, 710–713 (1968).
Chong, J., Leonard, K. J. & Salmeron, J. J. A North American system of nomenclature for Puccinia coronata f. sp. avenae. Plant Dis. 84, 580–585 (2000).
Sowa, S. & Paczos-Grzęda, E. Virulence structure of Puccinia coronata f. sp. avenae and effectiveness of Pc resistance genes in Poland during 2017–2019. Phytopathology 111, 1158–1165 (2021).
Sowa, S. & Paczos-Grzęda, E. Puccinia coronata f. sp. avenae virulence in south-eastern Poland in 2014. Folia Pomer. Univ. Technol. Stetin. Agric. Aliment. Pisc. Zootech. 336(43), 157–166 (2017).
Hsam, S. L. K. et al. Genetic studies of powdery mildew resistance in common oat (Avena sativa L.) I. Cultivars and breeding lines grown in Western Europe and North America. Euphytica 96, 421–427 (1997).
Sozoniuk, M., Parzymies, M., Kozak, D. & Ismael, B. S. Carex muskingumensis and osmotic stress: Identification of reference genes for transcriptional profiling by RT-qPCR. Genes 11, 1–14 (2020).
Dudziak, K. et al. Identification of stable reference genes for qPCR studies in common wheat (Triticum aestivum L.) seedlings under short-term drought stress. Plant Methods 16, 1–8 (2020).
Tajti, J., Pál, M. & Janda, T. Validation of reference genes for studying different abiotic stresses in oat (Avena sativa L.) by RT-qPCR. Plants 10, 1–19 (2021).
Gutierrez-Gonzalez, J. J., Tu, Z. J. & Garvin, D. F. Analysis and annotation of the hexaploid oat seed transcriptome. BMC Genomics 14, 66 (2013).
Pfaffl, M. W., Tichopad, A., Prgomet, C. & Neuvians, T. P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 26, 509–515 (2004).
Andersen, C. L., Jensen, J. L. & Ørntoft, T. F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250 (2004).
Vandesompele, J. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, 1–12 (2002).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408 (2001).
Chapman, J. R. & Waldenström, J. With reference to reference genes: A systematic review of endogenous controls in gene expression studies. PLoS ONE 10, e0141853 (2015).
Zhu, H., Ma, Y. & Guo, Q. Expression stability of internal reference gene in response to Trichoderma polysporum infection in Avena fatua L. Curr. Genet. https://doi.org/10.1007/s00294-021-01200-4 (2021).
Brunner, A. M., Yakovlev, I. A. & Strauss, S. H. Validating internal controls for quantitative plant gene expression studies. BMC Plant Biol. 4, 1–7 (2004).
Rego, E. C. S. et al. Stable reference genes for RT-qPCR analysis of gene expression in the Musa acuminata-Pseudocercospora musae interaction. Sci. Rep. 9, 66 (2019).
Satapathy, L., Kumar, D., Kumar, M. & Mukhopadhyay, K. Functional and DNA–protein binding studies of WRKY transcription factors and their expression analysis in response to biotic and abiotic stress in wheat (Triticum aestivum L). 3 Biotech 8, 40 (2018).
Zhou, Y., Hu, L., Wu, H., Jiang, L. & Liu, S. Genome-wide identification and transcriptional expression analysis of cucumber superoxide dismutase (SOD) family in response to various abiotic stresses. Int. J. Genomics 2017, 66 (2017).
Wei, L. et al. Abscisic acid enhances tolerance of wheat seedlings to drought and regulates transcript levels of genes encoding ascorbate-glutathione biosynthesis. Front. Plant Sci. 6, 66 (2015).
Prasad, P. et al. Evaluation of candidate reference genes for normalization of RT-qPCR data in wheat during pre-haustorial stages of Puccinia triticina infection. Trop. Plant Pathol. 45, 96–101 (2020).
Silver, N., Best, S., Jiang, J. & Thein, S. L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 7, 1–9 (2006).
Wang, B. et al. Systematic selection and validation of suitable reference genes for quantitative real-time PCR normalization studies of gene expression in Nitraria tangutorum. Sci. Rep. 10, 1–10 (2020).
Berka, M., Kopecká, R., Berková, V., Brzobohatý, B. & Černý, M. Regulation of heat shock proteins 70 and their role in plant immunity. J. Exp. Bot. https://doi.org/10.1093/jxb/erab549 (2022).
Author information
Authors and Affiliations
Contributions
S.S. Conceptualization, experimentation, data curation, resource managing, formal analysis, supervision, writing original draft, review and editing. M.S. Methodology, formal analysis, review and editing. J.T. Experimentation. K.K. Review and editing. E.P.G. Resources, review and editing. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sowa, S., Sozoniuk, M., Toporowska, J. et al. Validation of reference genes as an internal control for studying Avena sativa–Puccinia coronata interaction by RT-qPCR. Sci Rep 12, 14601 (2022). https://doi.org/10.1038/s41598-022-18746-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-18746-z
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.