Abstract
We investigated gluteal (GSAT) and abdominal subcutaneous adipose tissue (ASAT) DNA methylation of FKBP5 in response to a 12-week intervention in African women with obesity, as well as the effect of the rs1360780 single nucleotide polymorphism (SNP) on FKBP5 methylation, gene expression and post-exercise training adaptations in obesity and metabolic related parameters. Exercise (n = 19) participants underwent 12-weeks of supervised aerobic and resistance training while controls (n = 12) continued their usual behaviours. FKBP5 methylation was measured in GSAT and ASAT using pyrosequencing. SNP and gene expression analyses were conducted using quantitative real-time PCR. Exercise training induced FKBP5 hypermethylation at two CpG dinucleotides within intron 7. When stratified based on the rs1360780 SNP, participants with the CT genotype displayed FKBP5 hypermethylation in GSAT (p < 0.05), and ASAT displayed in both CC and CT carriers. CC allele carriers displayed improved cardiorespiratory fitness, insulin sensitivity, gynoid fat mass, and waist circumference (p < 0.05) in response to exercise training, and these parameters were attenuated in women with the CT genotype. These findings provide a basis for future studies in larger cohorts, which should assess whether FKBP5 methylation and/or genetic variants such as the rs1360780 SNP could have a significant impact on responsiveness to exercise interventions.
Similar content being viewed by others
Introduction
The obesity pandemic has been largely driven by the consumption of unhealthy, western diets coupled with sedentary lifestyles1. These metabolic stressors have been hypothesized to contribute to obesity by disrupting the hypothalamic–pituitary–adrenal (HPA) axis, a neuro-endocrine pathway with essential roles in the stress response2, through a combination of genetic and epigenetic modifications in HPA-axis regulatory genes3.
FK506-binding protein (FKBP5) is a stress responsive co-chaperone of the glucocorticoid receptor (GR) and negative regulator of the HPA axis4. Its expression is rapidly induced by glucocorticoids, after which it attenuates GR signalling by restricting hormone binding and translocation of the GR complex to the nucleus, thus forming an ultra-short negative feedback loop5. Numerous genetic variants of FKBP5 have been identified. Of these, the rs1360780 (C/T) single nucleotide polymorphism (SNP), located in intron 2, is the most extensively studied and has been demonstrated to alter FKBP5 gene expression by modulating DNA methylation dynamics and transcription factor binding at multiple intronic enhancer regions within the FKBP5 gene6.
While most FKBP5 research has examined its relationship with neuropsychiatric diseases, several recent studies have uncovered a role for this gene in metabolic control and obesity7. Preclinical studies in animal models have shown that gene deletion or pharmacological inhibition of FKBP5 leads to reduced white adipose tissue mass, and protection from diet-induced weight gain, insulin resistance and hepatic steatosis8,9. In humans, FKBP5 expression in abdominal subcutaneous adipose tissue was shown to positively associate with markers of insulin resistance and type 2 diabetes10. Moreover, the FKBP5 rs1360780 SNP has been associated with increased susceptibility to metabolic dysregulation, type 2 diabetes as well as decreased weight loss following bariatric surgery11,12. FKBP5 is also epigenetically regulated and two independent studies, conducted in peripheral blood and subcutaneous adipose tissue, reported an association between intronic FKBP5 hypermethylation and several obesity and cardiometabolic traits13,14.
The excessive accumulation and dysfunction of white adipose tissue is considered a primary hallmark of obesity15. White adipose tissue is localized throughout the body in distinct depots which have been shown to vary in their association with metabolic diseases16,17. It is widely accepted that central adipose depots (including visceral and abdominal subcutaneous fat) are associated with greater risk of obesity-related complications, while lower-body fat (gluteo-femoral) is thought to be protective against metabolic disease18. Differences in these associations have however been reported in Black African and African American individuals19,20, thus highlighting the need for additional studies in these populations. Although the molecular basis for adipose depot-specific differences remain elusive, numerous studies have shown that abdominal and gluteal fat depots display distinct epigenetic and transcriptome profiles which may account for their functional differences and consequences for disease21,22.
Exercise training has been widely recognized as a favourable intervention to reduce the risks of developing obesity and metabolic disease23,24. Within adipose tissue, exercise training has been shown to reduce adipocyte size, modulate the expression of pro-inflammatory adipokines, decrease the induction of reactive oxygen species and inflammation, and increase vasculature; all processes that are dysregulated during obesity16,25,26. As a result, exercise has been consistently used as a model to assess the underlying mechanisms of insulin resistance and obesity27. To our knowledge, no studies have explored the genetic and epigenetic effects of exercise training on insulin resistance in Black African women with obesity. Accordingly, we firstly aimed to investigate abdominal and gluteal adipose tissue DNA methylation of FKBP5 in response to a 12-week exercise intervention in Black African women with obesity. Secondly, we examined the depot-specific effect of the rs1360780 SNP on (1) FKBP5 methylation, (2) FKBP5 gene expression and in (3) response to the exercise intervention, as measured by obesity and metabolic parameters.
Results
Participant characteristics and response to the intervention
The characteristics of participants in response to the intervention have been previously described27 and are summarised in Table 1. In response to 12-weeks of exercise training there was a significant increase in peak oxygen consumption (VO2peak, p < 0.01 for interaction), an indicator of cardiorespiratory fitness, as well as insulin sensitivity (p = 0.049 for interaction). A modest but significant decrease in waist circumference (WC) was observed in the exercise group post intervention, whilst it increased in the control group (p < 0.001 for interaction). The control group also displayed increased body mass index (BMI, p = 0.006 for interaction) and weight (p = 0.006 for interaction) post intervention. Furthermore, exercise participants displayed a decrease in gynoid fat mass (FM, p = 0.003 for interaction) and an increase in circulating triglycerides levels (p = 0.001 for interaction) in response to the intervention.
Impact of exercise intervention on FKBP5 methylation
We employed targeted bisulfite pyrosequencing to analyse the impact of exercise training on DNA methylation of two CpG sites situated within intron 7 of FKBP5 (Fig. 1a). These CpG sites (CpG542, 137,847 base pairs from the transcriptional start site and CpG543, 137,872 base pairs from the transcriptional start site) span experimentally validated glucocorticoid response elements28, and we and others have previously implicated methylation of these sites in metabolic disorders13,14,28. At baseline and following the 12-week intervention, CpG542/3 methylation was investigated in gluteal (GSAT) and abdominal subcutaneous adipose tissue (ASAT), as representatives of lower body and central obesity, respectively. Pyrosequencing analysis of GSAT samples in response to exercise training revealed that DNA methylation of CpG542 and CpG543 increased by 4% and 1.8%, respectively (Fig. 1b). However, these differences were only statistically significant for CpG542 (p = 0.019). In ASAT, methylation of both CpG542 and CpG543 increased by 9% in response to exercise training (Fig. 1b). In contrast, no significant differences in CpG542 or CpG543 methylation were observed in GSAT of control participants (Fig. 1c). While CpG542/3 methylation in ASAT tended to decrease post-intervention, these differences were not statistically significant (p > 0.05).
(a) Schematic diagram of the FKBP5 gene, illustrating the rs1360780 SNP and CpG542/3 sites in introns 2 and 7, respectively. Pyrosequencing analysis of percentage methylation at FKBP5 CpG542 and CpG543 in gluteal adipose tissue (GSAT) and abdominal adipose tissue (ASAT) in exercise (n = 19) (b) and control (n = 12) (c) participants pre- and post-intervention. Data represented as mean ± SD, *p < 0.05, ***p < 0.001.
FKBP5 methylation associations with obesity and metabolic parameters
We next assessed associations between change in response to the intervention (Δ) in FKBP5 CpG542 and CpG543 methylation and measures of cardiorespiratory fitness, insulin sensitivity and body composition. As shown in Fig. 2, Δ in GSAT CpG542 methylation negatively correlated with Δ cardiorespiratory fitness (VO2peak, rs = −0.498, p = 0.03) and Δ CpG543 methylation positively associated with Δ WC (rs = 0.578; p = 0.01) in response to exercise training (Fig. 2a, b). In ASAT, Δ CpG543 methylation positively associated with Δ low-density lipoprotein (LDL) cholesterol levels (rs = 0.498, p = 0.03) in response to exercise training (Fig. 2c). Importantly, these associations were not significant in the control group (Fig. 2d-f). To control for socio-demographic and behavioural factors29,30, further linear regression models were adjusted for significant covariates (alcohol consumption and employment). Adjusted models showed that the associations between Δ CpG543 methylation and Δ WC (β = 1.26, p = 0.02) and Δ LDL (β = 3.51, p = 0.05) were maintained; however, the association between ΔCpG542 methylation and Δ cardiorespiratory fitness lost statistical significance (β = −0.01, p = 0.10).
Spearman’s correlation analysis between changes in response to exercise training (Δ) in depot specific FKBP5 methylation and Δ VO2peak (a) and Δ waist circumference (WC) (b) and Δ low-density lipoprotein (LDL) levels (c) (n = 12–19 per group). Linear regression lines used for descriptive purposes only. GSAT, gluteal subcutaneous adipose tissue; ASAT, abdominal subcutaneous adipose tissue.
Effects of rs1360780 SNP on FKBP5 methylation and gene expression
The rs1360780 SNP, located in intron 2 of FKBP5, has previously been associated with metabolic risk12,31,32 and has been experimentally demonstrated to modulate glucocorticoid receptor-induced DNA methylation of FKBP5 at multiple intronic CpG regions, including those in intron 728. Based on this, we hypothesized that the rs1360780 SNP may influence FKBP5 methylation and accordingly, gene expression levels.
To explore this hypothesis, we genotyped all study participants for the presence of either the CC “protective allele” or the CT/TT “risk allele”. Using the combined sample of participants from the control and exercise groups (n = 30), we determined the genotype frequencies for the CC and CT alleles to be 0.57 (n = 17) and 0.43 (n = 13), respectively. For the exercise group, the genotype frequencies for the CC and CT alleles were 0.61 (n = 11) and 0.39 (n = 8), respectively. No participants were carriers of the rare homozygous TT genotype. We did not observe a significant deviation from the Hardy Weinberg equilibrium in the combined (p = 0.784) or exercise (p = 0.809) sample populations (data not shown). When participants from the exercise group were stratified based on the rs1360780 SNP genotype, those with the CT genotype presented with significantly higher GSAT CpG542 (p = 0.027) and CpG543 (p = 0.018) methylation in response to exercise training, whereas methylation of these CpG sites remained unchanged by exercise in individuals with the CC genotype (Fig. 3a). Conversely, in ASAT, both CC and CT genotypes displayed CpG542 and CpG543 hypermethylation after exercise training (Fig. 3b). No significant DNA methylation differences were observed in GSAT of either CC or CT control participants post intervention (Fig. 3c). While CpG542/3 methylation appeared to be lower in ASAT post exercise-training, these differences were not significant (Fig. 3d).
Genotype effect of rs1360780 on FKBP5 CpG542 and CpG543 methylation in gluteal adipose tissue (GSAT) and abdominal adipose tissue (ASAT) in exercise participants (CC n = 11, CT n = 7) (a, b) and GSAT and ASAT in control participants (CC n = 6, CT n = 6) (c, d), pre- and post-intervention. Data represented as mean ± SD. *p < 0.05, **p < 0.01.
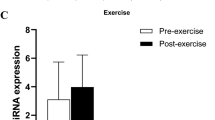
We next investigated FKBP5 messenger ribonucleic acid (mRNA) levels in GSAT and ASAT in response to exercise training, which was compared between rs1360780 SNP genotype groups. As shown in Fig. 4a, a modest but significant decrease in FKBP5 mRNA expression levels was observed in GSAT of individuals with the CC genotype after exercise training (p = 0.033), whilst individuals with the CT genotype displayed an apparent increase in FKBP5 expression, although these changes were not significant (p = 0.068). A comparison of FKBP5 mRNA expression in ASAT showed no significant differences between any of the groups in response to exercise training (Fig. 4b). In control participants, FKBP5 expression levels in GSAT increased slightly in individuals with the CT genotype, although this increase was not significant (p = 0.866) (Fig. S1). In CC participants, FKBP5 expression levels remained unchanged (Fig. S1).
Effect of rs1360780 SNP on post-exercise training parameters
Based on the results in Figs. 3 and 4, and to better describe and quantify the metabolic differences between CC and CT carriers, we reinvestigated the participant characteristics pre- and post-intervention (Table 1) after stratifying for the rs1360780 SNP. Figure 5 shows that CC carriers displayed significant improvements in cardiorespiratory fitness (Fig. 5a) and insulin sensitivity (Fig. 5b), and reduced gynoid FM % (Fig. 5c) and WC (Fig. 5d) resulting from the exercise intervention, while these parameters were attenuated in individuals with the CT genotype. Furthermore, an investigation of the stratified control group revealed that the increased BMI, weight, waist to hip ratio and waist circumference observed in this group after the 12-week intervention period, as reported in Table 1, were only significant in participants with the CT genotype (Fig. 5e–h). These findings were not influenced by any of the sociodemographic or behavioural factors recorded in the study, as these parameters were not statistically different between the stratified groups (data not shown).
Individual changes in VO2peak (a), waist circumference (WC) (b), gynoid fat mass % (FM) (c) and insulin sensitivity (SI) (d) in exercise group (n = 19, CC n = 11, CT n = 7) and body mass index (BMI) (e), weight (f), waist to hit ratio (WHR) (g) and WC (h) in control group (n = 12, CC n = 6, CT n = 6) pre- and post-intervention. Thick black line–median, shaded circles-CC genotype, empty circles–CT genotype. *p < 0.05, **p < 0.01, ***p < 0.001.
Discussion
A growing list of studies have implicated HPA axis dysregulation in obesity aetiology2,14,33,34. We present novel pilot data that shows exercise training induced genotype-specific hypermethylation of FKBP5, an important regulator of the HPA axis, in GSAT and ASAT from South African women with obesity. Importantly, we showed that the improvements in cardiorespiratory fitness, insulin sensitivity and adiposity were attenuated in carriers of the FKBP5 rs1360780 (C/T) SNP, although these findings need to be validated in a larger sample size.
Pyrosequencing analysis in this study revealed that 12 weeks of exercise training increased FKBP5 CpG542 methylation in GSAT and both CpG542 and CpG543 in ASAT. Interestingly, previously studies performed in our laboratory demonstrated that FKBP5 is hypermethylated in GSAT and ASAT of obese compared to normal-weight South African women13. While these findings seem contradictory to those reported here, it is important to note that the previous study was a cross-sectional comparison between FKBP5 methylation in normal weight compared to obese women, who did not undergo any intervention pre- and post- methylation analysis. In the current study, CpG542 methylation in GSAT associated with decreased cardiorespiratory fitness, an association which has, to the best of our knowledge, not been previously described. Moreover, depot specific FKBP5 CpG543 methylation was associated with increased WC (GSAT) and LDL levels (ASAT), which agrees with findings from a previous study by Ortiz and colleagues14, which showed that hypermethylation of FKBP5 within intron 2 in peripheral blood positively associated with WC and LDL in type 2 diabetic individuals. In line with this, we also previously reported a positive relationship between GSAT FKBP5 CpG542 and CpG543 methylation and WC in obese compared to normal-weight South African women13.
Studies on the FKBP5 functional rs1360780 SNP (C/T) have shown that the presence of the T allele may influence susceptibility to obesity and metabolic disease12. Indeed, carriers of the CT “risk allele” were shown to display higher insulin resistance, fasting insulin, circulating triglycerides and HOMA-IR values compared to wildtype CC carriers12. Furthermore, studies focusing on the role of the rs1360780 SNP in weight loss outcomes have shown that carriers of the T allele lose less weight in comparison to CC allele carriers following bariatric surgery31,35. In the current study, participants with the CC genotype showed significant improvements in cardiorespiratory fitness, insulin sensitivity and adiposity after exercise training, whilst these parameters were not significantly altered by exercise training in individuals with the CT genotype. Similarly, while the control group displayed significantly increased weight, BMI and waist circumference after the 12-week intervention period, stratification of these individuals according to the rs1360780 SNP revealed that these parameters were only significantly increased in CT carriers. Taken together, these finding agree with literature stating that CC carriers are more responsive to interventions associated with improvements in metabolic health compared to CT carriers, although we acknowledge that this requires validation in additional larger datasets12,31,35. Furthermore, although CT carriers did not display improvements in response to exercise training in the current study, we acknowledge that exercise may still be an appropriate intervention for the prevention of weight gain in these individuals.
Another important finding of the current study is that FKBP5 transcript levels were significantly downregulated by exercise in GSAT of CC allele carriers, while in contrast, CT carriers tended to display higher FKBP5 levels post-exercise intervention. We speculate that the gene expression changes for CT carriers may have not reached statistical significance due to the smaller sample size available for this genotype. Nonetheless, these findings agree with earlier reports showing that the rs1360780 (C/T) SNP is associated with higher FKBP5 induction by glucocorticoids36. Indeed, Klengel et al.36. showed that this is due to the T allele resulting in the formation of a putative TATA box, which interacts with the FKBP5 transcription start site through long-range chromatin interactions, thus enhancing FKBP5 expression36. The induction of FKBP5 in CT carriers observed here also agrees with previous preclinical studies showing that loss of FKBP5 in mice fed a high fat diet improves their metabolic health8, whilst higher FKBP5 levels in humans is associated with obesity and negative metabolic outcomes11. Importantly, while we did not observe any significant differences in baseline FKBP5 mRNA levels between CC and CT carriers, we note that other studies have similarly shown that FKBP5-mediated biological effects are evident only after the introduction of an environmental challenge, such as the exposure to a high fat diet4,8, bariatric surgery31,35 or psychological trauma and stress6,37,38. Interestingly, we did not observe any significant differences in FKBP5 gene expression in ASAT after exercise training. We suspect that this may be due to intrinsic transcriptome differences between GSAT and ASAT, as demonstrated in a previous study by Nono Nankam et al.22 which assessed these tissues from the same study population22.
The biological significance of FKBP5 gene expression changes in GSAT in response to exercise training was not experimentally explored in our study due to limited sample availability. However, it has been previously shown that FKBP5 knockdown in mature 3T3-L1 adipocytes reduced lipid accumulation and expression of adipogenic genes, and FKBP5 knockout in mice fed a high fat diet prevented white adipose tissue accumulation compared to wildtype controls7,9. This was mechanistically shown in the latter study to occur through FKBP5-mediated inhibition of the Akt-p38 MAPK pathway, which resulted in a reduction in the pro-lipolytic activity of GRα and promotion of lipogenesis by PPARγ, both of which are direct targets and reciprocally regulated by p38 MAPK phosphorylation7,9. According to this model, we propose that the reduction in FKBP5 in GSAT of CC participants in response to exercise training may have resulted in GRα activation via loss of Akt-p38 MAPK signaling, thus favouring and resulting in lipolysis of gynoid fat39. This in turn, may have contributed to the corresponding improvement in cardiorespiratory fitness and insulin sensitivity—a relationship previously demonstrated in this study population39. On the other hand, participants harbouring the rs1360780 (C/T) SNP retained elevated FKBP5 levels, and consequently, continued suppression of Akt-p38 MAPK signalling and sustained PPARγ-mediated adipogenic and lipogenic activity (Fig. 6). In the control group, suppression of the Akt-p38 MAPK pathway, and consequent adipogenic hyperactivity, may have been exacerbated in CT carriers, who continued their usual behaviours over the 12-week intervention period, and whose FKBP5 levels in GSAT appeared to increase compared to control participants harbouring the CC allele. We speculate that in this scenario, the rs1360780 (C/T) SNP may interact with an obesogenic environment to cause a genetic vulnerability to metabolic dysregulation, resulting in increased weight gain over time compared to CC carriers, as observed in this study (Fig. 6).
Schematic proposed model of study findings showing how exercise intervention can lead to intronic hypermethylation and downregulation of FKBP5 expression in individuals harbouring “protective” rs1360780 CC genotype. Reduced FKBP5 in turn may lead to re-activation of the Akt-p38 MAPK pathway, favouring GRα-induced lipolysis of gynoid fat mass (FM) and consequently improving insulin sensitivity and cardiorespiratory fitness. In contrast, individuals carrying the CT “risk” allele undergo sustained FKBP5 induction, causing a suppression of Akt-p38 MAPK signalling and promotion of lipogenesis through PPARγ activation. These molecular events lead to an attenuated response to exercise intervention. In the control group, the presence of the CT allele may result in an even greater suppression of the Akt-p38 MAPK pathway and adipogenic hyperactivity, resulting in increased weight gain over time compared to CC carriers. Diagram created in Bio-Render (https://biorender.com/).
Our study has several strengths. To the best of our knowledge, this is the first study investigating FKBP5 methylation, the rs1360780 SNP and gene expression changes in response to an exercise intervention in an exclusively African population of women with obesity. These findings are important since most available DNA methylation and transcriptome datasets have originated from studies performed in high-income countries and in European populations and may thus not be extrapolated to other nations with different ethnic and economic backgrounds. Furthermore, the current study investigated the impact of exercise on FKBP5 methylation in multiple adipose tissue depots, the primary affected organ during obesity development, compared to other studies which used peripheral blood14,31,35. As the participants in this study were advised to maintain their diet, no differences in energy consumption were found pre- and post-intervention, thus allowing for the exclusion of dietary influence on FKBP5 methylation. An additional strength of this study is the inclusion of both gluteal and abdominal subcutaneous adipose tissues in our analyses, thus enabling direct comparison between these depots and their associations with body composition and metabolic parameters.
Our study also has several limitations that need to be considered. Since the collection of adipose tissue biopsies is an invasive procedure, the study sample size is small27. Furthermore, due to a higher dropout rate in the control group, the sample sizes for the exercise and control groups were not equal and this is thus a limitation of the study27. Retrospective analysis of the statistical power for outcomes described in this pilot study, from which our main conclusions were drawn, showed that the power of these tests ranged between 60–90%. We therefore acknowledge that this study is underpowered to accurately assess the effects of FKBP5 methylation and the rs1360780 SNP on exercise outcomes. As a result, this study is a hypothesis generating pilot analysis, and future studies should investigate whether the associations reported here can be confirmed in additional, larger datasets. This study also only investigated an exercise intervention in South African women with obesity, and thus we cannot extrapolate our findings to men, or other ethnic populations. Finally, due to limited tissue availability, we could not measure FKBP5 protein levels in GSAT and ASAT, and therefore, cannot assess whether the changes in DNA methylation and gene expression were associated with functional FKBP5 protein levels.
In conclusion, we showed that a 12-week exercise intervention induced FKBP5 hypermethylation in a cohort of women with obesity. We found a significant relationship between FKBP5 methylation and measures of central obesity (WC), cardiorespiratory fitness (VO2peak) and hyperlipidaemia (LDL levels), which are important determinants of metabolic health. FKBP5 methylation, together with the effects of the rs1360780 SNP on the aforementioned parameters highlights the potential of dysregulated (epi)genetic processes to influence metabolic risk. These findings provide a basis for future studies in larger cohorts, which should assess whether FKBP5 methylation and/or genetic variants such as the rs1360780 SNP could have a significant impact on responsiveness to exercise interventions.
Methods
Study design
This is a pilot analyses of a larger randomized control trial (RCT), for which details of the study design, participant selection and methods have been described previously27. The study was approved by the UCT Human Research Ethics Committee (HREC REF: 054/2015) and was performed in accordance with principles of the Declaration of Helsinki (1964, amended in 2013). The trial was registered in the Pan African Clinical Trial Registry (Clinical trial number: PACTR201711002789113, registered on 21/11/2017). Written, informed consent was obtained from all participants prior to screening and participation. Sedentary, South African women with obesity were recruited through advertisements and selected based on the following inclusion criteria: (1) Black, South African women (based on parental Xhosa ancestry) between the ages of 20–35 years; (2) classified with obesity (BMI 30–40 kg/m2); (3) weight stable (no change in weight more than 5 kg/no change in clothing size over 6 months prior to selection); (4) sedentary (within the last 12 months had not participated in exercise training (more than 1 session lasting more than 20 min per week); (5) on injectable contraceptive (minimum 2 months; depot medroxyprogesterone acetate 400 mg); (6) no known metabolic/inflammatory disease; (7) no hypertension (≥ 140/90 mmHg), diabetes (random plasma glucose concentration > 11.1 mmol/l or HbA1C > 6.5%); (8) not currently on any medications; (9) non-smokers; (10) not currently pregnant/lactating; (11) no medical problems preventing participation in training; and (12) no surgical procedures 6 months prior to study. A total of 118 women were assessed for eligibility, of which 45 women partook in the study. Based on the availability of samples, 31 of these participants were included in our study, of which 12 and 19 participated in the control and exercise groups, respectively.
12-week intervention
The study participants were randomly divided into a control or experimental (exercise intervention) group. The control group was instructed to maintain their normal daily activities while the intervention group underwent 12 weeks of supervised aerobic/resistance training at moderate/vigorous intensity (60–70% maximal heart rate) for 40–60 min four times a week. Intensity of training was maintained throughout the study by adjusting exercise activity, which was monitored using heart rate monitors (Polar A300, Kempele, Finland). All participants were instructed to maintain their usual dietary habits throughout the duration of the study, which was assessed monthly as described previously39. All participants underwent pre- and post-testing; where anthropometric, metabolic and biochemical data were obtained, and adipose samples were collected, as previously described and detailed below27.
Pre and post intervention testing
Cardiorespiratory fitness
Peak oxygen consumption (VO2peak), a measure of cardiorespiratory fitness, was assessed as described previously27. Briefly, a walking treadmill-based (C, Quasar LE500CE, HP Cosmos, Nussdorf-Traunstein, Germany) graded exercise test was utilized, which involved an increase in treadmill gradient, followed by alternate increases in speed and gradient until volitional exhaustion was achieved. Pulmonary gas exchange was quantified by measuring the concentrations of O2 and CO2 and ventilation, from which VO2 consumption was calculated using a metabolic gas analysis system (CPET, Cosmed, Rome Italy).
Body composition
Basic anthropometric measurements (weight, height, waist and hip circumference) were measured. Whole body composition, including fat mass as well as regional body (gynoid and android) fat mass distribution, were assessed using dual-energy-x-ray absorptiometry (DXA) (Discovery-W, Software version 12.7.3.7; Hologic Inc., Medford, USA). MRI was used to determine visceral adipose tissue (VAT) and abdominal adipose tissue (SAT) using a 3 Tesla whole-body human MRI scanner (MAGNETOM Skyra; Siemens Medical Solutions). Region of interests (ROIs) were manually drawn using HOROS V 1.1.7 and volumes were determined by calculating the sum of the VAT and SAT areas from five images in a 15 cm region from the level of L1-5 and then multiplied by 340.
Frequently sampled intravenous glucose tolerance test
Following an overnight fast, blood samples were collected for the quantification of fasting glucose, insulin, triglycerides, total cholesterol, LDL and HDL. Thereafter, insulin sensitivity was calculated using an insulin-modified frequently sampled intravenous glucose tolerance test, as previously described41, from which the insulin sensitivity index (SI) was determined using Bergman’s minimal model of glucose kinetics42.
Biochemical analysis
Plasma glucose concentrations and serum lipids were measured using colorimetric assays (Randox, Gauteng, South Africa), and serum insulin was determined using immunochemiluminometric assays (IMMULITE 1000 immunoassay system, Siemens Healthcare, Midrand, South Africa). Plasma glucose and serum insulin concentrations were used to estimate insulin resistance, based on the homeostasis model of insulin resistance (HOMA-IR)43.
Adipose tissue biopsies
SAT biopsies (2–3 cm3) were obtained during both testing periods via mini liposuction following a 4–6 h fast44. Abdominal subcutaneous adipose tissue (ASAT) was obtained from the area directly above the umbilicus, whereas gluteal subcutaneous adipose tissue (GSAT) was obtained from the upper, right, outer quadrant of the gluteal region27.
DNA extraction
Genomic DNA from approximately 100 mg of GSAT and ASAT was isolated using the Qiagen DNeasy Blood & Tissue Kit (Qiagen, Valencia, USA) as per the manufacturer’s instructions. DNA purity and quantity were assessed using the Nanodrop Spectrophotometer (Nanodrop™ One, Thermo Fisher Scientific, Waltham, USA) and Qubit fluorometer (Life Technologies, CA, USA), respectively.
Bisulfite pyrosequencing
Bisulfite conversion and PCR amplification were carried out using the EpiTect Bisulfite Conversion kit (Qiagen, Hilden, Germany) and PyroMark PCR kit (Qiagen, Hilden, Germany), respectively, according to the manufacturer’s instructions (Qiagen, Hilden, Germany). FKBP5 (assay ID is ADS3828) primers were designed and purchased from EpigenDX (Worcester, MA, USA). The sequence of the primers was not disclosed by the company. The PCR reaction was carried out in the Veriti 96-well Thermal Cycler (ThermoFisher, Waltham, MA, USA) and the quality of amplicons was assessed by agarose gel electrophoresis. Amplicons were visualised using the ChemiDoc Imaging System (BioRad, Hercules, CA, USA). Pyrosequencing was conducted on the PyroMark Q96 using the PyroMark Gold Kit (Qiagen, Valencia, CA, USA), as previously described13. Each pyrosequencing run contained bisulfite conversion and PCR no template negative controls. Bisulfite conversion controls were also incorporated within each assay sequence to assess conversion efficiency. Assays were repeated if any of the inbuilt quality control measures failed. The nomenclature of the two FKBP5 CpGs are based on their sequencing identities from EpigenDX (Worcester, MA, USA).
FKBP5 genotyping
The FKBP5 rs1360780 (C/T) single nucleotide polymorphisms (SNPs) were genotyped as previously described13. Briefly, quantitative real-time PCR (qRT-PCR) was performed using Taqman genotyping assays (Applied Biosystems, Massachusetts, USA) on the QuantStudio™ 7 Flex Real-Time PCR System; with each reaction containing 9.4 ng of DNA, 5 µl of Taqman Master Mix and 0.25 µl of 40X TaqMan SNP genotyping assay in a total volume of 10 µl (Applied Biosystems, Massachusetts, USA). The PCR conditions were as follows: 10 min at 95 °C (initial denaturation/enzyme activation), 15 s at 95 °C (denaturation) and 60 s at 60 °C (annealing/extension) for 40 cycles. Eight randomly selected samples, genotyped in duplicate, were included as quality controls; with all plates including positive and negative controls.
RNA extraction, reverse transcription and quantitative real-time PCR
Total RNA was extracted from adipose tissue biopsies using the RNeasy/miRNeasy mini Kit (Qiagen, CA, United States) as previous described13, and assessed for quantity and purity using the NanoDrop ND-1000 Spectrophotometer (Agilent Technologies, Boeblingen, Germany). Complementary DNA (cDNA) was synthesized using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, CA, USA). Gene expression was quantified using TaqMan Universal PCR Master Mix and TaqMan gene expression assays (Applied Biosystems, CA, USA), according to the manufacturer’s instructions. Relative gene expression levels were calculated using the standard curve method, and FKBP5 expression levels were normalised to the RPLP0 endogenous control (identified as the best normalization gene using Normfinder)45.
Statistical analysis
This is a pilot analyses of a larger RCT, which has previously described the power analysis and sample size determination based on the primary outcomes relating to insulin sensitivity27. An adjustment for multiple testing was not considered due to the small sample size of this pilot analyses. Statistical analysis was conducted using GraphPad Prism (v.8.4.2) and STATA version 14.0 (StataCorp, College station, TX, USA). Data were assessed for normality using the Shapiro–Wilk test (p < 0.05) and are presented as the mean and standard deviation (SD) or the median and interquartile range (25th and 75th percentile) if normally or not normally distributed, respectively. Mixed-model analyses with main effects of time (pre- and post-intervention) and group (exercise and control) and interaction effects (group x time) were used to assess anthropometric, biochemical and metabolic parameters. DNA methylation and gene expression of FKBP5 were tested for significant changes pre- and post- exercise intervention using Paired Student T-Tests (normal distribution) or the non-parametric Wilcoxon Signed-Rank Tests (non-normal distribution), as appropriate. The correlation between the change (post-exercise training from baseline) in FKBP5 DNA methylation and metabolic parameters (cardiorespiratory fitness, body composition, glucose regulation and lipid profile) were determined using Pearson’s (normal distribution) or Spearman’s (non-normal distribution) correlation tests. Linear regression was used to explore any significant effects of social and behavioural factors on DNA methylation. Employment and alcohol consumption were determined to be significant covariates and were thus included in the adjusted model. Smoking was not considered as a covariate as no participants were current or previous smokers. Statistical significance was accepted as P < 0.05.
Ethics approval and consent to participate
This study was approved by the Research Ethics Committee of the Faculty of Health Sciences of the University of Cape Town and written informed consent to participate in the study was obtained from all subjects prior to participation in the study.
Data availability
The datasets generated and/or analysed in the current study are not publicly available as the study has only recently been completed. Data will be made publicly available after a two-year period; however, it can be made available from the corresponding author on reasonable request.
References
Kopp, W. How western diet and lifestyle drive the pandemic of obesity and civilization diseases. Diabetes Metab Syndrome Obesity: Targets Ther 12, 2221–2236. https://doi.org/10.2147/dmso.S216791 (2019).
Rutters, F. et al. The hypothalamic-pituitary-adrenal axis, obesity, and chronic stress exposure: foods and HPA axis. Curr. Obes. Rep. 1, 199–207. https://doi.org/10.1007/s13679-012-0024-9 (2012).
Argentieri, M. A., Nagarajan, S., Seddighzadeh, B., Baccarelli, A. A. & Shields, A. E. Epigenetic pathways in human disease: the impact of DNA methylation on stress-related pathogenesis and current challenges in biomarker development. EBioMedicine 18, 327–350. https://doi.org/10.1016/j.ebiom.2017.03.044 (2017).
Balsevich, G. et al. Interplay between diet-induced obesity and chronic stress in mice: potential role of FKBP51. J Endocrinol 222, 15–26. https://doi.org/10.1530/JOE-14-0129 (2014).
Zannas, A. S., Wiechmann, T., Gassen, N. C. & Binder, E. B. Gene-Stress-epigenetic regulation of FKBP5: clinical and translational implications. Neuropsychopharmacology 41, 261–274. https://doi.org/10.1038/npp.2015.235 (2016).
Binder, E. B. et al. Polymorphisms in FKBP5 are associated with increased recurrence of depressive episodes and rapid response to antidepressant treatment. Nat. Genet. 36, 1319–1325. https://doi.org/10.1038/ng1479 (2004).
Smedlund, K. B., Sanchez, E. R. & Hinds, T. D. Jr. FKBP51 and the molecular chaperoning of metabolism. Trends Endocrinol Metab 32, 862–874. https://doi.org/10.1016/j.tem.2021.08.003 (2021).
Balsevich, G. et al. Stress-responsive FKBP51 regulates AKT2-AS160 signaling and metabolic function. Nat Commun 8, 1725. https://doi.org/10.1038/s41467-017-01783-y (2017).
Stechschulte L.A., Q. B., Warrier M., Hinds T.D., Zhang M., Gu H. FKBP51 null mice are resistant to diet-induced obesity and the PPARγ agonist rosiglitazone. Endocrinology 157, 3888–3900 (2016).
Sidibeh, C. O. et al. FKBP5 expression in human adipose tissue: potential role in glucose and lipid metabolism, adipogenesis and type 2 diabetes. Endocrine 62, 116–128. https://doi.org/10.1007/s12020-018-1674-5 (2018).
Pereira, M. J. et al. FKBP5 expression in human adipose tissue increases following dexamethasone exposure and is associated with insulin resistance. Metabolism 63, 1198–1208. https://doi.org/10.1016/j.metabol.2014.05.015 (2014).
Fichna, M. et al. FKBP5 polymorphism is associated with insulin resistance in children and adolescents with obesity. Obes. Res. Clin. Pract. 12, 62–70. https://doi.org/10.1016/j.orcp.2016.11.007 (2018).
Willmer, T., Goedecke, J. H., Dias, S., Louw, J. & Pheiffer, C. DNA methylation of FKBP5 in South African women: associations with obesity and insulin resistance. Clin Epigenetics 12, 141. https://doi.org/10.1186/s13148-020-00932-3 (2020).
Ortiz, R., Joseph, J. J., Lee, R., Wand, G. S. & Golden, S. H. Type 2 diabetes and cardiometabolic risk may be associated with increase in DNA methylation of FKBP5. Clin Epigenetics 10, 82. https://doi.org/10.1186/s13148-018-0513-0 (2018).
Preis, S. R. et al. Abdominal subcutaneous and visceral adipose tissue and insulin resistance in the Framingham heart study. Obesity (Silver Spring) 18, 2191–2198. https://doi.org/10.1038/oby.2010.59 (2010).
Longo, M. et al. Adipose tissue dysfunction as determinant of obesity-associated metabolic complications. Int J Mol Sci 20, 2358. https://doi.org/10.3390/ijms20092358 (2019).
Pellegrinelli, V., Carobbio, S. & Vidal-Puig, A. Adipose tissue plasticity: how fat depots respond differently to pathophysiological cues. Diabetologia 59, 1075–1088. https://doi.org/10.1007/s00125-016-3933-4 (2016).
Kahn, B. B. & Flier, J. S. Obesity and insulin resistance. J. Clin. Investig. 106, 473–481. https://doi.org/10.1172/jci10842 (2000).
Goedecke, J. H. et al. Differential effects of abdominal adipose tissue distribution on insulin sensitivity in black and white South African women. Obesity (Silver Spring) 17, 1506–1512. https://doi.org/10.1038/oby.2009.73 (2009).
Goedecke, J. H. et al. Reduced gluteal expression of adipogenic and lipogenic genes in Black South African women is associated with obesity-related insulin resistance. J Clin Endocrinol Metab 96, E2029–E2033. https://doi.org/10.1210/jc.2011-1576 (2011).
Gehrke, S. et al. Epigenetic regulation of depot-specific gene expression in adipose tissue. PLoS ONE 8, e82516. https://doi.org/10.1371/journal.pone.0082516 (2013).
Nono Nankam, P. A. et al. Distinct abdominal and gluteal adipose tissue transcriptome signatures are altered by exercise training in African women with obesity. Sci Rep 10, 10240. https://doi.org/10.1038/s41598-020-66868-z (2020).
Hawley, J. A. & Lessard, S. J. Exercise training-induced improvements in insulin action. Acta Physiol (Oxf.) 192, 127–135. https://doi.org/10.1111/j.1748-1716.2007.01783.x (2008).
Sakurai, T. et al. The effects of exercise training on obesity-induced dysregulated expression of adipokines in white adipose tissue. Int J Endocrinol 2013, 801743. https://doi.org/10.1155/2013/801743 (2013).
Davegårdh, C. et al. Abnormal epigenetic changes during differentiation of human skeletal muscle stem cells from obese subjects. BMC Med. 15, 39. https://doi.org/10.1186/s12916-017-0792-x (2017).
Kolahdouzi, S., Talebi-Garakani, E., Hamidian, G. & Safarzade, A. Exercise training prevents high-fat diet-induced adipose tissue remodeling by promoting capillary density and macrophage polarization. Life Sci. 220, 32–43. https://doi.org/10.1016/j.lfs.2019.01.037 (2019).
Goedecke, J. H. et al. An exercise intervention to unravel the mechanisms underlying insulin resistance in a cohort of black South African Women: protocol for a randomized controlled trial and baseline characteristics of participants. JMIR Res Protoc 7, e75–e75. https://doi.org/10.2196/resprot.9098 (2018).
Yehuda, R. et al. Holocaust exposure induced intergenerational effects on FKBP5 methylation. Biol Psychiatry 80, 372–380. https://doi.org/10.1016/j.biopsych.2015.08.005 (2016).
Dogan, M. V., Lei, M. K., Beach, S. R., Brody, G. H. & Philibert, R. A. Alcohol and tobacco consumption alter hypothalamic pituitary adrenal axis DNA methylation. Psychoneuroendocrinology 66, 176–184. https://doi.org/10.1016/j.psyneuen.2016.01.018 (2016).
Lim, U. & Song, M. A. Dietary and lifestyle factors of DNA methylation. Methods Mol Biol 863, 359–376. https://doi.org/10.1007/978-1-61779-612-8_23 (2012).
Hartmann IB, Fries GR, Bücker J, Scotton E, von Diemen L, Kauer-Sant’Anna M. The FKBP5 polymorphism rs1360780 is associated with lower weight loss after bariatric surgery: 26 months of follow-up. Surg. Obes. Relat. Dis 12, 1554–1560 (2016).
Resmini, E. et al. Reduced DNA methylation of FKBP5 in Cushing’s syndrome. Endocrine 54, 768–777. https://doi.org/10.1007/s12020-016-1083-6 (2016).
Laryea, G., Schütz, G. & Muglia, L. J. Disrupting hypothalamic glucocorticoid receptors causes HPA axis hyperactivity and excess adiposity. Mol Endocrinol 27, 1655–1665. https://doi.org/10.1210/me.2013-1187 (2013).
Rosmond, R. & Bjorntorp, P. The hypothalamic-pituitary-adrenal axis activity as a predictor of cardiovascular disease, type 2 diabetes and stroke. J. Intern. Med. 247, 188–197. https://doi.org/10.1046/j.1365-2796.2000.00603.x (2000).
Peña, E. et al. Role of the FKBP5 polymorphism rs1360780, age, sex, and type of surgery in weight loss after bariatric surgery: a follow-up study. Surg. Obesity Related Dis. 16, 581–589. https://doi.org/10.1016/j.soard.2019.12.002 (2020).
Klengel, T. et al. Allele-specific FKBP5 DNA demethylation mediates gene-childhood trauma interactions. Nat Neurosci 16, 33–41. https://doi.org/10.1038/nn.3275 (2013).
Ising, M. et al. Polymorphisms in the FKBP5 gene region modulate recovery from psychosocial stress in healthy controls. Eur. J. Neurosci. 28, 389–398. https://doi.org/10.1111/j.1460-9568.2008.06332.x (2008).
Matosin, N., Halldorsdottir, T. & Binder, E. B. Understanding the molecular mechanisms underpinning gene by environment interactions in psychiatric disorders: the FKBP5 model. Biol Psychiatry 83, 821–830. https://doi.org/10.1016/j.biopsych.2018.01.021 (2018).
Clamp, L. D., Mendham, A. E., Kroff, J. & Goedecke, J. H. Higher baseline fat oxidation promotes gynoid fat mobilization in response to a 12-week exercise intervention in sedentary, obese black South African women. Appl. Physiol. Nutr. Metab. 45, 327–335, doi:https://doi.org/10.1139/apnm-2019-0460 (2020).
Fortuin-de Smidt, M. C. et al. Effect of exercise training on insulin sensitivity, hyperinsulinemia and ectopic fat in black South African women: a randomized controlled trial. European journal of endocrinology 183, 51–61, doi:https://doi.org/10.1530/eje-19-0957 (2020).
Goedecke, J. H. et al. Insulin response in relation to insulin sensitivity: an appropriate beta-cell response in black South African women. Diabetes Care 32, 860–865. https://doi.org/10.2337/dc08-2048 (2009).
Bergman, R. N., Bortolan, G., Cobelli, C. & Toffolo, G. Identification of a minimal model of glucose disappearance for estimating insulin sensitivity. IFAC Proc. Vol. 12, 883–890. https://doi.org/10.1016/S1474-6670(17)65505-8 (1979).
Matthews DR, H. J., Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 28, 412–419. (1985).
Evans, J. et al. Depot- and ethnic-specific differences in the relationship between adipose tissue inflammation and insulin sensitivity. Clin Endocrinol (Oxf) 74, 51–59. https://doi.org/10.1111/j.1365-2265.2010.03883.x (2011).
Vandesompele, J. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, Research0034, doi:https://doi.org/10.1186/gb-2002-3-7-research0034 (2002).
Acknowledgements
We thank the research volunteers for their participation in this study.
Funding
This work was supported by the South African Medical Research Council (SAMRC), the National Research Foundation professional development program (PDP) and (Grant number: 104987 awarded to TW), Thuthuka (Grant number: 129897 awarded to TW) and the International Atomic Energy agency. The content hereof is the sole responsibility of the authors and do not necessary represent the official views of the SAMRC or the funders.
Author information
Authors and Affiliations
Contributions
A.O. was responsible for laboratory work, data analysis and contributed to manuscript writing. T.W. contributed to the study design, laboratory work, data analysis, manuscript writing and manuscript revising. C.P. contributed to the study design, data analysis and corrected the manuscript. A.E.M. and J.G. were responsible for the initial study cohort design, exercise intervention, subject recruitment and sample/data collection. S.D. contributed to laboratory work. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Willmer, T., Oosthuizen, A., Dias, S. et al. A pilot investigation of genetic and epigenetic variation of FKBP5 and response to exercise intervention in African women with obesity. Sci Rep 12, 11771 (2022). https://doi.org/10.1038/s41598-022-15678-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-15678-6
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.