Abstract
Our research explores serum extracellular circulating miRNAs (ecmiRNAs) involved in dog stress response immediately after the search and rescue (SAR) of missing people. The experimental plan considers four arduous SAR simulations. The SAR dogs are trained by the Alpine School of the Military Force of Guardia di Finanza (Passo Rolle, Italy). The First SAR Trial analyzed dog serum samples at rest time (T0), and immediately after SAR performance (T1) using the miRNome-wide screening next-generation sequencing (NGS). T1 versus T0 NGS results revealed a different expression level of let-7a and let-7f. Subsequently, in a large sample size including: 1st (n = 6), 2nd (n = 6), 3rd (n = 6), and 4th (n = 4) trials, let-7a and let-7f were validated by qPCR. Bioinformatics analysis with TarBase (v.8) and the Diana-mirPath (v.3) revealed a functional role of let-7a and let-7f in the p53 pathway to restore cellular homeostasis. Let-7a and let-7f, highly expressed at T1, could stop MDMs-p53 inhibition inducing the p53 increase in level. In addition, let-7a and let-7f, via p53 post-transcriptional regulation, buffers p53 transcription spikes. During SAR stress, the possibility of p53 preconditioning could explain the phenomenon of “stress hardening” where the tolerance of particular stress increases after preconditioning.
Similar content being viewed by others
Introduction
Stress is essential for existence. Physiological stress responses are an adaptive system to cope with environmental changes, but a metabolic imbalance can be incurred if stress severity increases. The best strategies for animal stress counteracting rely on early diagnosis of stress-induced harms1.
Recent research suggests that stress is implicated in sensory biology2,3. Olfactory detection is a process exhibiting plasticity4, and a part of this plasticity depends on exposure to environmental stress conditions. Stressful experiences lead to alterations in dog olfaction affecting their performance when scent-based work is required.
Dogs are macrosmatic animals recruited for smell-based work in detecting cancer cells5, viral infections6, explosives7, drugs8, or missing people9. Search and rescue dogs are valuable after natural disasters, mass-casualty incidents, and for locating missing people. SAR dogs explore a large open-space, particularly shaped terrain in chaos and various weather conditions. Though SAR dogs must have an excellent physical status and are trained to work with an apt mental focus, SAR actions cause extreme physical and mental exhaustion10,11 in the dog. When the dog searches, he focuses on the smell of missing people, and while mild stress helps him focus on the goal, prolonged or too intense stress could negatively affect his performance or compromise his health. Bearing in mind that detection dog applications continue to grow, knowledge of the molecular mechanism regulating SAR dog stress to improve dog training and protect dog well-being is urgently needed.
In recent years, advances in epigenetic research, concerning heritable changes in environment-related gene expression that do not represent variations in the DNA sequence, have identified a new class of molecules known as microRNAs (miRNAs). MiRNAs are small single-stranded non-coding RNAs of approximately 22 nucleotides that can regulate gene expression in the post-transcriptional stage through interaction with target mRNAs, leading to translational inhibition or gene silencing. MiRNAs seem to have a high impact on stress signaling to restore homeostasis in sudden environmental changes, de facto stress conditions, altering miRNA biogenesis, modifying the expression of miRNA targets, and buffering transcription peaks12. MiRNAs bind to their target mRNAs and down-regulate translation thereby inhibiting protein production. Each miRNA can potentially regulate several hundred mRNAs, and tens of miRNAs may regulate each targeted mRNA.
Extracellular miRNAs (ecmiRNAs) secreted in body fluids, including blood plasma, urine, saliva, and semen, are excellent biomarkers. EcmiRNAs may mediate paracrine and endocrine communications between tissues and modulate distal cell survival and differentiation13. EcmiRNAs are exported and imported from cells and travel in biofluids protected by extracellular vesicles (EV)14 or bound to proteins (Argonaute-2 or high-density lipoproteins). Exosomes and microvesicles15 are the main EVs that contribute to miRNA transfer to facilitate intercellular communication with function as protective packaging for the delivery of controlled concentrations of miRNAs and guide targeting towards their destination. Exosome-mediated miRNA delivery is the most endogenous transport generated under physiological or pathological conditions.
EcmiRNAs were correlated with differences in lifestyle activities, such as exercise and environmental stressors, suggesting that ecmiRNAs may serve as circulating signatures of metabolic homeostasis16. Of late, ecmiRNAs significantly impacted medical research generating new molecular diagnostics hopes as biomarkers for homeostatic imbalance and disease states due to their abundance, cell-type specificity, and stability in most solid and liquid clinical specimens17.
Our research aim was to examine the SAR dog ecmiRNAs abilities to regulate homeostatic response to SAR stress. We planned a high-stress SAR simulation that required many of the skills dogs utilize when searching for missing persons after a natural disaster. The working dogs enrolled in this study belong to the Italian Military Forces of the Guardia di Finanza (GdF) and possess skills acquired during high professional training courses. Becoming a SAR dog requires immense expertise, and not every dog is up to this task. The NGS strategy was used to sequence dog serum miRNome at rest time and after SAR simulation.
We believe few observations on the role of ecmiRNAs as regulators of working dog stress are found in the literature. Nonetheless, in view of this powerful molecular approach, we have pioneered the diagnostics of working dog stress through the quantitative detection of minimally invasive ecmiRNA biomarkers that could improve training in the working dog as well as dog well-being.
Methods
Study approval was provided by the Research Ethics Committee of the University of Perugia (report n.2018-21 of 11/12/2018) according to Italian Ministry of Health legislation18. All methods were carried out following relevant guidelines and regulations and the study was carried out in compliance with the ARRIVE guidelines. Informed consent is not required as no human subjects were included in the study.
Animal enrolment
The dogs enrolled in the study were tested physically and behaviorally18. The day before SAR trials, all dogs were tested for routine health control by analyzing some blood biochemical parameters. The experiment included the dogs refer to the respective table (SM 1. SAR dog characteristics). Dog-handler teams were all specialized in the avalanche, surface, and rubble search at least by specific and increasing challenging training courses at the GdF Dog Breeding and Training Centre18 (Castiglione del Lago, Perugia, Italy) and the SAR Alpine School (Passo Rolle, Trento, Italy). All SAR dogs lived and worked with their handlers all year round to strengthen the dog-handler relationship, one of the most essential requirements for the success of a rescue operation.
SAR trial
The simulated SAR trial required dogs to find a target odor (a hidden operator and his breath, simulating a missing person) on a rubble field (30 × 35 m), within a maximum time of 15 min. Before starting SAR trials dog handlers ignored where the operator was hidden. During the search, both dog and the handler were allowed to enter the field area. Simulated SAR performance included some potential stress factors, such as the presence of people working on the rubble area, shouting and making loud noises with hammer and shovel, and sources of smoke to simulate a real post-earthquake scenery. The SAR trial is considered successful when the handler raises his arm to communicate that his dog has signaled the target odor. If the dog does not find the hidden operator within the assigned time, the SAR trial is considered a failure. To safeguard the welfare of the SAR dogs, during all trials, the authors observed if the dogs showed signs of stress or anxiety according to the codes described in our previous studies19,20. The results of these observations were only used for practical monitoring of the level of stress during the field search and interrupting it if necessary.
Sample collections time and experimental flowchart
Time points
All dog measures were evaluated in two experimental time points: T0—basal value at rest, immediately before the SAR trial; T1—immediately after the SAR trial. The points T0 and T1 allowed a comparison before and after the SAR trial.
Experimental flowchart
The research plan was divided into two experimental blocks; NGS SAR Trial and QPCR SAR Cluster, and both experimental blocks included T0 and T1 sampling times. NGS SAR Trial comprehended the 1st SAR trial, while QPCR SAR Cluster comprehended four SAR trials: 1st, 2nd, 3rd, and 4th. The four trials were held in different sessions, with different GdF dog-handler teams but the same experimental. The four trials included 22 dogs distributed respectively: 6 in the 1st, 6 in the 2nd, 6 in the 3rd, and 4 in the 4th Trial (see SM 1. SAR dog characteristics).
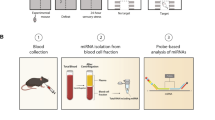
NGS SAR Trial investigation was planned to evaluate dog serum miRNome and identify differently expressed ecmiRNAs between T0 and T1. QPCR SAR Cluster validated in large sample size, including the 1st, 2nd, 3rd, and 4th trials, ecmiRNAs with different expression profiles between T0 and T1 (according to the data obtained from NGS SAR Trial) (Fig. 1).
Research flowchart. The upper part of figure (a) shows the six dogs included in the NGS SAR Trial. The serum of six dogs was collected at two experimental time points (T0 and T1), RNA was extracted and processed for next-generation sequencing miRNome. The figure below (b) shows the second part of the research plan referred to as QPCR SAR Cluster. This part of the study includes blood dog sampling at T0 and T1 of the 2nd, 3rd, and 4th SAR trials. Serum RNA was extracted, reverse transcribed, and only the microRNAs differently expressed, detected in NGS SAR Trial, were validated by qPCR. The figure is created by G. Guelfi using PowerPoint 2021, Microsoft Corporation, USA.
The day before the SAR, blood parameters were evaluated in all dogs included in the research. At experimental times, T0 and T1, heart rate, rectal temperature, serum cortisol level, and differential levels of serum microRNA expression were evaluated. The performance of the SAR dogs was examined throughout the SAR operation.
Physiological parameter
The GdF veterinarian monitored heart rate (HR) in beats per minute with a stethoscope (3 M LITTMANN, Classic II SE, Milano, Italia) and measured rectal body temperature with a digital thermometer (Reckitt Benckiser SPA, MB Termo 7126500, Milano, Italia) at T0 and T1, in each SAR dog belonging to First SAR Trial, and QPCR SAR Cluster.
Blood sampling and serum RNA purification
Blood sampling was taken following the routine health check protocol in the Gdf training program. During the GdF veterinarian procedures, the handler asked the dog to stand and stay still for 1 min while gently manipulating and distracting it. Next, the handler asked the dog to sit; simultaneously, the veterinarian collected the 3 mL of blood sample via the radial vein into Vacuette Z Serum Sep Clot Activator (GREINER BIO-ONE). After centrifugation (2000 × g, 10 min), serum was obtained and stored at − 80 °C until use. Hemolysis was controlled in all serum samples to prevent the release of microRNA contained in the blood cells altered ecmiRNA profile (SM 2. Hemolysis assessment during sample preparations). Each serum sample was separated into two collection tubes: one tube (300 µL) for analyzing biochemical parameters and one (200 µL) to explore differential miRNA expression profiles. Total RNA, including ecmiRNAs, was extracted from 200 µL of serum using the miRNeasy Serum/Plasma Kit (QIAGEN CLC bio, Aarhus, Denmark) according to the manufacturer’s instructions, with an elution volume of 14 µL (SM 3. RNA extraction and Spike-in for qPCR validations) and then stored at − 80 °C until use. MiRNA concentration was assessed using the Qubit Fluorometer 4 and the Qubit microRNA Assay Kit (THERMOFISHER SCIENTIFIC, Kandel, Germany).
Spike-in to monitor RNA extraction, cDNA, and qPCR technical quality
Spike-in was used to perform quality control (QC) of ecmiRNA from biofluid samples. The QC assessment is essential because it enables obtaining sensitive and reliable microRNA data from low RNA content samples. During sample preparation, the spike-in control oligonucleotide was added to observe the sample QC of the NGS SAR Trial and the QPCR SAR Cluster (Fig. 2). To assess the technical reproducibility and linearity of the mapped NGS reads, in the samples targeted for NGS, 1 µl of 52 QIAseq miRNA Library QC Spike-in (QIAGEN CLC bio, Aarhus, Denmark) was added during extraction, as suggested by QIAGEN NGS Services proprietary protocol. Whereas, in the QPCR SAR Cluster samples, the spike-ins UniSp2 and UniSp4 were added during the RNA extraction phase to assess the efficiency and yield of the RNA isolation as recommended by the manufacturer. UniSp6 spike-in was included in reverse-transcription reaction to monitor cDNA synthesis performance and check the presence of PCR inhibitors.
Quality control (QC) workflow. The NGS QC (left panel) was performed through a panel of 52 Spike-ins. The 52 RNA spike-in mix was added during RNA isolation to identify contamination and measure and validate assay parameters. In QPCR QC (right panel), 2 spike-in controls were added during RNA extraction to monitor the yield of RNA isolation, and 1 spike-in control was added to cDNA synthesis to monitor the efficiency of RT reactions. In addition, 3 spike-ins control PCR amplification. Spike-in synthetic oligonucleotides fragments were amplified and quantified by QPCR thanks to the respective primer pair. The figure is created by G. Guelfi using PowerPoint 2021, Microsoft Corporation, USA.
MiRNA library preparation
EcmiRNA sequencing experiments and data analysis were conducted by QIAGEN Genomic Services (Hilden, Germany). Library preparation is the first step of next-generation sequencing. Once libraries were prepared, the following workflow step will be high-throughput sequencing. The RNA used in this step was extracted (as described in SM 1) by adding to the serum 1 μL of a 52 miRNA Library QC Spike-In mix (QIAGEN CLC bio, Aarhus, Denmark) (SM 4. NGS serum miRNA Spike-in). The library preparation was done using the QIAseq miRNA Library Kit. A total of 5 µl RNA was converted into miRNA NGS libraries. Adapters containing UMIs (Unique Molecular Identifiers) were ligated to the RNA. Then RNA was converted to cDNA. The cDNA was amplified using qPCR (22 cycles), and during the qPCR, indices were added. After qPCR, the samples were purified. Library preparation was quality controlled using capillary electrophoresis (Agilent DNA 1000 Chip). Based on the quality of the inserts and the concentration measurements, the libraries were pooled in equimolar ratios. The library pools were quantified using qPCR. The library pool was then sequenced on a NextSeq (Illumina) to obtain 19 M 1 × 75 bp reads. Raw data was de-multiplexed, and FASTQ files for each sample were generated using the bcl2fastq (Illumina).
Next-generation sequencing
NGS Data analysis was performed via QIAGEN CLC Genomics Server v20.0.4 (Hilden, Germany). The miRNA-seq counts were normalized by a TMM (trimmed mean of M values) method21 to calculate the effective library sizes, which were then used as part of the per-sample normalization. Adapters containing Unique Molecular Identifiers (UMIs) were ligated to the RNA, and the RNA was then converted to cDNA. The 12 nt UMIs between the linker sequence and Illumina adapter sequence were extracted from each sample-demultiplexed sequencing reads, and the corresponding UMI annotated the reads. After removing all artificial sequences, we discarded reads with < 15 nt or > 55 nt. Reads with the same or 1 nt differences in their UMIs were grouped as UMI grouped reads, and reads from each UMI group were merged to produce a consensus sequence representing a single molecule from the starting RNA sample. These consensus reads were then aligned to miRBase v22 with a maximum mismatch of 2 nt, and the unmapped reads were subsequently mapped to the human genome GRCh38 (ENSEMBL).
Quantitative validation of ecmiRNAs by RT-qPCR
RT-qPCR reactions were performed to validate the expression profiling of selected miRNAs following the NGS results filtered with max group means more than 10, FDR p-value less than 0.05, and standard p-value significance at < 0.05.
A first-strand cDNA synthesis reaction was conducted using miRCURY LNA RT Kit (QIAGEN CLC bio, Aarhus, Denmark). Purified RNA (10 ng) was reverse-transcribed, including artificial RNA Spike-in control UniSp6. RT reaction was carried out according to the manufacturer’s guidelines (SM 5. Reverse transcription reaction). QPCR amplifications were executed utilizing miRCURY LNA SYBR Green PCR Kits (QIAGEN CLC bio, Aarhus, Denmark), in a final volume of 20 μL according to the manufacturer’s recommendations (SM 6. QPCR ecmiRNA validations). QPCR reaction was performed with 3 μL cDNA (diluted 1:50), and 9 specific pair primers (QIAGEN CLC bio, Aarhus, Denmark): 3 kinds of spike-in control (SM 7. Spike-ins Quality control); 4 kinds of potentials endogenous control (EC) of serum samples (SM 8. Selection of potential EC miRNAs); and 2 target miRNAs. The 9 pair primers are listed in Table 1. The amplification was performed in the StepOnePlus real-Time PCR system (APPLIED BIOSYSTEMS, California, USA). Three technical replicates were performed for each biological sample, and the average Cq (Quantification cycle according to the MIQE guidelines22) the averaged values of triplicate Cq were calculated; no-template controls were included in the analysis check contamination. The StepOnePlus Real-Time PCR software plotted the fluorescence intensity against the number of cycles and provided the Cq value using the automatic method. The 2^-ΔCq method was used to calculate the relative expression of the target miRNAs23.
Analysis of reference miRNA stability
MiRNA qPCR analysis requires data normalization with the best EC to minimize data variation that can mask or exaggerate biological changes. Among the analyzed EC miRNAs: miR-320, miR-148a, miR-24, miR-23a, the best ECs were estimated thanks to four algorithms: GeNorm, Normfinder, BestKeeper, and the comparative method ΔCt (Cq is the recent nomenclature of Ct); RefFinder integration tool was then used to compare and integrate the four algorithms results.
Statistical analysis
RT-qPCR reactions were performed to validate the expression profiling of selected miRNAs following the NGS results filtered with max group means higher than 10, FDR p-value less than 0.05, and standard p-value significance at < 0.05. The quantification and statistical tests used were described in the figure legends and the methods section. No data were excluded from our studies. GraphPad Prism 8 (GraphPad Software Inc.) was used to plot all of the graphs and calculate statistical significance using a two-tailed Student’s t-test.
Results
Biochemical blood parameters and physiological data
All blood biochemical parameters are included within the canine reference range values (SM 9. Biochemical blood parameters). These results allow us to exclude a pathological state that limits the enrollment of some subjects in the study. The stress levels were monitored by measuring SAR dog cortisol blood level; all dogs showed at T1 higher cortisol levels than at T0 (140.33 ± 4.52 ng/mL and 80.61 ± 1.38 ng/mL, respectively). At T0, the mean heart rate (70–155 bpm) and rectal temperature (38.6–39.1 °C) were within the normality range.
SAR dog performance evaluations
All trained dogs detected the presence of a missing person and his breath within the maximum time allowed (15 min). No significant differences were found in detection time between the trials.
Serum RNA quality and quantity evaluation
The mean RNA integrity number (RIN) was 8.9 (range 8.5–9.0), the 260/280 ratio was 1.9 (range 1.88–2.00), and the 260/230 ratio was 2.1 (range 2.00–2.20) indicating the high purity of RNA preparation in every sample. Total RNA yield was not significantly different among samples. RNA concentrations ranged from 9 to 12 ng/µL. Minimal variations in total RNA content were corrected during reverse transcription using fixed RNA input.
MiRNA-next generation sequencing data
NGS evaluated miRNA differential expression profiles at two different times: before SAR stress (T0) and after SAR stress (T1); the authors declare that all relevant data supporting the NGS findings are provided as a supplemental file (see the supplemental file—NGS data analysis). Differential expression analysis of ecmiRNAs revealed that only cfa-let-7f and cfa-let-7a have significantly upregulated expression levels at T1 versus T0 (Table 2).
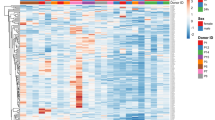
The Volcano Plot (Fig. 3) was applied to identify differentially expressed ecmiRNAs with statistical significance. The heatmap (Fig. 4) showed the graphical representation of the fifteen miRNAs with the highest FDR p-value (T1 versus T0).
Volcano plot. The figure shows the relationship between the magnitude of the difference in expression values (fold change) and statistical p-values. The lower difference in expression, the nearer the point will be to the x-axis. The more significant the difference, the smaller the p-value, and the higher -log10 p-value. Thus, points for features with highly significant differences will lie high in the plot. The cfa-let-7f (0.000002 p-value) and cfa-let-7a in the plot (red) represent the differentially expressed ecmiRNAs with statistical significance (0.000253 p-value).
Heat map. Fifteen miRNAs with the highest FDR p-value across samples (T1 versus T0) were selected. Each row represents one miRNA, and each column represents one sample. The color represents the difference of the count value to the row mean. They are also called centered values. Red indicates high relative expression, and green indicates low relative expression.
Two-level control strategy of qPCR data
When the goal is to identify the expression level accurately, technical variability sources should be reduced. For this reason, we have adopted a two-level control strategy of qPCR data analysis: controlling the quality of the sample (hit 1) and identifying the best EC for data normalization (hit 2). By inspecting the amplification curves of UniSp 2, UniSp 4, UniSp 6, it was possible to identify and remove the outliers (Cq > 35), deriving from an incorrect technical procedure. Only two samples showed higher UniSp2 (Cq = 38), suggesting a problem in one of the RNA isolation procedure steps but repeating the isolation, the two samples yielded higher-purity RNA (hit 1).
According to MIQE guidelines, we normalized qPCR data with the most stable EC miRNA to reduce experimental variability linked to the different quantities of the starting RNA. The best EC miRNA was identified using Delta Ct, BestKeeper, NormFinder, and GeNorm mathematical approaches. Subsequently, the data were analyzed using the four algorithms with RefFinder, which assigned a final ranking, identifying the best EC for qPCR data in the miR-320 (SM 10. Bioinformatics approaches for qPCR Endogenous miRNA selection). Target ecmiRNAs let-7a and let-7f Cq were normalized as follows: 2^ − ΔCq = 2^ − (Cq miRNA target – Cq miR-320) (hit 2) (Fig. 5).
Control of qPCR data. In all serum sample, qPCR reaction was performed by amplifying 9 targets: let-7a and let-7f differentially expressed miRNAs, UniSp 2, 4, 6 Spike-in, and the EC miR-191, miR-423a, miR-93, and miR-425. UniSp2, UniSp4, UniSp6 enable confident interpretation, first data control (1 red). Spike-ins were compared, and the outlier was detected for sample exclusion or re-isolation. UniSp Cq was interpreted as described by QIAGEN in “Guidelines for Profiling Biofluid miRNA” (summarized in the table). The second data control was miRNAs normalization with the most stable miRNA EC (identified as mir-320). The figure is created by G. Guelfi using PowerPoint 2021, Microsoft Corporation, USA.
QPCR validation of let-7a and let-7f ecmiRNAs
The differential NGS miRNAs of First Trial dogs were chosen after removing the miRNAs that did not meet the NGS selection criteria (max group means higher than 10, FDR p-value less than 0.05, and standard p-value significance at < 0.05), and which had a low read count (≤ 10 reads) in paired samples. Then the p-value and FDR value of miRNA reads were calculated, and the ecmiRNAs with FDR < 0.05 were selected. At last, we obtained the relevant ecmiRNAs with onefold or more change. So, NGS revealed two ecmiRNAs: cfa-let-7a (FDR p-value < 0.05; applying Bonferroni correction 0.054397) and cfa-let-7f (FDR p-value < 0.001 applying Bonferroni correction 0.000493) significantly upregulated after SAR-related stress (T1) compared to the expression levels of rest time (T0). The NGS results of cfa-let-7a and cfa-let-7f were validated by qPCR in n = 22 including: First Trial (n = 6), 2nd (n = 6), 3rd (n = 6), and 4th (n = 4) Trial. The qPCR normalized expression values (2^ − ΔCq) of let-7a and let-7f ecmiRNAs showed a significant increase at T1 (immediately after SAR trial) compared to T0 (p ≤ 0.001) (Fig. 6).
EcmiRNAs expression levels. The normalized expression values (y-axis) of the let-7a and let-7f ecmiRNAs were calculated with the 2^-ΔCq. The let-7a and let-7f graph show a white box (T0; 22 dog serum samples) and a grey box (T1; 22 dog serum samples). In the box plots, the thick central line represents the median; the top and bottom lines of the box represent the third quartile and the first quartile; whiskers indicate the variability in the data outside the upper and lower quartile. The figure shows the statistical significance of the overlap between the two groups determined using a two-tailed Student’s t-test; ***p ≤ 0.001.
In silico microRNA target and pathway prediction
MiRNA target prediction and functional analysis of let-7a and let-7f were performed with TarBase (v.8) and Diana-mirPath tool (v.3) implemented in the Kyoto Encyclopedia of Genes and Genomes (KEGG24,25,26), and StarBase v2.027 revealing that these ecmiRNAs were involved in cell growth, proliferation, death, and cell survival (FDR ≤ 0.05, Fisher’s test). Our attention was focused on the p53 pathway (let-7a and let-7f ecmiRNAs union Diana-mirPath merge results showed p-value 0.0027432, and 23 gene interactions). The combinatorial effect of let-7a and let-7f in the p53 pathway is illustrated in Fig. 7. Let-7a and let-7f block the production of proteins transcribed by the target MDM2 and MDM4 and TP53 genes. The NMDs non-translation removes NMD-p53 inhibition restoring the p53 pathway. We like to underline that by merging in Diana-mirPath let-7a and let-7b, all the two ecmiRNAs have as mRNA targets: MDM2, MDM4, and TP53 (highlighted in orange in Fig. 7) (SM 11. Diana MiRPath and P53 KEGG pathway analysis).
Pathway and targets prediction. The figure shows the associations between let-7a and let-7f ecmiRNAs and their putative p53 signaling pathway (p-value 0.0027432, and 23 gene interactions). The server offers two-gene labeling levels: yellow (gene targeted by 1 selected miRNA, let-7a), orange (gene targeted by 2 selected ecmiRNAs, let-7a and let-7f). Green color indicates genes that are not targeted by let-7a and let-7f Pathways analysis was performed using DIANA-miRPath v3.
Discussion
Dogs are able to identify individuals by scent, although the exact molecular process is not fully understood. Dog variability in olfactory detection threshold is the product of genetic factors28 (olfactory receptor polymorphisms), physiological factors (hormone levels), life environment history (previous exposure to odorants, specific training29), and stress levels30. Recent research suggests that stress affects several aspects of olfactory detection30; odor perception especially is a process exhibiting plasticity4 altered by various endogenous signaling stress modulation. In light of this new evidence, our research hypothesizes that the SAR working dog may adapt the expression levels of ecmiRNAs as an epigenetic mechanism to balance SAR performance stress. SAR dogs are trained to find missing people, and their work can be time-consuming and complicated by endogenous and exogenous environmental stress factors. Dogs store these stressors inadvertently as they stay focused on their goal.
Based on dog serum next-generation sequencing, the first part of our study revealed let-7a and let-7f as ecmiRNAs capable of significantly differentiating dogs at rest and dogs immediately after SAR performance. In the second part of the study, involving a larger sample size of SAR dogs, we performed qPCR validation of let-7a and let-7f serum ecmiRNAs. The stringent filtering methods to improve the NGS data quality and the accurate qPCR data analysis allowed us to emphasize how let-7a and let-7f are involved in SAR stress response. Furthermore, we can underlie how from let-7a and let-7f ecmiRNA serum expression levels, it is possible to discriminate dogs that have performed SAR from those at rest. Concerning SAR stress, we focused our attention on ecmiRNAs expression profiling because, in humans, it was proven that in conditions of the stress response, cells restore their gene expression patterns modulating miRNA levels31. During unfavorable conditions, in an attempt to adapt to a reversible period of stress, a cell controls gene expression by post-transcriptional regulation32. Recent studies using genome-wide approaches and single-cell transcription measurements posit that in eukaryotic cells, miRNAs are ideal for buffering transcript surges during stress to restore gene expression programs via epigenetic regulation33. MiRNAs could ensure a steady gene expression level unless the stress signal is sustained long enough to increase the transcripts over a certain threshold12. For outlined reasons in recent years, there has been an enormous interest in investigating circulating miRNAs in blood plasma/serum as potential biomarkers for early diagnoses. Mori et al. (2019)16 suggests that ecmiRNAs may serve as circulating indicators of the physiological status and a tool for precision medicine targeted treatment.
Our research evaluated stress-mediated ecmiRNA levels as part of a natural physiological process, recovered in a short time, that occurs in SAR dogs. When SAR dogs spring into action, endogenous and exogenous stressors SAR-related temporarily affect canine physiological and behavioral status. Dog stress symptoms vary widely, including changes in the relationship with the handler, failure to perform work-related tasks, depression, and general signs of fear and anxiety34. Over the past decade, many open field tests have been conducted to identify the relationship between stress levels and the attitudinal and behavioral changes of the working dog. We are convinced that focusing on the molecular mechanism would help improve dog welfare, perform more efficient SAR, and probably identify a biomarker. This research is the first to investigate the implication of serum ecmiRNA in SAR performance stress modulation. However, a limitation of this study is not being able to provide detailed information on the quality and reproducibility assessment of NGS because, although accurately assessed, they are considered proprietary to the sequencing company. Our experimental study demonstrates that the physiological response to SAR stress through modulation of let-7a and let-7f levels contribute to the homeostatic recovery mechanism. Given the lack of knowledge on dog miRNA profiling and the high degree of evolutionary conservation of many miRNAs, in this scientific contribution, we disentangle the potential role of let-7a and let-7f in SAR stress based on scientific observations in humans35 and on in silico prediction of miRNA target and pathway. DIANA-miRPath v3.0 targets and pathway analysis implemented in the KEGG database showed that most of the let-7a and let-7f target genes were identified in the p53 stress signaling pathway. A crucial protein in the stress response is p53; the alteration of the TP53 gene or post-translational modification in the p53 protein can alter the response to cellular stress36. Canine p53 family members, like their human counterparts, are expressed at low levels under no stress conditions and rapidly upregulated under a stress condition37.
The p53-linked stress-sensing mechanism is activated by various intrinsic and extrinsic stress signals to promote cell cycle arrest and repair if stress damage is not severe38. Scientific research over the past decade suggests that the significant p53 stress response negative regulators are MDM2 and MDM4 homologs. In non-stress conditions, MDM2 and MDM4 inhibit p53 activity. Conversely, in a state of stress, when a p53 response is required to protect the cell, MDM2 and MDM4 stop p53 inhibition. In silico prediction of let-7a and let-7f targets revealed that these ecmiRNAs are involved in MDM2 and MDM4 post-transcriptional regulation. The upregulation of let-7a and let-7f, blocking MDM2 and MDM4, stops MDM-p53 inhibition inducing pro-stress signaling39,40. Let-7a and let-7f stop MDM-p53 inhibition and p53 increase in level depending on the degree of stress and preconditioning.
Post-translational modification of MDM2 to regulate the MDM2-p53 signaling axis to govern p53 activities in mammalian cellular stress was previously described by Malonia et al. (2015), which invites pharmaceutical control of the p53 pathway to protect against cellular stress41. On the other hand, it is proved that p53 can induce transcription of MDM2, generating a negative feedback loop42.
In addition, let-7a and let-7f, via p53 post-transcriptional regulation, buffering p53 transcription spikes. Therefore, dog expertise could represent an environmental pretreatment that through preconditioning of p53, creates a cellular need-based gain that helps the dog during a stressful SAR (Fig. 8).
We suppose that SAR training developed the dog skills to exert a stress-protective epigenetic preconditioning mechanism by adapting the let-7a and let-7f miRNA levels. Epigenetic regulation is the basis of long-term changes, not written in the genome but directly associated with the environment. We assume the theory that let-7a and let-7f upregulation rapidly set the p53 preconditioning by triggering physiological stress reactions. The induction of miRNA expression upon stress might partly explain the phenomenon of “stress hardening”. Leung et al.12 and Kültz43 describe the physiological processes of miRNA upregulation under stress, to explain the phenomenon of “stress hardening” where the tolerance of same as previous increases after preconditioning. It should be emphasized that the long half-lives of ecmiRNAs, enable long-lasting gene regulation12. In the manuscript, we identify the SAR trial as a single stressor without distinguishing the importance of the components of the stressor. As Kültz argues, the cellular stress response is not stress-specific because a cell reacts to stress in response to the macromolecular damage sustained. Very few cellular responses directed at re-establishing homeostasis are stressor-specific43.
This research represents biomolecular advancement in understanding how working dogs adapt to SAR stress; it is not an in vitro functional study. Only reliable computational prediction information on the probable target genes and pathways involved in post-transcriptional let-7a let-7f regulation can be added. We preferred to evaluate single aspects, leaving further hypotheses to future works that will surely deepen comprehension of the functional role of let-7a and let-7f ecmiRNAs in physiological stress conditions concerning time recovery and performance complexity. In our opinion, it is fundamental to recognize the stressor outcome in working dogs to understand the behavioral consequence. It is critical to avoid that dogs30.
Concluding remarks
NGS analysis and in silico prediction of SAR dog serum suggest that the let-7a and let-7f upregulation stop MDM-p53 inhibition, thus inducing p53 pro-stress signaling. The let-7a and let-7f upregulation could operate a double regulation, one inducing p53 pro-stress signaling activation and the other, in opposition, buffering p53 transcription spike. The balance of these two opposing molecular signals could mediate p53 preconditioning to the recovery of the homeostasis. We believe that SAR dogs, thanks to enriched environment experience of constant training, are able to control p53 preconditioning allowing behavior control and physiological stress recovery in arduous SAR performances. We hope this study will provide the cue to suggest interest in further research to improve dog well-being and dog SAR performance and, maybe explore the use of a circulating saliva miRNA as a non-invasive stress biomarker. This research is to be considered a first step in gaining further knowledge on how to build resilience in working dogs and better preserve their welfare.
References
Dhama, K. et al. Biomarkers in stress related diseases/disorders: diagnostic, prognostic, and therapeutic values. Front. Mol. Biosci. 6, 91 (2019).
Manella, L. C., Alperin, S. & Linster, C. Stressors impair odor recognition memory via an olfactory bulb-dependent noradrenergic mechanism. Front. Integr. Neurosci. 7, 97 (2013).
Wojtaś, J., Karpiński, M. & Zieliński, D. Salivary cortisol levels in search and rescue (SAR) dogs under rescue examination conditions. J. Vet. Behav. 42, 11–15 (2021).
Wilson, D. A., Best, A. R. & Sullivan, R. M. Plasticity in the olfactory system: Lessons for the neurobiology of memory. Neuroscientist 10, 513–524 (2004).
Pirrone, F. & Albertini, M. Olfactory detection of cancer by trained sniffer dogs: A systematic review of the literature. J. Vet. Behav. 19, 105–117 (2017).
Else, H. Can dogs smell COVID? Here’s what the science says. Nature 587, 530–531 (2020).
Furton, K. The scientific foundation and efficacy of the use of canines as chemical detectors for explosives. Talanta 54, 487–500 (2001).
Menchetti, L. et al. Benefits of dietary supplements on the physical fitness of German shepherd dogs during a drug detection training course. PLoS ONE 14, e0218275 (2019).
Diverio, S. et al. A simulated avalanche search and rescue mission induces temporary physiological and behavioural changes in military dogs. Physiol. Behav. 163, 193–202 (2016).
Slensky, K. A., Drobatz, K. J., Downend, A. B. & Otto, C. M. Deployment morbidity among search-and-rescue dogs used after the September 11, 2001, terrorist attacks. J. Am. Vet. Med. Assoc. 225, 868–873 (2004).
Ahrens, F., Knies, K., Schneider, M., Köhler, F. & Erhard, M. H. Influence of different training and outdoor conditions on plasma histamine and cortisol concentrations in search-and-rescue dogs. Inflamm. Res. 54, S34–S35 (2005).
Leung, A. K. L. & Sharp, P. A. MicroRNA functions in stress responses. Mol. Cell 40, 205–215 (2010).
Sohel, M. H. Extracellular/circulating microRNAs: Release mechanisms, functions and challenges. Achieve Life Sci. 10, 175–186 (2016).
Weber, J. A. et al. The microRNA spectrum in 12 body fluids. Clin. Chem. 56, 1733–1741 (2010).
Boilard, E. Extracellular vesicles and their content in bioactive lipid mediators: More than a sack of microRNA. J. Lipid Res. 59, 2037–2046 (2018).
Mori, M. A., Ludwig, R. G., Garcia-Martin, R., Brandão, B. B. & Kahn, C. R. Extracellular miRNAs: From biomarkers to mediators of physiology and disease. Cell Metab. 30, 656–673 (2019).
Gustafson, D., Tyryshkin, K. & Renwick, N. microRNA-guided diagnostics in clinical samples. Best Pract. Res. Clin. Endocrinol. Metab. 30, 563–575 (2016).
Guelfi, G. et al. A cross-talk between blood-cell neuroplasticity-related genes and environmental enrichment in working dogs. Sci. Rep. 9, 6910 (2019).
Diverio, S. et al. Dogs’ coping styles and dog-handler relationships influence avalanche search team performance. Appl. Anim. Behav. Sci. 191, 67–77 (2017).
Diverio, S. et al. Non-invasive assessment of animal exercise stress: Real-time PCR of GLUT4, COX2, SOD1 and HSP70 in avalanche military dog saliva. Animal 9, 104–109 (2015).
Robinson, M. D. & Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25 (2010).
Bustin, S. A. et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622 (2009).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001).
Kanehisa, M. & Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Vol. 4.
Kanehisa, M., Furumichi, M., Sato, Y., Ishiguro-Watanabe, M. & Tanabe, M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 49, D545–D551 (2021).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951 (2019).
Li, J.-H., Liu, S., Zhou, H., Qu, L.-H. & Yang, J.-H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein–RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 42, D92–D97 (2014).
Ignatieva, E. V., Levitsky, V. G., Yudin, N. S., Moshkin, M. P. & Kolchanov, N. A. Genetic basis of olfactory cognition: extremely high level of DNA sequence polymorphism in promoter regions of the human olfactory receptor genes revealed using the 1000 Genomes Project dataset. Front. Psychol. 5, 247 (2014).
Jenkins, E. K., DeChant, M. T. & Perry, E. B. When the nose doesn’t know: Canine olfactory function associated with health, management, and potential links to microbiota. Front. Vet. Sci. 5, 56 (2018).
Bombail, V. Perception and emotions: On the relationships between stress and olfaction. Appl. Anim. Behav. Sci. 212, 98–108 (2019).
Mendell, J. T. & Olson, E. N. MicroRNAs in stress signaling and human disease. Cell 148, 1172–1187 (2012).
Kucherenko, M. M. & Shcherbata, H. R. miRNA targeting and alternative splicing in the stress response – events hosted by membrane-less compartments. J. Cell Sci. 131, jcs202002 (2018).
Raj, A. & van Oudenaarden, A. Nature, nurture, or chance: Stochastic gene expression and its consequences. Cell 135, 216–226 (2008).
Bergeron, S. Modified open-field test with odor search stimulus: Anticipated canine motivation and behavioral outcomes between anxious and non-anxious dogs. (2019).
Lee, H., Han, S., Kwon, C. S. & Lee, D. Biogenesis and regulation of the let-7 miRNAs and their functional implications. Protein Cell 7, 100–113 (2016).
Perwez Hussain, S. & Harris, C. C. p53 biological network: At the crossroads of the cellular-stress response pathway and molecular carcinogenesis. J. Nippon Med. Sch. 73, 54–64 (2006).
Zhang, J., Chen, X., Kent, M. S., Rodriguez, C. O. & Chen, X. Establishment of a dog model for the p53 family pathway and identification of a novel isoform of p21 cyclin-dependent kinase inhibitor. Mol. Cancer Res. 7, 67–78 (2009).
Holmberg Olausson, K., Nistér, M. & Lindström, M. p53-dependent and -independent nucleolar stress responses. Cells 1, 774–798 (2012).
Carr, M. I. & Jones, S. N. Regulation of the Mdm2-p53 signaling axis in the DNA damage response and tumorigenesis. Transl. Cancer Res. 5, 707–724 (2016).
Liu, J., Zhang, C., Zhao, Y. & Feng, Z. MicroRNA control of p53: MicroRNAs and p53. J. Cell. Biochem. 118, 7–14 (2017).
Malonia, S. K., Dutta, P., Santra, M. K. & Green, M. R. F-box protein FBXO31 directs degradation of MDM2 to facilitate p53-mediated growth arrest following genotoxic stress. Proc. Natl. Acad. Sci. 112, 8632–8637 (2015).
Huun, J. et al. The functional roles of the MDM2 splice variants P2-MDM2-10 and MDM2-∆5 in breast cancer cells. Transl. Oncol. 10, 806–817 (2017).
Kültz, D. Molecular and evolutionary basis of the cellular stress response. Annu. Rev. Physiol. 67, 225–257 (2005).
Acknowledgements
We would like to extend our sincere gratitude and appreciation for all of the work and dedication provided by the military GdF personnel Walter Di Mari, Massimo Teodoro, Michele Matteo Santoro, Stefano Murari, Sergio Lancerin, Giorgia Rossi, Alessandro Alberioli, Valter Levis and the SAGF rescue detection dogs and their trainers Davide Buzzo, Filippo Cagnati, Simone Catalini, Angelo Centi, Piero Chatrian, Jody Conti, Michele D'andrea, Simone Di Cataldo, Francesco D’Urso, Michele Gaio, Damiano Fontanive, Giovanni Gatti, Nicola Leo, Andrea Marana, Raffaele Marazzi, Norbert Niederkofler, Daniele Ollier, Nereo Ongaro, Simone Pizzi, Giuseppe Randine, Marco Rinaldo, Lucia Rocchi, Maurizio Simoni, Marco Terroni, Oscar Verban, Cristian Venneri, Vito Ventre, Nicola Zarbo, Gerry De Zolt, Agostino Zortea, Alfredo Zus for their invaluable assistance and outstanding contributions. Without their collaboration this work could not be possible. Medical writing assistance was provided by Diana Cella, writer on CLA University of Perugia We gratefully acknowledge the Fondazione Cassa di Risparmio di Perugia for supporting our scientific research.
Funding
This study was funded by Fondazione Cassa di Risparmio di Perugia, COD. PROG.: 2017.0210.021.
Author information
Authors and Affiliations
Contributions
S.D., G.G. and M.M.S. conceived and planned the experiments. S.D., M.I., A.S., and G.G. carried out the experiments. G.G. and C.C. contributed to bioinformatics analysis and figure design. The manuscript was drafted by G.G. and S.D. All authors discussed the results and contributed to the final manuscript. Funding from Fondazione Cassa di Risparmio di Perugia COD. PROG.: 2017.0210.021 was obtained by S.D. The sponsor had no role in study design, collection, analysis, interpretation of data, or writing the manuscript and the decision to submit for publication.
Corresponding authors
Ethics declarations
Competing interests
The authors declare that the research was conducted in the absence of any financial and non-financial competing of interests that could be construed as a potential conflict of interest, including specific interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Guelfi, G., Iaboni, M., Sansone, A. et al. Extracellular circulating miRNAs as stress-related signature to search and rescue dogs. Sci Rep 12, 3213 (2022). https://doi.org/10.1038/s41598-022-07131-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-07131-5
This article is cited by
-
Effects of fecal microbial transplantation on police performance and transportation stress in Kunming police dogs
Applied Microbiology and Biotechnology (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.