Abstract
The lamprey represents the oldest group of living vertebrates and has been a key organism in various research fields such as evolutionary developmental biology and neuroscience. However, no knock-in technique for this animal has been established yet, preventing application of advanced genetic techniques. Here, we report efficient generation of F0 knock-in lampreys by CRISPR-Cas9-mediated genome editing. A donor plasmid containing a heat-shock promoter was co-injected with a short guide RNA (sgRNA) for genome digestion, a sgRNA for donor plasmid digestion, and Cas9 mRNA. Targeting different genetic loci, we succeeded in generating knock-in lampreys expressing photoconvertible protein Dendra2 as well as those expressing EGFP. With its simplicity, design flexibility, and high efficiency, we propose that the present method has great versatility for various experimental uses in lamprey research and that it can also be applied to other “non-model” organisms.
Similar content being viewed by others
Introduction
The lamprey belongs to the basal-most group of vertebrates, the cyclostomes, and retains ancestral characteristics, such as lack of a jaw and paired fins. Consequently, this animal has attracted attention from a wide range of researchers, including evolutionary developmental biologists and neuroscientists1,2,3,4,5,6. However, its long, complex life cycle7 prevents establishment of genetic lines, and thus it had been practically impossible to apply modern genetic techniques to the lamprey until quite recently. Now, F0 knock-out lamprey mutants can be generated effectively using the CRISPR-Cas9 system for the sea lamprey Petromyzon marinus8, Northeast Chinese lamprey Lethenteron mori9, and Arctic/Japanese lamprey L. camtschaticum10, allowing loss-of-function analysis in these species. However, many techniques, including in vivo imaging, photoconversion, calcium imaging, and optogenetics, require a knock-in method. Although I-SceI meganuclease-mediated tissue-specific transient transgenesis has been reported, its transgene integration into the lamprey genome was not either specifically targeted or determined11,12. In this study, we succeeded in generating F0 knock-in lampreys (for the Arctic/Japanese lamprey L. camtschaticum) expressing reporter genes, the genomic integration of which was determined by PCR and sequencing. This method is simple and versatile for various experimental aims. In addition, the efficiency of obtaining transgenic lampreys is very high (20–35% of survivors). Based on these results, we believe that this CRISPR-Cas9-mediated versatile knock-in method opens up new research horizons using lamprey and possibly other “non-model” organisms in the sense of molecular genetics.
Results
Strategy for generating knock-in lampreys with the LcHsp70A promoter
For CRISPR-Cas9-mediated knock-in via non-homologous end joining (NHEJ) in the lamprey, we used an experimental strategy previously established in zebrafish13 and medaka14 with minor modification: co-injection of sgRNA1 (for genome digestion), sgRNA2 (for plasmid digestion), donor plasmids, Cas9 mRNA, and fast green (for visualization of the injection cocktail) (Fig. 1A).
CRISPR-Cas9-mediated knock-in strategy using LcHsp70A promoter. (A) For the generation of knock-in lamprey, sgRNA1 (for genome digestion), sgRNA2 (for plasmid digestion), the donor plasmids having a bait sequence, and Cas9 mRNA in distilled water containing Fast Green to aid visualization of the spread of the injection are co-injected into lamprey zygotes. (B) After injection, the CRISPR-Cas9-mediated concurrent cleavage occurs in the genome at the site upstream (approximately, 200–600 bp) of the target gene and in the donor plasmid at the bait sequence. This leads to a homology independent DNA repair, resulting in the integration of the donor plasmid into the targeted locus. Here, both forward and reverse integrations can occur. Cis-regulatory DNA sequences for the target gene expression act on the LcHsp70A promoter (enhancer-trapping), resulting in the expression of the reporter gene in cells that express the target gene.
Species-specific heat shock protein 70 (Hsp70) promoters work effectively as a minimal promoter in zebrafish and medaka13,14. Thus, we first aimed to isolate a lamprey Hsp70-like promoter. We found two homologs of Hsp70-like genes in the P. marinus and L. camtschaticum genomes, one of which we named LcHsp70A. Phylogenetic analysis suggests that those two homologs are the result of lineage-specific duplication in the lamprey (Fig. S1A). The LcHsp70A sequence was also found in the transcriptomes of L. camtschaticum embryos obtained previously15. In this study, we extracted a ~ 0.2 kb sequence of the LcHsp70A promoter for plasmid construction (Fig. S1B).
The donor plasmid contains a bait sequence (Tbait, a 23 bp sequence derived from the mouse Tet1 gene13, upstream of the insertion cassette for sgRNA2-guided DNA cleavage (Fig. 1B). This bait sequence was selected because the corresponding sgRNA (sgT) appears to have no potential off-target binding sites except for those with three or more mismatches in the L. camtschaticum genome (Table S1). Tbait is followed by the LcHsp70A promoter, which we expect to work as a minimal promoter (Fig. 1B). Finally, the LcHsp70A promoter is followed by reporter genes (in this study, EGFP or Dendra2).
We set the target site for genome digestion approximately 200–600 bp upstream of the predicted transcriptional-start site of the target genes (Fig. 1B; the sequences used are shown in Table S2). Concurrent digestion of the genome and plasmid DNA (guided by sgRNA1 and sgRNA2, respectively) with Cas9 would result in the integration of the donor plasmid into the genome via NHEJ (Fig. 1B). Then, cis-regulatory sequences for tissue-specific expression of the target gene would act on the LcHsp70A promoter, resulting in expression of a transgene in cells that express the target gene (enhancer-trapping).
Generation of Bra:EGFP and MA2:EGFP knock-in lampreys
To examine the effect of the CRISPR-Cas9-mediated knock-in system, we first targeted brachyury (Bra) and muscle actin 2 (MA2) genes for an EGFP reporter assay (hereafter, we call the knock-in animals Bra:EGFP and MA2:EGFP knock-in lampreys, respectively). In the lamprey, a T-box family transcription factor Bra is expressed in the dorsal protostome (axial mesoderm progenitor), the notochord, and the tailbud16,17, while MA2 is a muscle marker18,19,20. We tested two sgRNAs for Bra and one for MA2 (Table S2). Each sgRNA was co-injected with sgRNA for Tbait (sgT), the donor plasmids, and Cas9 mRNA into zygotes. We incubated the injected embryos and investigated their EGFP expressions during development (Figs. 2 and 3). In this investigation procedure, we screened and counted embryos or prolarvae with specific EGFP expression at stage 16 for Bra:EGFP and stage 26 for MA2:EGFP knock-in lampreys (Table S3).
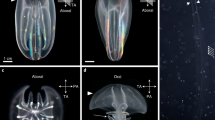
Bra:EGFP knock-in lampreys generated by microinjection of Bra-sg1. (A) At stage 16, in dorsal view (A) and posterior view (A’). EGFP is expressed in the axial mesodermal cells (am). A, D, P, V indicate anterior, dorsal, posterior, and ventral, respectively. Scale bar: 500 μm. (B) At stage 21, in lateral view (B) and dorsal view (B’). EGFP is persistently expressed in the axial mesodermal cells (am). L, P, R, V indicate left, posterior, right, and ventral, respectively. Scale bar: 500 μm. (C) At stage 25, in lateral view. The EGFP expression was restricted mostly in the tailbud and anal regions. In the york, some non-specific signals (by unintegrated plasmids) were observed (arrows). A, D, P, V indicate anterior, dorsal, posterior, and ventral, respectively. Scale bar: 500 μm.
MA2:EGFP knock-in lampreys. (A) A MA2:EGFP knock-in lamprey showing EGFP expression in the head region at stage 30, in lateral view. The head region is magnified in (A’). The asterisk (*) indicates the eyeball. EGFP is expressed both somatic and pharyngeal muscles (m.). In the york, some non-specific signals (by unintegrated plasmids) were observed (arrows). A, D, P, V indicate anterior, dorsal, posterior, and ventral, respectively. Scale bar: 500 μm. (B) A MA2:EGFP knock-in lamprey showing EGFP expression in the trunk region at stage 30, in lateral view. A part of the trunk region is magnified in (B’). Both EGFP-positive and EGFP-negative somatic muscle cells are observed in the same somite. In the york, some non-specific signals (by unintegrated plasmids) were observed (arrows). A, D, P, V indicate anterior, dorsal, posterior, and ventral, respectively. Scale bar: 500 μm.
With Bra-sg1, we observed axial mesoderm progenitor-specific EGFP expression (see Fig. 2A as an example) in 34% (37 of 108) of the surviving animals (Table S3). The remaining 66% showed either sparse, widespread non-specific EGFP expression or no expression. The efficiency was not improved by injecting Cas9 protein instead of mRNA (Table S4). To determine the injection damage and toxicity, we also performed sgRNA1-removed cocktail injection and water injection, comparing the result with the no-injection control. The water injection control showed slightly higher survival rate (28.5%) than that of the sgRNA1-removed cocktail injection control (18.5%), which was within the range of the experimental group (21.0%) (Table S4). As the survival rate of the no injection control was far higher (90.5%) than water injection control, both injection damage and RNA toxicity affect to the survival rate.
At stage 16 (late gastrula), EGFP expression was observed in axial mesoderm-progenitor cells (Fig. 2A) and it was persistently observed in those cells at stage 21 (late neurula) (Fig. 2B). The expression was confirmed histologically (Fig. S2). At stage 25 (late pharyngula), the EGFP expression was restricted mostly to the tailbud and anal regions with some non-specific signals (by unintegrated plasmids) observed in the yolk (Fig. 2C). Likewise, in the case of Bra-sg2, we found specific EGFP expression in 23% (25 of 110) of the surviving animals (Table S3). The expression patterns were identical to those observed in Bra-sg1-injected animals (Fig. S3). These expression patterns were consistent with reports based on in situ hybridization analysis16,17. However, the specific EGFP expression was observed only on one side of the animal body in all examined embryos/prolarvae, suggesting that the integration of the donor plasmid occurred only in some of the blastomeres, causing mosaicism.
In the case of MA2-sg1, we observed muscle-specific EGFP expression in 21% (25 of 119) of the surviving animals (Table S3). The EGFP signals were persistently detected in muscle cells of stage 30 ammocoetes larvae (~ 40 days after fertilization; Fig. 3). In all prolarvae examined, the EGFP expression was again restricted to some cells only on the left or right side of the animal body, as observed in the Bra:EGFP knock-in lampreys described above. In the animals that expressed EGFP in somatic muscle cells, mosaicism was found even in the same somite (Fig. 3B). Both EGFP-positive and -negative muscle cells were present in these somites, suggesting heterogeneity of mesodermal progenitor cells in somitogenesis. In the animals that expressed EGFP in the head region, the EGFP signals were detected in pharyngeal derivatives, including the upper lip muscle, ventral labial elevator, buccal constrictor, superficial branchial constrictors, interbranchial muscles, and branchial sphincters (Fig. 3A’; nomenclature based on previous description21,22).
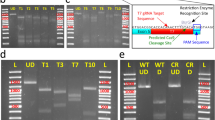
These results strongly suggest that CRISPR-Cas9-mediated knock-in occurred in some of the blastomeres in the injected animals. To confirm that the integration of the donor plasmid indeed occurred in the targeted loci of the genome, we performed insertion mapping of the MA2:EGFP knock-in lampreys (Figs. S4A and S4B). This indicated that the targeted integrations occurred both in the forward (#1 and #2 in Fig. S4B) and reverse (#3 in Fig. S4B) directions. Further evidence of the targeted integration was obtained by the sequencing the transgene-integrated animals (Fig. S4C).
Generation and photoconversion of SoxE3:Dendra2 knock-in lampreys
To explore the potential versatility of our knock-in strategy, we next generated SoxE3-targeted knock-in lampreys using donor plasmids that contained the coding sequence for the photoconvertible fluorescent protein Dendra223. In the lamprey, a Sox family transcription factor SoxE3 is expressed in neural crest cells (NCCs) and plays key roles in pharyngeal development24,25. The migration patterns of NCCs have been investigated from the viewpoint of jaw evolution in cell-labeling experiments using lipophilic tracers, such as DiI20,26,27. However, these dyes cannot selectively label NCCs, causing inevitable artifact signals. To overcome this problem, we planned to generate SoxE3:Dendra2 knock-in lampreys and to perform cell type-specific lineage tracing by selectively highlighting SoxE3-positive NCCs.
For this purpose, sgRNA SoxE3-sg1 (Table S2) was co-injected with sgRNA for Tbait (sgT), the donor plasmids containing the Dendra2 sequence, and Cas9 mRNA into zygotes. We incubated the injected embryos and investigated them for their native (green) Dendra2 expression at stage 21. We observed NCC-specific expression in 31% (18 of 59) of the surviving animals (Table S3). We then selectively highlighted some of Dendra2-positive cells by photoconversion with region-specific application with ultraviolet light at this stage (Fig. 4A). We further incubated the photoconversion-treated embryos and re-examined them at stage 24. In these animals, some photoconverted cells (originally situated at the neural tube level on the dorsoventral axis) were observed in the pharyngeal region, suggesting that those cells migrated ventrally (Fig. 4B). These results indicate the usefulness of the CRISPR-Cas9-mediated knock-in system in this kind of lineage tracing analysis.
Cell lineage analysis of photoconverted SoxE3:Dendra2 knock-in lampreys. (A) Photoconversion experiments were performed in the head region of SoxE3:Dendra2 knock-in lampreys at stage 21. Native green fluorescence, photoconverted red fluorescence (shown in magenta), and the merged image are shown in (A), (A’), and (A’’), respectively. The midbrain–hindbrain boundary (MHB) is indicated with dashed lines. Scale bar: 500 μm. (B) The same animal shown in (A) is raised to the stage 24 and reexamined. Some photoconverted cells are observed in the mandibular arch (ma), suggesting ventral migration of these NCC cells (arrows). Native green fluorescence, photoconverted red fluorescence (shown in magenta), and the merged image are shown in (B), (B’), and (B’’), respectively. The midbrain–hindbrain boundary (MHB) is indicated with dashed lines. Scale bar: 200 μm.
Discussion
Although the lamprey is an important organism in evolutionary biology and neuroscience research, no knock-in technique for this animal had not been established. By contrast, knock-in transgenic zebrafish and medaka can be efficiently generated via NHEJ by co-injection of two sgRNAs (one to digest the genome and the other to digest donor plasmids), donor plasmids, and Cas9 mRNA. Here, we showed that this method is perfectly applicable to generate F0 knock-in lamprey.
One major impediment in applying modern genetic techniques to the lamprey was that the establishment of transgenic lines is practically impossible due to its long and complex life cycle. This problem would be serious in the situation if we had only low-efficiency genetic engineering techniques. In this study, however, we showed that F0 knock-in lampreys can be generated with high efficiency (20–35% of survivors). The higher efficiency compared with zebrafish (around 5%13) could possibly result from the slower development of lamprey embryos compared to zebrafish28,29. In addition, trained experimenters can perform thousands of microinjections per day30. The high efficiency and producibility allow researchers to obtain many knock-in lampreys without the huge effort or time needed to establish transgenic lines. This efficiency was not improved by injecting Cas9 protein (Table S4), although the knock out efficiency approached 99% with co-injection of Cas9 protein and a tyrosinase sgRNA in a previous study8. It is possible that the plasmids need to be freshly digested in the cell for efficient knock-in.
Furthermore, sgRNAs design appears to influence the knock-in efficiency. A study reported that the indel-inducing efficiency of each sgRNA correlated with the levels of transient expressions in knock-in zebrafish13. As we applied the same method here, we expect that the same correlation will be observed when generating F0 knock-in lampreys using this method.
The mosaicism observed broadly in the F0 knock-in lampreys imposes an obstacle on the application of this method to some specific experiments. Nevertheless, we have shown that the reporter gene expression in the injected embryos remained until the late developmental period (stage 30 ammocoetes larva, ~ 40 days after fertilization), allowing researchers to perform long-term in vivo experiments in lamprey. This long retention of transgene expression is a major advantage of the CRISPR-Cas9-mediated knock-in method. Furthermore, we have shown that our knock-in system is also applicable to lineage-tracing analysis by generating and photoconverting SoxE3:Dendra2 knock-in lampreys. The transgene in the donor plasmid can also be replaced with genes encoding other various proteins such as calcium indicators and channelrhodopsins, enabling researchers to perform calcium imaging, optogenetics, and so on. In our system, the same donor plasmid can be used for different target genes by co-injecting corresponding sgRNAs. The reverse is also true: the same sgRNA for a target gene can be co-injected with different donor plasmids according to various experimental purposes. This flexibility is another significant advantage compared to previous methods such as simple plasmid injection and I-SceI meganuclease-mediated transient transgenesis11,12,31. In addition to in vivo imaging and lineage tracing, the method described here has great versatility for various experimental uses in lamprey research.
Finally, we presume that the same strategy can be applied to the vast majority of animals for which one-cell stage embryos are available. As mentioned above, lampreys appear to be particularly amenable to gene editing using the CRISPR/Cas9 system, possibly due to their slow early development time. In addition, the relative abundance of PAM sites due to the high GC content of the lamprey genome32 facilitates sgRNA design for genes of interest. Nevertheless, the method established here opens up new research horizons using animals that have previously been regarded as “non-model” organisms in the sense of modern molecular genetics. Although an annotated genome is a de facto requirement for CRISPR/Cas9 genome editing, the present method may conversely facilitate genomic research in “non-model” species.
Methods
Ethics approval
This study is reported in accordance with ARRIVE guidelines (https://arriveguidelines.org). All procedures in this study were performed in compliance with the guidelines approved by the animal care and use committees of the National Institutes of Natural Sciences (approved project no. 19A041) and University of Tsukuba (specific approval is not needed for experimentation on fishes at University of Tsukuba, under the Japanese law, Act on Welfare and Management of Animals).
Animals and embryos
Adult lampreys (Lethenteron camtschaticum) were collected from the Shiribeshi-Toshibetsu River, Hokkaido, Japan. In the next spawning season (May to June), the animals were anesthetized in ethyl,3-amino-benzoate methanesulfonate (MS-222). Mature eggs and sperm were squeezed from adults and fertilized in vitro. Embryos and prolarvae were maintained at 16 °C and staged according to Tahara28’s description.
Identification lamprey heat shock protein 70-like sequences
A BLASTN search was performed to identify heat shock protein 70 homologs in a gene model GRAS-LJ33 for the L. camtschaticum genome assembly LetJap1.0 (BioProject: PRJNA192554) and in transcriptome data obtained from stage 25–26 L. camtschaticum larvae15 (the raw reads were deposited in the DDBJ Sequence Reads Archives as with an accession number DRA007317), using the zebrafish hsp70l-1 sequence (ENSDARG00000055723) as a query.
Phylogenetic analysis of Hsp70 amino acid sequences
For other species, we surveyed Hsp70 sequences from available genomic/transcriptomic databases. The accession/reference numbers for each sequence are shown in Fig. S1. The sequences were aligned using MAFFT34 (Katoh and Toh 2008). The best-fitting amino acid substitution model and maximum likelihood (ML) tree were inferred using RAxML software (ver. 8.2.0)35,36. The bootstrap values were calculated using 1,000 replicates.
Isolation of LcHsp70A promoter region and construction of donor DNA for knock-in
L. camtschaticum genomic DNA was extracted from sperm obtained from a matured L. camtschaticum male using DNeasy Blood & Tissue Kit (Qiagen). To amplify LcHsp70A promoter region, genomic PCR was performed with the forward primer containing a Not I restriction enzyme recognition sequence and a Tbait sequence (shown in lowercase letters), agagcggccgggctgctgtcagggagctcatggGAAACAAAAGTCGCGCGAGA, and the revers primer containing a BamH I restriction enzyme recognition sequence (shown in lowercase letters), aggatccTGTTACCCCAAACCCTTCGA. The donor plasmid containing Tbait, LcHsp70A promoter, and EGFP (Tbait-LcHsP-EGFP) was synthesized from a plasmid described previously13, which contains zebrafish heat shock promoter (Tbait-DrHsP-EGFP): Tbait-DrHsP-GFP was digested with Not I and BamH I restriction enzymes and then ligated with the isolated LcHsp70A promoter region (Fig. S5). Here, Tbait (GGCTGCTGTCAGGGAGCTCATGG) sequence13 was used as a bait sequence in donor plasmids. The plasmid containing Tbait, LcHsp70A promoter, and Dendra2 (Tbait-LcHsP-Dendra2) was similarly synthesized from Tbait-LcHsP-EGFP and tetO-Dendra2 plasmid described previously37 by BamH I and Xba I restriction enzyme digestion.
Preparation of sgRNAs and Cas9 mRNAs
The target sites for genome digestion were set approximately 200–600 bp upstream of the predicted transcriptional-start site of the target genes, with the sequence 5′-GA(17 N or 18 N)NGG-3′ or 5′-GG(17 N or 18 N)NGG-3′ with 50–70% GC content. Template DNA for sgRNA synthesize was PCR-amplified from pDR27438 with the forward primer, ATTTAGGTGACACTATAgaxxxxxxxxxxxxxxxxxxGTTTTAGAGCTA GAAATAGC (for SP6 polymerase) or TAATACGACT- CACTATAggxxxxxxxxxxxxxxxxxxGTTTTAGAGCTAGAAATAGC (for T7 polymerase), and the reverse primer, AAAAGCACCGACTCGGTGCC. The lowercase letters correspond to genome-targeting sequences in sgRNAs. The genome-targeting sequences in sgRNAs used in this study are shown in Table S2. After PCR amplification with Prime Star Taq polymerase (Takara, Otsu, Japan), PCR product was purified using a QIAquick PCR Purification Kit (Qiagen, Hilden, Germany). Template DNA thus obtained was used for the in vitro transcription of sgRNAs using a MAXIscript T7 kit (Life Technologies, Carlsbad, USA). pCS2-hSpCas9 (gifted from M. Kinoshita and F. Zhang39) was digested with NotI, and Cas9 mRNA was transcribed using an mMESSAGEmMACHINE SP6 kit (Life Technologies). sgRNAs and Cas9 mRNA were purified using a RNeasy Mini kit (Qiagen).
Genomic location of potential off-target sites of sgT
Genomic locations of potential off-target sites of sgT were identified by using the Cas-OFFinder algorithm40).
Microinjection for knock-in
sgRNAs and Cas9 mRNA/protein (Guide-it Recombinant Cas9, 3 µg/µl, Takara Bio USA, Inc., Mountain View, CA, USA) were co-injected into lamprey zygotes with Qiagen miniprep (Qiagen) purified donor DNA in distilled water containing Fast Green to aid visualization of the spread of the injection. Each zygote was injected with ~ 5 nl solution containing ~ 45 pg sgRNA for digesting Tbait, ~ 90 pg sgRNA for digesting genome DNA, ~ 1000 pg Cas9 mRNA/protein, and ~ 45 pg donor plasmid as described previously14.
For Microinjection, 2% agar plates were prepared and the agar layer was made holes by a 1000 μl disposal pipette tip, where zygotes were placed. Microinjection was performed under a stereomicroscope, using a microinjector FemtoJet (Eppendolf, Hamburg, Germany) connected to a fine glass pipet placed on a mechanical manipulator.
Insertion mapping
For insertion mapping, fluorescent F0 knock-in lampreys were collected, and genomic DNA was extracted with standard protocols. The insertion status was examined on either the 5′ side or the 3′ side of the insertion (Fig. S3A). For example, to examine the 5′ side of the insertion, a PCR reaction was performed using a 5′ primer that was specific to each gene (upstream of the expected insertion site) and a 3′ primer that was specific to the donor plasmid (sequence within the LcHsp70A promoter for detecting the forward insertion, and sequence within pBluescriptSK for detecting the inverse insertion) as described previously13. Some of the amplified fragments were sequenced to examine the joint regions of the insertions.
Photoconversion
Photoconversion was carried out by illuminating violet-blue light for 30 s using an X-Cite Exacte XCT10A fluorescence microscope illuminater system (Lumen Dynamics, Mississauga, Canada) and a 400–410 nm band-pass filter mounted in a fluorescence microscope BX51WI (Olympus, Tokyo, Japan).
Histological sections
For histological analysis, specimens were fixed with 4% paraformaldehyde in PBS for 1 h, washed with PBS, and mounted Optimal Cutting Temperature (OCT) compound. Then, the specimens were sectioned using a CM3050 III (Leica) and mounted in SlowFade Diamond Antifade Mountant with DAPI (Thermo Fisher Scientific, Waltham, MA, USA).
Imaging
Images were taken using a fluorescence stereomicroscope MVX10 (Olympus, Tokyo, Japan).
References
Auclair, F. & Dubuc, R. Neural control of swimming in lampreys. The Neural Control of Movement (Elsevier Inc., 2020). https://doi.org/10.1016/B978-0-12-816477-8.00005-3
Grillner, S. et al. Intrinsic function of a neuronal network-a vertebrate central pattern generator. Brain Res. Rev. 1, 184–197 (1998).
Kuratani, S., Kuraku, S. & Murakami, Y. Lamprey as an evo-devo model: lessons from comparative embryology and molecular phylogenetics. Genesis 34, 175–183 (2002).
McCauley, D. W., Docker, M. F., Whyard, S. & Li, W. Lampreys as diverse model organisms in the genomics era. Bioscience 65, 1046–1056 (2015).
Osório, J. & Rétaux, S. The lamprey in evolutionary studies. Science 5, 221–235. https://doi.org/10.1007/s00427-008-0208-1 (2008).
Shimeld, S. M. & Donoghue, P. C. J. Evolutionary crossroads in developmental biology: cyclostomes (lamprey and hagfish). Dev. 139, 2091–2099 (2012).
Evans, T. M., Janvier, P. & Docker, M. F. The evolution of lamprey (Petromyzontida) life history and the origin of metamorphosis. Rev. Fish Biol. Fish. 28, 825–838 (2018).
Square, T. et al. CRISPR/Cas9-mediated mutagenesis in the sea lamprey Petromyzon marinus: a powerful tool for understanding ancestral gene functions in vertebrates. Development 142, 4180–4187 (2015).
Zu, Y. et al. Biallelic editing of a lamprey genome using the CRISPR/Cas9 system. Sci. Rep. 6, 23496 (2016).
Kusakabe, R. et al. Novel developmental bases for the evolution of hypobranchial muscles in vertebrates. BMC Biol. 18, 120 (2020).
Parker, H. J., Sauka-spengler, T., Bronner, M. & Elgar, G. A reporter assay in lamprey embryos reveals both functional conservation and elaboration of vertebrate enhancers. Science 9, 21–79 (2014).
Hockman, D. et al. A genome-wide assessment of the ancestral neural crest gene regulatory network. Nat. Commun. 10, 29 (2019).
Kimura, Y., Hisano, Y., Kawahara, A. & Higashijima, S. I. Efficient generation of knock-in transgenic zebrafish carrying reporter/driver genes by CRISPR/Cas9-mediated genome engineering. Sci. Rep. 4, 1–7 (2014).
Watakabe, I. et al. Highly efficient generation of knock-in transgenic medaka by CRISPR/Cas9-mediated genome engineering. Zool. Lett. 4, 1–11 (2018).
Yokoyama, H., Morino, Y. & Wada, H. Identification of a unique lamprey gene with tandemly repeated sequences and pharyngeal chondrocyte-specific expression. Gene 701, 9–14 (2019).
Sauka-Spengler, T., Baratte, B., Lepage, M. & Mazan, S. Characterization of Brachyury genes in the dogfish S. canicula and the lamprey L. fluviatilis: insights into gastrulation in a chondrichthyan. Dev. Biol. 263, 296–307 (2003).
Takeuchi, M., Takahashi, M., Okabe, M. & Aizawa, S. Germ layer patterning in bichir and lamprey; an insight into its evolution in vertebrates. Dev. Biol. 332, 90–102 (2009).
Kusakabe, R., Takechi, M., Tochinai, S. & Kuratani, S. Lamprey contractile protein genes mark different populations of skeletal muscles during development. J. Exp. Zool. B. Mol. Dev. Evol. 302, 121–133 (2004).
Suzuki, D. G. et al. Comparative morphology and development of extra-ocular muscles in the lamprey and gnathostomes reveal the ancestral state and developmental patterns of the vertebrate head. Zool. Lett. 2, 10 (2016).
Yokoyama, H., Yoshimura, M., Suzuki, D. G., Higashiyama, H. & Wada, H. Development of the lamprey velum and implications for the evolution of the vertebrate jaw. Dev. Dyn. 250, 88–98 (2021).
Mallatt, J. Ventilation and the origin of jawed vertebrates: a new mouth. Zool. J. Linn. Soc. 117, 329–404 (1996).
Ziermann, J. M., Miyashita, T. & Diogo, R. Cephalic muscles of Cyclostomes (hagfishes and lampreys) and Chondrichthyes (sharks, rays and holocephalans): comparative anatomy and early evolution of the vertebrate head muscles. Zool. J. Linn. Soc. 172, 771–802 (2014).
Gurskaya, N. G. et al. Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Zool. J. Linn. Soc. 24, 461–465 (2006).
Ohtani, K. et al. Expression of Sox and fibrillar collagen genes in lamprey larval chondrogenesis with implications for the evolution of vertebrate cartilage. J. Exp. Zool. B. Mol. Dev. Evol. 310, 596–607 (2008).
Yao, T., Ohtani, K., Kuratani, S. & Wada, H. Development of lamprey mucocartilage and its dorsal-ventral patterning by endothelin signaling, with insight into vertebrate jaw evolution. J. Exp. Zool. B. Mol. Dev. Evol. 316, 339–346 (2011).
Mccauley, D. W. & Bronner-fraser, M. Neural crest contributions to the lamprey head. Science 11, 2317–2327. https://doi.org/10.1242/dev.00451 (2003).
Shigetani, Y. et al. Heterotopic shift of epithelial-mesenchymal interactions in vertebrate jaw evolution. Science (80-). 296, 1316–1319 (2002).
Tahara, Y. Normal stages of development in the lamprey, Lampetra reissneri (Dybowski). Zoolog. Sci. 5, 109–118 (1988).
Kimmel, C. B., Ballard, W. W., Kimmel, S. R., Ullmann, B. & Schilling, T. F. Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310 (1995).
Square, T. A day in the life of a lamprey lab. the NODE: the community site for and by developmental biologists (2015).
Kusakabe, R., Tochinai, S. & Kuratani, S. Expression of foreign genes in lamprey embryos: an approach to study evolutionary changes in gene regulation. J. Exp. Zool. Part B Mol. Dev. Evol. 296, 87–97 (2003).
Mehta, T. K. et al. Evidence for at least six Hox clusters in the Japanese lamprey (Lethenteron japonicum). Proc. Natl. Acad. Sci. U. S. A. 110, 16044–16049 (2013).
Kadota, M. et al. CTCF binding landscape in jawless fish with reference to Hox cluster evolution. Sci. Rep. 7, 112 (2017).
Katoh, K. & Toh, H. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 9, 286–298 (2008).
Stamatakis, A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Sci. Rep. 22, 2688–2690 (2006).
Stamatakis, A., Hoover, P. & Rougemont, J. A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 57, 758–771 (2008).
Uemura, Y. et al. Neuronal circuits that control rhythmic pectoral fin movements in Zebrafish. J. Neurosci. 40, 6678–6690 (2020).
Hwang, W. Y. et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31, 227–229 (2013).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Bae, S., Park, J. & Kim, J. S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30, 1473–1475 (2014).
Acknowledgements
This study is supported by a grant from the Narishige Zoological Science Award to DGS and by Grant-in-Aid from Japan Society for the Promotion of Science (18J00045 and 20K15855 to DGS).
Author information
Authors and Affiliations
Contributions
D.G.S. and S.H. conceived the project. D.G.S. designed and performed the experiments. D.G.S. and H.W. captured and maintained the lampreys. D.G.S. analyzed the data. D.G.S., H.W., and S.H. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Suzuki, D.G., Wada, H. & Higashijima, Si. Generation of knock-in lampreys by CRISPR-Cas9-mediated genome engineering. Sci Rep 11, 19836 (2021). https://doi.org/10.1038/s41598-021-99338-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-99338-1
This article is cited by
-
Exploring the origin of a unique mutant allele in twin-tail goldfish using CRISPR/Cas9 mutants
Scientific Reports (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.