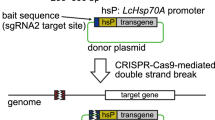
Abstract
Despite long-standing experimental interest in ctenophores due to their unique biology, ecological influence and evolutionary status, previous work has largely been constrained by the periodic seasonal availability of wild-caught animals and difficulty in reliably closing the life cycle. To address this problem, we have developed straightforward protocols that can be easily implemented to establish long-term multigenerational cultures for biological experimentation in the laboratory. In this protocol, we describe the continuous culture of the Atlantic lobate ctenophore Mnemiopsis leidyi. A rapid 3-week egg-to-egg generation time makes Mnemiopsis suitable for a wide range of experimental genetic, cellular, embryological, physiological, developmental, ecological and evolutionary studies. We provide recommendations for general husbandry to close the life cycle of Mnemiopsis in the laboratory, including feeding requirements, light-induced spawning, collection of embryos and rearing of juveniles to adults. These protocols have been successfully applied to maintain long-term multigenerational cultures of several species of pelagic ctenophores, and can be utilized by laboratories lacking easy access to the ocean. We also provide protocols for targeted genome editing via microinjection with CRISPR–Cas9 that can be completed within ~2 weeks, including single-guide RNA synthesis, early embryo microinjection, phenotype assessment and sequence validation of genome edits. These protocols provide a foundation for using Mnemiopsis as a model organism for functional genomic analyses in ctenophores.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on request.
References
Dunn, C. W., Leys, S. P. & Haddock, S. H. D. The hidden biology of sponges and ctenophores. Trends Ecol. Evol. 30, 282–291 (2015).
Neff, E. P. What is a lab animal? Lab Anim. 47, 223–227 (2018).
Ryan, J. F., Schnitzler, C. E. & Tamm, S. L. Meeting report of Ctenopalooza: the first international meeting of ctenophorologists. Evodevo 7, 19 (2016).
Chun, C. Die Ctenophoren des Golfes von Neapel und der angrenzenden Meeres-Abschnitte. (W. Engelmann, 1880).
Hyman, L. H. in The Invertebrates: Protozoa through Ctenophora vol. 1 662–695 (McGraw-Hill, 1940).
Harbison, G. R., Madin, L. P. & Swanberg, N. R. On the natural history and distribution of oceanic ctenophores. Deep Sea Res. I 25, 233–256 (1978).
Harbison, G. R. in The Origins and Relationships of Lower Invertebrates (eds. Morris, S. C., George, J. D., Gibson, R. & Platt, H. M.) 78–100 (Oxford Univ. Press, 1985).
Mills, C. E. & Haddock, S. H. D. in Light and Smith’s Manual: Intertidal Invertebrates of the Central California Coast (ed. Carlton, J. T.) 47–49 (Univ. California Press, 2007).
Pang, K. & Martindale, M. Q. Ctenophores. Curr. Biol. 18, R1119–R1120 (2008).
Dunn, C. W. et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature 452, 745–749 (2008).
Hejnol, A. et al. Assessing the root of bilaterian animals with scalable phylogenomic methods. Proc. Biol. Sci. 276, 4261–4270 (2009).
Philippe, H. et al. Phylogenomics revives traditional views on deep animal relationships. Curr. Biol. 19, 706–712 (2009).
Pick, K. S. et al. Improved phylogenomic taxon sampling noticeably affects nonbilaterian relationships. Mol. Biol. Evol. 27, 1983–1987 (2010).
Ryan, J. F. et al. The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science 342, 1242592 (2013).
Moroz, L. L. et al. The ctenophore genome and the evolutionary origins of neural systems. Nature 510, 109–114 (2014).
Pisani, D. et al. Genomic data do not support comb jellies as the sister group to all other animals. Proc. Natl. Acad. Sci. USA. 112, 15402–15407 (2015).
Telford, M. J., Budd, G. E. & Philippe, H. Phylogenomic insights into animal evolution. Curr. Biol. 25, R876–R887 (2015).
Shen, X.-X., Hittinger, C. T. & Rokas, A. Contentious relationships in phylogenomic studies can be driven by a handful of genes. Nat. Ecol. Evol. 1, 126 (2017).
Whelan, N. V. et al. Ctenophore relationships and their placement as the sister group to all other animals. Nat. Ecol. Evol. 1, 1737–1746 (2017).
Li, Y., Shen, X.-X., Evans, B., Dunn, C. W. & Rokas, A. Rooting the animal tree of life. Mol. Biol. Evol. 38, 4322–4333 (2021).
Afzelius, B. A. The fine structure of the cilia from ctenophore swimming-plates. J. Biophys. Biochem. Cytol. 9, 383–394 (1961).
Tamm, S. L. Mechanisms of ciliary co-ordination in ctenophores. J. Exp. Biol. 59, 231–245 (1973).
Tamm, S. L. Cilia and the life of ctenophores. Invertebr. Biol. 133, 1–46 (2014).
Abbott, J. F. The morphology of Coeloplana. Zool. Jahrb. Abt. Anat. Ontog. Tiere 24, 41–70 (1907).
Bargmann, W., Jacob, K. & Rast, A. Über Tentakel und Colloblasten der Ctenophore Pleurobrachia pileus. Z. Zellforsch. 123, 121–152 (1972).
von Byern, J., Mills, C. E. & Flammang, P. in Biological Adhesive Systems: From Nature to Technical and Medical Application (eds. von Byern, J. & Grunwald, I.) 29–40 (Springer, 2010).
Leonardi, N. D., Thuesen, E. V. & Haddock, S. H. D. A sticky thicket of glue cells: a comparative morphometric analysis of colloblasts in 20 species of comb jelly (phylum Ctenophora). Cienc. Mar. 46, 211–225 (2020).
Horridge, G. A. Relations between nerves and cilia in ctenophores. Am. Zool. 5, 357–375 (1965).
Tamm, S. L. Formation of the statolith in the ctenophore Mnemiopsis leidyi. Biol. Bull. 227, 7–18 (2014).
Jokura, K. & Inaba, K. Structural diversity and distribution of cilia in the apical sense organ of the ctenophore Bolinopsis mikado. Cytoskeleton 77, 442–455 (2020).
Jager, M. et al. New insights on ctenophore neural anatomy: immunofluorescence study in Pleurobrachia pileus (Müller, 1776). J. Exp. Zool. B 316B, 171–187 (2011).
Moroz, L. L. & Kohn, A. B. Independent origins of neurons and synapses: insights from ctenophores. Philos. Trans. R. Soc. Lond. B 371, 20150041 (2016).
Horridge, G. A. The giant mitochondria of ctenophore comb-plates. J. Cell Sci. s3-105, 301–310 (1964).
Pett, W. et al. Extreme mitochondrial evolution in the ctenophore Mnemiopsis leidyi: insight from mtDNA and the nuclear genome. Mitochondrial DNA 22, 130–142 (2011).
Kohn, A. B. et al. Rapid evolution of the compact and unusual mitochondrial genome in the ctenophore, Pleurobrachia bachei. Mol. Phylogenet. Evol. 63, 203–207 (2012).
Christianson, L. M., Johnson, S. B., Schultz, D. T. & Haddock, S. H. D. Hidden diversity of Ctenophora revealed by new mitochondrial COI primers and sequences. Mol. Ecol. Resour. 22, 283–294 (2022).
Hernandez-Nicaise, M. L., Mackie, G. O. & Meech, R. W. Giant smooth muscle cells of Beroë. Ultrastructure, innervation, and electrical properties. J. Gen. Physiol. 75, 79–105 (1980).
Hernandez-Nicaise, M. L. & Amsellem, J. Ultrastructure of the giant smooth muscle fiber of the ctenophore Beroe ovata. J. Ultrastruct. Res. 72, 151–168 (1980).
Hernandez-Nicaise, M.-L., Nicaise, G. & Malaval, L. Giant smooth muscle fibers of the ctenophore Mnemiopsis leidyi: ultrastructural study of in situ and isolated cells. Biol. Bull. 167, 210–228 (1984).
Mackie, G. O., Mills, C. E. & Singla, C. L. Structure and function of the prehensile tentilla of Euplokamis (Ctenophora, Cydippida). Zoomorphology 107, 319–337 (1988).
Vandepas, L. E., Warren, K. J., Amemiya, C. T. & Browne, W. E. Establishing and maintaining primary cell cultures derived from the ctenophore Mnemiopsis leidyi. J. Exp. Biol. 220, 1197–1201 (2017).
Presnell, J. S. et al. The presence of a functionally tripartite through-gut in Ctenophora has implications for metazoan character trait evolution. Curr. Biol. 26, 2814–2820 (2016).
Haddock, S. H. D. & Case, J. F. Not all ctenophores are bioluminescent: Pleurobrachia. Biol. Bull. 189, 356–362 (1995).
Bessho-Uehara, M. et al. Evidence for de novo biosynthesis of the luminous substrate coelenterazine in ctenophores. iScience 23, 101859 (2020).
Martindale, M. Q. The onset of regenerative properties in ctenophores. Curr. Opin. Genet. Dev. 40, 113–119 (2016).
Edgar, A., Mitchell, D. G. & Martindale, M. Q. Whole-body regeneration in the lobate ctenophore Mnemiopsis leidyi. Genes 12, (2021).
Ramon-Mateu, J., Ellison, S. T., Angelini, T. E. & Martindale, M. Q. Regeneration in the ctenophore Mnemiopsis leidyi occurs in the absence of a blastema, requires cell division, and is temporally separable from wound healing. BMC Biol. 17, 80 (2019).
Chun, C. Die Dissogonie, eine neue Form der geschlechtlichen Zeugung. Festsch. zum siebensigsten Geburtstage Rudorf Leuckarts. Engelmarm, Leipzig 77–108 (1892).
Martindale, M. Q. Larval reproduction in the ctenophore Mnemiopsis mccradyi (order Lobata). Mar. Biol. 94, 409–414 (1987).
Hirota, J. Laboratory culture and metabolism of the planktonic ctenophore, Pleurobrachia bachei A. Agassiz. in Biological oceanography of the northern North Pacific Ocean (ed. Takenouti, A. Y.) 465–484 (Idemitu Shoten, 1972).
Edgar, A., Ponciano, J. M. & Martindale, M. Q. Ctenophores are direct developers that reproduce continuously beginning very early after hatching. Proc. Natl Acad. Sci. 119, e2122052119 (2022).
Agassiz, A. Embryology of the Ctenophorae. Mem. Am. Acad. Arts Sci. 10, 357–398 (1874).
Hertwig, R. Über den Bau der Ctenophoren (Fischer, G, 1880).
Driesch, H. & Morgan, T. H. Zur Analysis der ersten Entwickelungsstadien des Ctenophoreneies. Wilhelm. Roux Arch. Entwickl. Mech. Org. 2, 204–215 (1895).
Fischel, A. Experimentelle Untersuchungen am Ctenophorenei. Arch. Entwickelungsmech. Organismen 6, 109–130 (1897).
Yatsu, N. Observations and experiments on the ctenophore egg: II. Notes on early cleavage stages and experiments on cleavage. Annot. Zool. Jpn 7, 333–346 (1911).
Podar, M., Haddock, S. H., Sogin, M. L. & Harbison, G. R. A molecular phylogenetic framework for the phylum Ctenophora using 18S rRNA genes. Mol. Phylogenet. Evol. 21, 218–230 (2001).
Simion, P., Bekkouche, N., Jager, M., Quéinnec, E. & Manuel, M. Exploring the potential of small RNA subunit and ITS sequences for resolving phylogenetic relationships within the phylum Ctenophora. Zoology 118, 102–114 (2015).
Yatsu, N. Observations and experiments on the ctenophore egg: III. Experiments on germinal localization of the egg of Beroe ovata. Annot. Zool. Jpn 8, 5–13 (1912).
Franc, J.-M. Etude ultrastructurale de la spermatogenèse du Cténaire Beroe ovata. J. Ultrastruct. Res. 42, 255–267 (1973).
Carré, D. & Sardet, C. Fertilization and early development in Beroe ovata. Dev. Biol. 105, 188–195 (1984).
Carré, D., Rouvière, C. & Sardet, C. In vitro fertilization in ctenophores: sperm entry, mitosis, and the establishment of bilateral symmetry in Beroe ovata. Dev. Biol. 147, 381–391 (1991).
Goudeau, M. & Goudeau, H. Successive electrical responses to insemination and concurrent sperm entries in the polyspermic egg of the ctenophore Beroe ovata. Dev. Biol. 156, 537–551 (1993).
Houliston, E., Carré, D., Johnston, J. A. & Sardet, C. Axis establishment and microtubule-mediated waves prior to first cleavage in Beroe ovata. Development 117, 75–87 (1993).
Rouvière, C., Houliston, E., Carré, D., Chang, P. & Sardet, C. Characteristics of pronuclear migration in Beroe ovata. Cell Motil. Cytoskelet. 29, 301–311 (1994).
Jokura, K. et al. CTENO64 is required for coordinated paddling of ciliary comb plate in ctenophores. Curr. Biol. 29, 3510–3516.e4 (2019).
Derelle, R. & Manuel, M. Ancient connection between NKL genes and the mesoderm? Insights from Tlx expression in a ctenophore. Dev. Genes Evol. 217, 253–261 (2007).
Jager, M., Quéinnec, E., Chiori, R., Le Guyader, H. & Manuel, M. Insights into the early evolution of SOX genes from expression analyses in a ctenophore. J. Exp. Zool. B 310, 650–667 (2008).
Alié, A. et al. Somatic stem cells express Piwi and Vasa genes in an adult ctenophore: ancient association of “germline genes” with stemness. Dev. Biol. 350, 183–197 (2011).
Dayraud, C. et al. Independent specialisation of myosin II paralogues in muscle vs. non-muscle functions during early animal evolution: a ctenophore perspective. BMC Evol. Biol. 12, 107 (2012).
Jager, M. et al. Evidence for involvement of Wnt signalling in body polarities, cell proliferation, and the neuro-sensory system in an adult ctenophore. PLoS ONE 8, e84363 (2013).
Freeman, G. The effects of altering the position of cleavage planes on the process of localization of developmental potential in ctenophores. Dev. Biol. 51, 332–337 (1976).
Freeman, G. The role of cleavage in the localization of developmental potential in the ctenophore Mnemiopsis leidyi. Dev. Biol. 49, 143–177 (1976).
Freeman, G. The establishment of the oral–aboral axis in the ctenophore embryo. Development 42, 237–260 (1977).
Martindale, M. Q. The ontogeny and maintenance of adult symmetry properties in the ctenophore, Mnemiopsis mccradyi. Dev. Biol. 118, 556–576 (1986).
Martindale, M. Q. & Henry, J. Q. Diagonal development: establishment of the anal axis in the ctenophore Mnemiopsis leidyi. Biol. Bull. 189, 190–192 (1995).
Martindale, M. Q. & Henry, J. Q. Development and regeneration of comb plates in the ctenophore Mnemiopsis leidyi. Biol. Bull. 191, 290–292 (1996).
Martindale, M. Q. & Henry, J. Q. Reassessing embryogenesis in the Ctenophora: the inductive role of e1 micromeres in organizing ctene row formation in the “mosaic” embryo, Mnemiopsis leidyi. Development 124, 1999–2006 (1997).
Martindale, M. Q. & Henry, J. Q. Intracellular fate mapping in a basal metazoan, the ctenophore Mnemiopsis leidyi, reveals the origins of mesoderm and the existence of indeterminate cell lineages. Dev. Biol. 214, 243–257 (1999).
Henry, J. Q. & Martindale, M. Q. Regulation and regeneration in the ctenophore Mnemiopsis leidyi. Dev. Biol. 227, 720–733 (2000).
Henry, J. Q. & Martindale, M. Q. Multiple inductive signals are involved in the development of the ctenophore Mnemiopsis leidyi. Dev. Biol. 238, 40–46 (2001).
Henry, J. Q. & Martindale, M. Q. Inductive interactions and embryonic equivalence groups in a basal metazoan, the ctenophore Mnemiopsis leidyi. Evol. Dev. 6, 17–24 (2004).
Fischer, A. H., Pang, K., Henry, J. Q. & Martindale, M. Q. A cleavage clock regulates features of lineage-specific differentiation in the development of a basal branching metazoan, the ctenophore Mnemiopsis leidyi. Evodevo 5, 4 (2014).
Babonis, L. S. et al. Integrating embryonic development and evolutionary history to characterize tentacle-specific cell types in a ctenophore. Mol. Biol. Evol. 35, 2940–2956 (2018).
Yamada, A. & Martindale, M. Q. Expression of the ctenophore Brain Factor 1 forkhead gene ortholog (ctenoBF-1) mRNA is restricted to the presumptive mouth and feeding apparatus: implications for axial organization in the Metazoa. Dev. Genes Evol. 212, 338–348 (2002).
Yamada, A., Pang, K., Martindale, M. Q. & Tochinai, S. Surprisingly complex T-box gene complement in diploblastic metazoans. Evol. Dev. 9, 220–230 (2007).
Pang, K. & Martindale, M. Q. Developmental expression of homeobox genes in the ctenophore Mnemiopsis leidyi. Dev. Genes Evol. 218, 307–319 (2008).
Layden, M. J., Meyer, N. P., Pang, K., Seaver, E. C. & Martindale, M. Q. Expression and phylogenetic analysis of the zic gene family in the evolution and development of metazoans. Evodevo 1, 12 (2010).
Pang, K. et al. Genomic insights into Wnt signaling in an early diverging metazoan, the ctenophore Mnemiopsis leidyi. Evodevo 1, 10 (2010).
Pang, K., Ryan, J. F., Baxevanis, A. D. & Martindale, M. Q. Evolution of the TGF-β signaling pathway and its potential role in the ctenophore, Mnemiopsis leidyi. PLoS ONE 6, e24152 (2011).
Reitzel, A. M. et al. Nuclear receptors from the ctenophore Mnemiopsis leidyi lack a zinc-finger DNA-binding domain: lineage-specific loss or ancestral condition in the emergence of the nuclear receptor superfamily? Evodevo 2, 3 (2011).
Schnitzler, C. E. et al. Genomic organization, evolution, and expression of photoprotein and opsin genes in Mnemiopsis leidyi: a new view of ctenophore photocytes. BMC Biol. 10, 107 (2012).
Simmons, D. K., Pang, K. & Martindale, M. Q. Lim homeobox genes in the ctenophore Mnemiopsis leidyi: the evolution of neural cell type specification. Evodevo 3, 2 (2012).
Schnitzler, C. E., Simmons, D. K., Pang, K., Martindale, M. Q. & Baxevanis, A. D. Expression of multiple Sox genes through embryonic development in the ctenophore Mnemiopsis leidyi is spatially restricted to zones of cell proliferation. Evodevo 5, 15 (2014).
Reitzel, A. M., Pang, K. & Martindale, M. Q. Developmental expression of “germline”- and “sex determination”-related genes in the ctenophore Mnemiopsis leidyi. Evodevo 7, 17 (2016).
Presnell, J. S. & Browne, W. E. Krüppel-like factor gene function in the ctenophore Mnemiopsis leidyi assessed by CRISPR/Cas9-mediated genome editing. Development 148, dev199771 (2021).
Lowe, S., Browne, M., Boudjelas, S. & De Poorter, M. 100 of the World’s Worst Invasive Alien Species: A Selection from the Global Invasive Species Database (Hollands Printing, 2000).
Kideys, A. E. Ecology. Fall and rise of the Black Sea ecosystem. Science 297, 1482–1484 (2002).
Costello, J. H., Bayha, K. M., Mianzan, H. W., Shiganova, T. A. & Purcell, J. E. Transitions of Mnemiopsis leidyi (Ctenophora: Lobata) from a native to an exotic species: a review. Hydrobiologia 690, 21–46 (2012).
Jaspers, C. et al. Ocean current connectivity propelling the secondary spread of a marine invasive comb jelly across western Eurasia. Glob. Ecol. Biogeogr. 27, 814–827 (2018).
Jaspers, C. et al. Invasion genomics uncover contrasting scenarios of genetic diversity in a widespread marine invader. Proc. Natl Acad. Sci. USA 118, e2116211118 (2021).
Colin, S. P., Costello, J. H., Hansson, L. J., Titelman, J. & Dabiri, J. O. Stealth predation and the predatory success of the invasive ctenophore Mnemiopsis leidyi. Proc. Natl Acad. Sci. USA 107, 17223–17227 (2010).
Gemmell, B. J., Colin, S. P., Costello, J. H. & Sutherland, K. R. A ctenophore (comb jelly) employs vortex rebound dynamics and outperforms other gelatinous swimmers. R. Soc. Open Sci. 6, 181615 (2019).
Jaspers, C., Titelman, J., Hansson, L. J., Haraldsson, M. & Ditlefsen, C. R. The invasive ctenophore Mnemiopsis leidyi poses no direct threat to Baltic cod eggs and larva. Limnol. Oceanogr. 56, 431–439 (2011).
Jaspers, C., Møller, L. F. & Kiørboe, T. Salinity gradient of the Baltic Sea limits the reproduction and population expansion of the newly invaded comb jelly Mnemiopsis leidyi. PLoS ONE 6, e24065 (2011).
Jaspers, C., Møller, L. F. & Kiørboe, T. Reproduction rates under variable food conditions and starvation in Mnemiopsis leidyi: significance for the invasion success of a ctenophore. J. Plankton Res. 37, 1011–1018 (2015).
Jaspers, C., Marty, L. & Kiørboe, T. Selection for life-history traits to maximize population growth in an invasive marine species. Glob. Chang. Biol. 24, 1164–1174 (2018).
Reeve, M. R., Syms, M. A. & Kremer, P. Growth dynamics of a ctenophore (Mnemiopsis) in relation to variable food supply. I. Carbon biomass, feeding, egg production, growth and assimilation efficiency. J. Plankton Res. 11, 535–552 (1989).
Jaspers, C. et al. Resilience in moving water: effects of turbulence on the predatory impact of the lobate ctenophore Mnemiopsis leidyi. Limnol. Oceanogr. 63, 445–458 (2018).
Jaspers, C., Costello, J. H. & Colin, S. P. Carbon content of Mnemiopsis leidyi eggs and specific egg production rates in northern Europe. J. Plankton Res. 37, 11–15 (2015).
Winnikoff, J. R., Haddock, S. H. D. & Budin, I. Depth- and temperature-specific fatty acid adaptations in ctenophores from extreme habitats. J. Exp. Biol. jeb.242800 (2021).
Jaspers, C. et al. Microbiota differences of the comb jelly Mnemiopsis leidyi in native and invasive sub-populations. Front. Mar. Sci. 6, 635 (2019).
Sutherland, K. R., Costello, J. H., Colin, S. P. & Dabiri, J. O. Ambient fluid motions influence swimming and feeding by the ctenophore Mnemiopsis leidyi. J. Plankton Res. 36, 1310–1322 (2014).
Colin, S. P. et al. Elevating the predatory effect: sensory-scanning foraging strategy by the lobate ctenophore Mnemiopsis leidyi. Limnol. Oceanogr. 60, 100–109 (2015).
Parker, G. H. The movements of the swimming-plates in ctenophores, with reference to the theories of ciliary metachronism. J. Exp. Zool. 2, 407–423 (1905).
Baker, L. D. & Reeve, M. R. Laboratory culture of the lobate ctenophore Mnemiopsis mccradyi with notes on feeding and fecundity. Mar. Biol. 26, 57–62 (1974).
Reeve, M. R., Walter, M. A. & Ikeda, T. Laboratory studies of ingestion and food utilization in lobate and tentaculate ctenophores. Limnol. Oceanogr. 23, 740–751 (1978).
Swanberg, N. The feeding behavior of Beroe ovata. Mar. Biol. 24, 69–76 (1974).
Haddock, S. H. D. Comparative feeding behavior of planktonic ctenophores. Integr. Comp. Biol. 47, 847–853 (2007).
Mayer, A. G. Ctenophores of the Atlantic Coast of North America (Carnegie Institution of Washington, 1912).
Seravin, L. N. The systematic revision of the genus Mnemiopsis (Ctenophora, Lobata). 2. Species attribution of Mnemiopsis from the Black Sea and the species composition of the genus Mnemiopsis. Zool. Zh. 73, 19–34 (1994).
Bayha, K. M. et al. Worldwide phylogeography of the invasive ctenophore Mnemiopsis leidyi (Ctenophora) based on nuclear and mitochondrial DNA data. Biol. Invasions 17, 827–850 (2015).
Costello, J. H., Sullivan, B. K., Gifford, D. J., Van Keuren, D. & Sullivan, L. J. Seasonal refugia, shoreward thermal amplification, and metapopulation dynamics of the ctenophore Mnemiopsis leidyi in Narragansett Bay, Rhode Island. Limnol. Oceanogr. 51, 1819–1831 (2006).
Pang, K. & Martindale, M. Q. Ctenophore whole-mount in situ hybridization. CSH Protoc. 2008, db.prot5087 (2008).
Salinas-Saavedra, M. & Martindale, M. Q. Improved protocol for spawning and immunostaining embryos and juvenile stages of the ctenophore Mnemiopsis leidyi. Protoc. Exchange https://doi.org/10.1038/protex.2018.092 (2018).
Dieter, A. C., Vandepas, L. E. & Browne, W. E. in Whole-Body Regeneration: Methods and Protocols (eds. Blanchoud, S. & Galliot, B.) (Springer, 2022).
Yamada, A., Martindale, M. Q., Fukui, A. & Tochinai, S. Highly conserved functions of the Brachyury gene on morphogenetic movements: insight from the early-diverging phylum Ctenophora. Dev. Biol. 339, 212–222 (2010).
Moreland, R. T. et al. A customized Web portal for the genome of the ctenophore Mnemiopsis leidyi. BMC Genomics 15, 316 (2014).
Moreland, R. T., Nguyen, A.-D., Ryan, J. F. & Baxevanis, A. D. The Mnemiopsis Genome Project Portal: integrating new gene expression resources and improving data visualization. Database 2020, baaa029 (2020).
Davidson, P. L. et al. The maternal–zygotic transition and zygotic activation of the Mnemiopsis leidyi genome occurs within the first three cleavage cycles. Mol. Reprod. Dev. 84, 1218–1229 (2017).
Sebé-Pedrós, A. et al. Early metazoan cell type diversity and the evolution of multicellular gene regulation. Nat. Ecol. Evol. 2, 1176–1188 (2018).
Levin, M. et al. The mid-developmental transition and the evolution of animal body plans. Nature 531, 637–641 (2016).
Sachkova, M. Y. et al. Neuropeptide repertoire and 3D anatomy of the ctenophore nervous system. Curr. Biol. https://doi.org/10.1016/j.cub.2021.09.005 (2021).
Fidler, A. L. et al. Collagen IV and basement membrane at the evolutionary dawn of metazoan tissues. eLife 6, (2017).
Draper, G. W., Shoemark, D. K. & Adams, J. C. Modelling the early evolution of extracellular matrix from modern ctenophores and sponges. Essays Biochem. 63, 389–405 (2019).
Ryan, J. F. et al. The homeodomain complement of the ctenophore Mnemiopsis leidyi suggests that Ctenophora and Porifera diverged prior to the ParaHoxozoa. Evodevo 1, 9 (2010).
Maxwell, E. K., Ryan, J. F., Schnitzler, C. E., Browne, W. E. & Baxevanis, A. D. MicroRNAs and essential components of the microRNA processing machinery are not encoded in the genome of the ctenophore Mnemiopsis leidyi. BMC Genomics 13, 714 (2012).
Traylor-Knowles, N., Vandepas, L. E. & Browne, W. E. Still enigmatic: innate immunity in the ctenophore Mnemiopsis leidyi. Integr. Comp. Biol. 59, 811–818 (2019).
Felsenstein, J. Phylogenies and the comparative method. Am. Nat. 125, 1–15 (1985).
Dunn, C. W., Giribet, G., Edgecombe, G. D. & Hejnol, A. Animal phylogeny and its evolutionary implications. Annu. Rev. Ecol. Evol. Syst. 45, 371–395 (2014).
Giribet, G. Morphology should not be forgotten in the era of genomics—a phylogenetic perspective. Zool. Anz. 256, 96–103 (2015).
Patry, W. L., Bubel, M., Hansen, C. & Knowles, T. Diffusion tubes: a method for the mass culture of ctenophores and other pelagic marine invertebrates. PeerJ 8, e8938 (2020).
Baker, L. D. The Ecology of the Ctenophore Mnemiopsis mccradyi Mayer, in Biscayne Bay, Florida (Rosenstiel School of Marine and Atmospheric Science, 1973).
Raskoff, K. A., Sommer, F. A., Hamner, W. M. & Cross, K. M. Collection and culture techniques for gelatinous zooplankton. Biol. Bull. 204, 68–80 (2003).
Greve, W. The “planktonkreisel”, a new device for culturing zooplankton. Mar. Biol. 1, 201–203 (1968).
Ward, W. W. Aquarium systems for the maintenance of ctenophores and jellyfish and for the hatching and harvesting of brine shrimp (Artemia salina) larvae. Chesap. Sci. 15, 116–118 (1974).
Kremer, P. Effect of food availability on the metabolism of the ctenophore Mnemiopsis mccradyi. Mar. Biol. 71, 149–156 (1982).
Kremer, P. & Reeve, M. R. Growth dynamics of a ctenophore (Mnemiopsis) in relation to variable food supply. II. Carbon budgets and growth model. J. Plankton Res. 11, 553–574 (1989).
Harbison, G. R., Biggs, D. C. & Madin, L. P. The associations of Amphipoda Hyperiidea with gelatinous zooplankton—II. Associations with Cnidaria. Ctenophora Radiolaria. Deep Sea Res. I 24, 465–488 (1977).
Laval, P. Hyperiid amphipods as crustacean parasitoids associated with gelatinous zooplankton. Oceanogr. Mar. Biol. Annu. Rev. 18, 11–56 (1980).
Yip, S. Y. Parasites of Pleurobrachia pileus Müller, 1776 (Ctenophora), from Galway Bay, western Ireland. J. Plankton Res. 6, 107–121 (1984).
Martorelli, S. R. Digenea parasites of jellyfish and ctenophores of the southern Atlantic. Hydrobiologia 451, 305–310 (2001).
Moss, A. G., Estes, A. M., Muellner, L. A. & Morgan, D. D. Protistan epibionts of the ctenophore Mnemiopsis mccradyi Mayer. Hydrobiologia 451, 295–304 (2001).
Reitzel, A. M. et al. Ecological and developmental dynamics of a host–parasite system involving a sea anemone and two ctenophores. J. Parasitol. 93, 1392–1402 (2007).
Zeidler, W. & Browne, W. E. A new Glossocephalus (Crustacea: Amphipoda: Hyperiidea: Oxycephalidae) from deep-water in the Monterey Bay region, California, USA, with an overview of the genus. Zootaxa 4027, 408–424 (2015).
Reitzel, A. M., Daly, M., Sullivan, J. C. & Finnerty, J. R. Comparative anatomy and histology of developmental and parasitic stages in the life cycle of the lined sea anemone Edwardsiella lineata. J. Parasitol. 95, 100–112 (2009).
Pang, K. & Martindale, M. Q. Mnemiopsis leidyi spawning and embryo collection. CSH Protoc. 2008, db.prot5085 (2008).
Freeman, G. & Reynolds, G. T. The development of bioluminescence in the ctenophore Mnemiopsis leidyi. Dev. Biol. 31, 61–100 (1973).
Martindale, M. Q. & Henry, J. J. Experimental analysis of tentacle formation in the ctenophore Mnemiopsis leidyi. Biol. Bull. 193, 245–247 (1997).
Noda, N. & Tamm, S. L. Lithocytes are transported along the ciliary surface to build the statolith of ctenophores. Curr. Biol. 24, R951–R952 (2014).
Tamm, S. L. & Moss, A. G. Unilateral ciliary reversal and motor responses during prey capture by the ctenophore Pleurobrachia. J. Exp. Biol. 114, 443–461 (1985).
Salinas-Saavedra, M. & Martindale, M. Q. Par protein localization during the early development of Mnemiopsis leidyi suggests different modes of epithelial organization in the metazoa. eLife 9, (2020).
Technau, U. Brachyury, the blastopore and the evolution of the mesoderm. Bioessays 23, 788–794 (2001).
Papaioannou, V. E. The T-box gene family: emerging roles in development, stem cells and cancer. Development 141, 3819–3833 (2014).
Yasuoka, Y., Shinzato, C. & Satoh, N. The mesoderm-forming gene brachyury regulates ectoderm-endoderm demarcation in the coral Acropora digitifera. Curr. Biol. 26, 2885–2892 (2016).
Servetnick, M. D. et al. Cas9-mediated excision of Nematostella brachyury disrupts endoderm development, pharynx formation and oral–aboral patterning. Development 144, 2951–2960 (2017).
Jinek, M. et al. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Xiao, A. et al. CasOT: a genome-wide Cas9/gRNA off-target searching tool. Bioinformatics 30, 1180–1182 (2014).
Hsu, P. D. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832 (2013).
Gagnon, J. A. et al. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS ONE 9, e98186 (2014).
Kistler, K. E., Vosshall, L. B. & Matthews, B. J. Genome engineering with CRISPR–Cas9 in the mosquito Aedes aegypti. Cell Rep. 11, 51–60 (2015).
Varshney, G. K. et al. High-throughput gene targeting and phenotyping in zebrafish using CRISPR/Cas9. Genome Res. 25, 1030–1042 (2015).
Schultz, D. T. et al. A chromosome-scale genome assembly and karyotype of the ctenophore Hormiphora californensis. G3 https://doi.org/10.1093/g3journal/jkab302 (2021).
Acknowledgements
We thank the anonymous reviewers for their time and generous feedback.
Author information
Authors and Affiliations
Contributions
Conceptualization: husbandry, W.P. and W.E.B.; genome editing, W.E.B. Methodology: husbandry, W.P. and W.E.B.; genome editing, J.S.P. and W.E.B. Investigation: husbandry, all authors; genome editing, J.S.P. and W.E.B. Validation: husbandry, all authors; genome editing, J.S.P. and W.E.B. Visualization: J.S.P. and W.E.B. Resources: husbandry, W.P. and W.E.B.; genome editing, W.E.B. Writing original draft: J.S.P., W.P. and W.E.B. Writing review and editing: all authors. Supervision: W.P. and W.E.B. Project administration: husbandry, W.P. and W.E.B.; genome editing, W.E.B. Funding acquisition: husbandry, W.P. and W.E.B.; genome editing, W.E.B.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Presnell, J. S. et al. Curr. Biol. 26, 2814–2820 (2016): https://doi.org/10.1016/j.cub.2016.08.019
Bessho-Uehara, M. et al. iScience 23, 101859 (2020): https://doi.org/10.1016/j.isci.2020.101859
Presnell, J. S. & Browne, W. E. Development 148, dev199771 (2021): https://doi.org/10.1242/dev.199771
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2
Supplementary Video 1
Demonstration of target feeding a Mnemiopsis leidyi adult. Zebrafish larvae (Danio rerio) that have been pre-washed with ASW are delivered directly to the tentacle lined feeding grooves via plastic pipette where they become ensnared in adhesive material produced by colloblast cells lining the tentilla. The trapped fish larvae are then transported orally, and engulfed
Supplementary Video 2
Juvenile Mnemiopsis leidyi cydippid capturing prey in tentacles. During juvenile stages, the two main tentacles extend into the surrounding water column from which side branching tentilla are then deployed to form a dense network of sticky colloblast cells used to ensnare nearby plankton
Supplementary Video 3
Juvenile Mnemiopsis leidyi cydippid characteristic tentacle deployment behavior. Cydippids typically swim a looping pattern, saturating the local water column with colloblast lined tentacles and tentilla to maximize prey capture
Rights and permissions
About this article
Cite this article
Presnell, J.S., Bubel, M., Knowles, T. et al. Multigenerational laboratory culture of pelagic ctenophores and CRISPR–Cas9 genome editing in the lobate Mnemiopsis leidyi. Nat Protoc 17, 1868–1900 (2022). https://doi.org/10.1038/s41596-022-00702-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-022-00702-w
This article is cited by
-
The ctenophore Mnemiopsis leidyi deploys a rapid injury response dating back to the last common animal ancestor
Communications Biology (2024)
-
Extracellular DNA traps in a ctenophore demonstrate immune cell behaviors in a non-bilaterian
Nature Communications (2024)
-
Origins and diversification of animal innate immune responses against viral infections
Nature Ecology & Evolution (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.