Abstract
Gut barrier dysfunction is often implicated in pathology following alcohol intoxication and burn injury. MicroRNAs (miRNAs) are negative regulators of gene expression that play a central role in gut homeostasis, although their role after alcohol and burn injury is poorly understood. We performed an integrated analysis of miRNA and RNA sequencing data to identify a network of interactions within small intestinal epithelial cells (IECs) which could promote gut barrier disruption. Mice were gavaged with ~ 2.9 g/kg ethanol and four hours later given a ~ 12.5% TBSA full thickness scald injury. One day later, IECs were harvested and total RNA extracted for RNA-seq and miRNA-seq. RNA sequencing showed 712 differentially expressed genes (DEGs) (padj < 0.05) in IECs following alcohol and burn injury. Furthermore, miRNA sequencing revealed 17 differentially expressed miRNAs (DEMs) (padj < 0.1). Utilizing the miRNet, miRDB and TargetScan databases, we identified both validated and predicted miRNA gene targets. Integration of small RNA sequencing data with mRNA sequencing results identified correlated changes in miRNA and target expression. Upregulated miRNAs were associated with decreased proliferation (miR-98-3p and miR-381-3p) and cellular adhesion (miR-29a-3p, miR-429-3p and miR3535), while downregulated miRNAs were connected to upregulation of apoptosis (Let-7d-5p and miR-130b-5p) and metabolism (miR-674-3p and miR-185-5p). Overall, these findings suggest that alcohol and burn injury significantly alters the mRNA and miRNA expression profile of IECs and reveals numerous miRNA–mRNA interactions that regulate critical pathways for gut barrier function after alcohol and burn injury.
Similar content being viewed by others
Introduction
Burn injury is one of the leading causes of accidental injury in the United States, contributing to approximately half a million cases and 40,000 hospitalizations each year1. Sepsis and multiple organ failure, resulting from gut barrier disruption and bacterial translocation, are the most prominent cause of death among patients with severe burn injury2,3,4,5. Alcohol use is a common confounding factor in many traumas, as it increases the risk of accidental injury6. Similar to other traumas, nearly half of reported burn injuries occur under the influence of alcohol7. Compared to patients with similar burn size and depth, individuals intoxicated at the time of burn injury require longer hospital stays and surgical procedures. Additionally, they have an increased risk of infection, sepsis, multiple organ failure, and ultimately higher mortality rates6,7,8,9,10. Therefore, alcohol intoxication at the time of burn injury is not only prevalent, but also significantly contributes to worsened patient outcomes. Consequently, the mechanisms underlying this worsened pathology are an important facet of trauma research that requires further study. To investigate how alcohol exacerbates burn pathophysiology, our laboratory utilizes a well-established mouse model of acute alcohol intoxication and burn injury. Using this model, our laboratory and others have shown that alcohol worsens intestinal inflammation, gut barrier disruption and bacterial dysbiosis after burn injury, which can contribute to the increased risk of sepsis seen in patients8,9,10.
MicroRNAs (miRNAs) are small noncoding RNAs which post-transcriptionally regulate gene expression via complementary binding to the 3′ untranslated region (UTR) of their target mRNAs. This interaction negatively impacts gene expression by either translational repression or mRNA degradation11,12. Over half of the genome is estimated to be regulated by miRNAs which regulate key signaling networks in a wide variety of cell types13,14. Recent studies demonstrate that miRNAs play a crucial role in regulating intestinal homeostasis and inflammation15,16. In addition, dysregulation of numerous miRNAs has been linked to worsened disease in models of chronic intestinal inflammation, including Inflammatory Bowel Disease (IBD) and colorectal cancer17,18,19. Far less is known about the roles of miRNA in models of acute injury and inflammation, such as trauma and burn injury. Moreover, the contributions of miRNA in regulating intestinal barrier dysfunction following alcohol and burn injury are poorly understood. Previous studies in our laboratory show decreased expression of miR-150 in intestinal epithelial cells (IECs) after alcohol and burn injury20. Additionally, we see decreased expression of Drosha and Argonaute 2 (Ago2) in intestinal epithelial cells after alcohol and burn injury20. Both Drosha and Ago2 are components of the miRNA biogenesis pathway and changes in their expression could have a significant impact on the levels of numerous miRNAs. Furthermore, studies looking at intestinal knockout of Dicer-1, an essential miRNA processing enzyme, show disruption of tight junction protein expression and localization, increased apoptosis, and increased intestinal inflammation15. This indicates that global changes in miRNA expression can adversely affect intestinal inflammation and barrier integrity and highlights the need for further research into the impact of miRNA on post-burn intestinal dysfunction.
To assess the global profile of miRNA and begin to evaluate miRNA as a molecular mechanism behind intestinal barrier disruption after alcohol and burn injury, we performed an integrated analysis of miRNA and mRNA expression in intestinal epithelial cells using our well-established mouse model. Small RNA sequencing analysis, also known as miRNA-seq, was used to generate a miRNA expression profile of intestinal epithelial cells one day following alcohol intoxication and burn injury. In addition, we performed mRNA sequencing (RNA-seq) in parallel to assess gene expression. To understand how changes in miRNA expression would impact diverse networks of gene expression, we then integrated miRNA-seq data with the RNA sequencing data to assess correlated changes in miRNA expression and their gene targets. Our findings suggest that IEC miRNA expression is considerably and globally impacted by alcohol and burn injury and is therefore likely to significantly contribute to post-burn pathogenesis. Furthermore, integrated analysis performed using correlated changes in gene and miRNA expression alongside databases of both validated and predicted miRNA gene targets, allowed us to identify mRNA–miRNA interactions relevant to intestinal barrier function, which could play a critical role in perpetuating gut barrier disruption following alcohol exposure and burn injury.
Results
Altered miRNA expression in intestinal epithelial cells after ethanol intoxication and burn injury
To examine the overall impact of ethanol intoxication and burn injury on miRNA expression in the gut, we performed miRNA-seq analysis of small intestinal epithelial cells isolated 24 h after either vehicle treatment and sham injury (n = 5) or ethanol treatment and burn injury (n = 5). Preliminary differential expression analysis (p < 0.1) yielded 65 microRNAs with altered expression in ethanol burn mice compared to sham vehicle (Fig. 1A). To further narrow our results, we adjusted p values using Benjamin–Hochburg to control for multiple testing. Using this method, we identified 17 differentially expressed microRNA (DEMs), 11 of which were significantly upregulated and 6 of which were significantly downregulated (padj < 0.1) (Fig. 1B). To elucidate the potential role of these DEMs in intestinal homeostasis after ethanol and burn injury, we utilized the miRNet Database to assess validated gene targets. The database found at miRNet is a powerful online tool that incorporates data from two widely utilized databases for experimentally validated miRNA gene targets, miRTarBase and TarBase (v8.0)21,22. Out of the 17 DEMs, 12 miRNAs (miR-1964-3p, miR-501-3p, Let-7d-5p, miR-185-5p, miR-30b-5p, miR-29a-3p, miR-429-5p, miR181d-5p, miR-429-3p, miR-26b-5p, miR-181a-5p, and miR-381-3p) were present in the miRNet Database. In total, the 5,068 validated gene targets were identified for these 12 DEMs. We then performed functional enrichment analysis on these targets using miRNet’s built in hypergeometric testing with both the KEGG and Gene Ontology (GO) databases. Overall, KEGG pathway analysis showed that our DEMs are associated with several important signal transduction pathways including forkhead box family proteins (Foxo), mitogen-activated protein kinases (MAPK), phosphatidylinositol 3-kinases (PI3K), Akt (also known as protein kinase B or PKB), tumor necrosis factor (TNF), hypoxia-inducible factor-1 (HIF-1), Janus kinases (JAKs) and signal transducer and activator of transcription proteins (STATs). In addition, we found enrichment of genes associated with endocytosis, focal adhesions, regulation of the actin cytoskeleton, ubiquitin mediated proteolysis and the cell cycle (Fig. 1C). To distinguish between pathways that are potentially upregulated or downregulated after alcohol and burn injury, we separated the upregulated DEMs from those that were downregulated. Then, we performed functional enrichment analysis on the validated gene targets identified via miRNet using GO biological process (BP) terms. We found the gene targets specific to upregulated miRNAs were highly associated with proliferation and cell differentiation, as well as the cytoskeleton and actin organization (Fig. 2A). Predicted gene targets for significantly downregulated miRNAs were most associated with metabolism involving proteins, lipids and carbohydrates, cell–cell junction organization, cell cycle arrest and cell death (Fig. 2B).
Small RNA sequencing analysis to identify differentially expressed microRNAs in small intestinal epithelial cells 1 day after alcohol and burn injury. (A) Volcano plot of differentially expressed microRNAs (p < 0.1) where each point on the graph represents an individual microRNA. Log2 fold change between ethanol burn and sham vehicle treated mice is plotted on the x-axis and the − log10 of the padj value is plotted on the y-axis. Overall, small RNA sequencing analysis identified 65 differentially expressed microRNAs (including 37 upregulated and 28 downregulated). (B) Heat map of fold change expression (log scale) of 17 differentially expressed miRNAs (DEMs) in each ethanol burn sample (EB1-5) relative to sham vehicle controls. Each row shows the fold change (log scale) of an individual DEM (padj < 0.1), ranging from downregulated (green) to upregulated (red) (C) KEGG pathway analysis of validated microRNA gene targets utilizing miRnet. Each bar represents a KEGG pathway, with the number of associated gene targets in parentheses, found to be significantly enriched (p value < 0.05) among the validated gene targets of the 17 DEMs outlined in (B).
Gene Ontology Enrichment Analysis of Upregulated and Downregulated DEMs. Validated miRNA gene targets for differentially expressed miRNAs were separated into those associated with upregulated DEMs versus downregulated DEMs. GO-BP gene set enrichment analysis was then performed using miRnet built in hypergeometric testing. Significantly enriched (p value < 0.1) pathways of interest are depicted in the bar graphs above for gene targets of either (A) significantly upregulated or (B) significantly downregulated microRNAs. Numbers in parentheses for each pathway represent to number of identified gene targets associated with the GO-BP pathway term.
Impact of ethanol and burn injury on transcriptional profile of intestinal epithelial cells
To further delineate the impact of alcohol and burn injury on gene expression, we performed RNA-seq alongside the miRNA sequencing, using the same samples to again compare small intestinal epithelial cells from sham vehicle mice (n = 5) and ethanol burn mice (n = 5). Sequencing achieved an average of 45.7 million cleaned reads per sample and an average mapping rate of 94.9%. In total, we identified 11,809 expressed genes (FPKM > 1), of which 11,078 were expressed by both treatment groups (93.8%). Differential gene expression analysis was then performed using DESeq2 and identified 712 differentially expressed genes (DEGs), including 349 genes significantly upregulated and 363 genes significantly downregulated in ethanol burn mice compared to sham vehicle (padj < 0.05) (Fig. 3A). To assess the functional impact of these gene expression changes and the differences between downregulated and upregulated genes, we utilized KEGG pathway analysis. KEGG pathway analysis of upregulated DEGs demonstrates significant enrichment (padj < 0.05) of metabolic pathways, including oxidative phosphorylation, fatty acid degradation, cytochrome p450 drug metabolism and the peroxisome (Fig. 3B). On the other hand, downregulated DEGs showed significant enrichment (padj < 0.05) in a wide variety of pathways, including cell adhesion molecules, regulation of the actin cytoskeleton, and chemokine signaling (Fig. 3C).
mRNA sequencing analysis of small intestinal epithelial cells 1 day after alcohol and burn injury. (A) Volcano plot of differentially expressed genes (padj < 0.05) where each point on the graph represents an individual gene. Log2 fold change between ethanol burn and sham vehicle treated mice is plotted on the x-axis and the − log10 of the padj value is plotted on the y-axis. Overall, mRNA sequencing analysis identified 712 differentially expressed genes (including 349 upregulated genes and 363 downregulated genes). (B,C) KEGG Pathway enrichment analysis of differentially expressed genes. Each bar represents a KEGG pathway found to be significantly enriched (p < 0.05) in either (B) upregulated or (C) downregulated genes. All significant pathways (p < 0.05) are shown.
Analysis of miRNA–mRNA interactions via integration of sequencing data
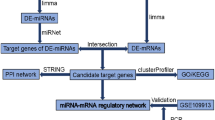
Complex networks of miRNA and their gene targets regulate a wide range of signaling pathways. An individual miRNA can have several different mRNA targets and, therefore, regulate a diverse selection of pathways. In addition, a single mRNA transcript can be targeted by multiple miRNA molecules. Consequently, networks of miRNAs cooperate to significantly impact gene expression and subsequent cellular signaling. To visualize the global range of miRNA and gene target interactions most impacted by alcohol and burn injury, we constructed an interaction network that integrates both our miRNA and RNA sequencing data. To begin, we assessed the 5068 validated gene targets identified previously for our DEMs (Fig. 1) utilizing the miRNet database for overlap with genes identified as differentially expressed by RNA sequencing. Of the 712 DEGs present in our sequencing data, 188 (26.4%) genes were validated targets of 9 of our differentially expressed miRNAs. A miRNA–mRNA network was then generated via miRNet to visualize the interactions between dysregulated genes and miRNAs after alcohol and burn injury (Fig. 4). The miRNet database provides a several measurements of network connectivity and interactions for each gene and miRNA node, including node degree, or the number of nodes directly connected to a particular node, and betweenness centrality, which is a more global look at network structure measuring the number of shortest paths through a node (Supplementary Table ST1). To assess the functional impact of the most connected, core DEMs and their gene targets, we performed GO-BP enrichment analysis on central network genes (degree > 3, betweenness > 100) using g:Profiler with g:SCS multiple testing correction method23. Significantly enriched pathways (padj < 0.05) include cell adhesion, proliferation, metabolism, and signaling pathways associated with stress and inflammatory responses (Table 1).
Integrated network of miRNAs and their predicted gene targets which are differentially expressed in small intestinal epithelial cells one day after alcohol and burn injury. miRNA–mRNA interaction network constructed using miRNet containing DEMs (blue squares) and their validated miRNA gene targets which are differentially expressed genes (DEGs) via mRNA sequencing (red circles).
As an alternative approach to elucidate potentially important gene targets for individual DEMs after alcohol and burn injury, we performed a separate integrated analysis of each DEM with our RNA sequencing data. As some of our DEMs did not have experimentally validated gene targets in the miRnet database, we expanded our analysis to include predicted gene targets for each DEM from the TargetScan and miRDB databases24,25. We then identified which targets exhibited negatively correlated differential expression via RNA sequencing analysis (i.e. for downregulated DEMs we extracted significantly upregulated genes). The results from this data aggregation is shown in Supplementary Tables ST2 and ST3. Biological pathways associated with the identified interactions for each DEM were then assessed by KEGG pathway enrichment analysis via g:Profiler with g:SCS multiple testing correction. To obtain a broad understanding of the regulatory network, we began by combining all identified targets for downregulated versus upregulated DEMs. For upregulated miRNAs, we found a total of 167 predicted gene targets that were downregulated by RNA sequencing. The significantly enriched KEGG pathways for these genes included Rap1 signaling, cellular adhesion, calcium signaling, and hormone signaling, including aldosterone, parathyroid hormone and apelin (Fig. 5). On the other hand, we identified 121 upregulated predicted gene targets for downregulated miRNAs, which were linked to metabolic pathways, TNF signaling, IL-17 signaling, and genes associated with colorectal cancer (Fig. 5). In order to gain a deeper understanding of the role each miRNA could play in gut barrier dysfunction after alcohol and burn injury, we also isolated the predicted gene targets with correlated expression for individual DEMs (Supplementary Tables ST2 and ST3) and again performed pathway enrichment analysis. Supplementary Table ST4 demonstrates that downregulated DEMs miR-674-3p and miR-185-5p exhibited the highest number of significantly associated KEGG pathways. In particular, upregulated gene targets of miR-185-5p were significantly associated with pathways that could disrupt intestinal barrier integrity, including bacterial invasion of epithelial cells and TNF signaling. Other downregulated DEMs exhibited either no significant pathway enrichment among upregulated gene targets, or just one significantly enriched pathway. For example, the peroxisome pathway was the only significantly enriched KEGG term found for Let-7d-5p associated genes. Supplementary Table ST5 demonstrates the plethora of KEGG pathways significantly associated with the downregulated gene targets of our upregulated DEMs. The most common KEGG pathways associated with upregulated DEMs included cell adhesion and tight junctions (miR-29a-3p, miR-429-3p, and miR-3535), Ras signaling (miR-429-3p and miR-26b-5p) and the cell cycle (miR-98-3p and miR-381-3p). In addition, we saw significant enrichment in hormone signaling pathways associated with GI motility and mucosal function, including oxytocin (miR-181a-5p, miR-98-3p and miR-381-3p), aldosterone (miR-429-5p, miR-181a-5p and miR-98-3p), and parathyroid hormone (miR-98-3p and miR-381-3p).
Pathway enrichment analysis using integrated analysis of differentially expressed miRNAs and gene targets with correlated expression changes. Validated and predicted gene targets for each identified DEM were collected from miRNet, TargetScan, and miRDB databases. Gene target lists were then integrated with gene expression data from mRNA sequencing. Gene targets with significantly upregulated expression were extracted for each downregulated DEM, while significantly downregulated gene targets were extracted for each upregulated DEM. The resulting upregulated and downregulated gene targets were compiled separately, and KEGG pathway enrichment analysis was performed using G:profiler with g:SCS multiple testing correction. All significantly enriched (padj < 0.05) pathways are shown in the bar graph above.
Discussion
Disruption of the gut barrier after alcohol intoxication and burn injury is known to contribute to worsened patient outcomes, including sepsis, multiple organ failure and ultimately death2,3,4,5,6,7,8. Although previous studies demonstrate reduced expression of key miRNA processing enzymes Drosha and Ago2 in intestinal epithelial cells after alcohol and burn injury, the influence of miRNA dysregulation on gut barrier function following acute trauma is understudied. The goal of this report was to assess the global impact of alcohol and burn injury on intestinal epithelial cell miRNA expression and gain deeper insight into the potential mechanisms by which miRNA could contribute to gut barrier disruption following combined injury. We found widespread changes in miRNAs and their target gene expression within intestinal epithelial cells after alcohol and burn injury. Moreover, integration of miRNA and mRNA sequencing data revealed several interactions which are highly associated with intestinal barrier integrity and provide evidence that changes in miRNA expression following alcohol intoxication and burn injury are an important component of post-burn pathogenesis.
Intestinal epithelial cells form a crucial physical barrier, which maintains homeostasis between pathogens present in the microbiome and the host defense response3,26. Disruption of this barrier promotes severe complications after burn injury, including sepsis and multiple organ failure2,4. Following alcohol and burn injury, intestinal barrier disruption is primarily instigated by heightened inflammation, loss of tight junction proteins, reduced IEC proliferation and increased IEC apoptosis27,28. In line with the literature, our RNA sequencing data shows significant association between downregulated genes and cellular adhesion pathways (Fig. 3C). Under normal conditions, intestinal homeostasis is maintained, in part, through complex interactions between miRNAs and their gene targets, which coordinate signaling pathways critical for the regulation of IEC apoptotic and inflammatory signaling, in addition to the preservation of tight junction stability15. Our findings show that widespread changes in miRNA expression after alcohol intoxication and burn injury likely contribute to disruption of normal gut homeostasis. Utilizing miRNet, a database of experimentally validated miRNA gene targets, we found that several of our differentially expressed miRNAs regulate gene targets associated with intestinal barrier integrity (Fig. 1C). Specifically, pathways associated with cellular adhesion, including organization of the actin cytoskeleton and Rho-GTPase signaling, were enriched among predicted gene targets of our upregulated DEMs (Fig. 2A). Additionally, predicted gene targets associated with negative regulation of cytoskeleton organization are enriched among downregulated DEMs (Fig. 2B). While cell adhesion is critical for maintaining an intact intestinal barrier, intestinal epithelial cells are constantly being sloughed off and replaced. This process makes the balance between cellular proliferation and cell death very important. We found that downregulated DEMs were significantly associated with cell cycle arrest and programmed cell death, including apoptosis (Fig. 2B), while cellular division and the cell cycle were among the enriched pathways for the gene targets of upregulated DEMs (Fig. 2A). As miRNAs inhibit the translation of their mRNA targets, we anticipated that these identified changes to the miRNA expression profile of IECs after alcohol and burn injury would consequently promote cell death while reducing proliferation and cellular adhesion. To further elucidate which of these DEMs are most likely contributing to intestinal dysfunction after alcohol and burn injury, we utilized two separate methods to assess correlated changes in the expression of DEMs and their gene targets. From this, we identified numerous interactions related to regulation of gut barrier integrity, including interactions which would reduce cellular proliferation (increased miR-98-3p and miR-381-3p), and adhesion (increased miR-29a-3p, miR-429-3p and miR-3535) while increasing inflammation (reduced miR-130b-5p, miR-674-3p, miR-185-5p and Let-7d-5p) and cell death (Let-7d-5p and miR-130b-5p) (Table 1, Fig. 5, and Supplementary Tables ST4 and ST5).
Following the acute phase of injury, burn patients must content with systemic and chronic hypermetabolic responses for up to years after injury. This hypermetabolic state is characterized by increased energy expenditure and metabolic rates, full body catabolism, muscle protein degradation, elevated adipose lipolysis, and insulin resistance29,30,31. Studies suggest this process is mediated by increases in both plasma catecholamine and corticosteroid levels and elevated system pro-inflammatory mediators29,32,33. It is also known that increased inflammation following chronic alcohol consumption negatively impacts insulin signaling, thereby also contributing to increases in metabolic processes33,34. Although both burn injury and chronic alcohol consumption promote hypermetabolism, there are few studies which explore the impact of combined alcohol intoxication and burn injury on metabolic outcomes. One recent study revealed significant alterations in metabolism after combined insult, including hyperglycemia and changes in serum insulin, glucagon and gastrointestinal hormones in alcohol and burn mice compared to sham or burn alone35. In our study, almost all KEGG pathways significantly associated with upregulated genes in IECs after alcohol and burn injury were metabolic in nature (Fig. 3B). The gastrointestinal tract is the initial sight of nutrient absorption and metabolism in the body. Proper metabolic function of intestinal epithelial cells is critical for both gut homeostasis and barrier function. For example, glucagon-like peptide-2 (GLP-2) is a hormone produced in the gastrointestinal epithelium that is important for nutrient absorption, lipid metabolism and energy homeostasis. However, studies also show GLP-2 promotes intestinal barrier integrity by stimulating IEC proliferation, inhibiting apoptosis, and increasing tight junction protein expression36,37. In addition to the levels of a variety of nutrients, lipid and bile acid metabolism have also been shown to impact gut barrier function and inflammation38,39,40,41. Taken together, these findings suggest that intestinal hypermetabolism after alcohol and burn injury likely plays a major role in gut dysfunction. In the last decade, miRNAs have emerged as crucial regulators of metabolism in the liver, pancreas, muscle, and adipose tissue. Dysregulation of miRNA expression has been linked to several metabolic disorders, including obesity and diabetes42,43. More recent studies demonstrate a clear relationship between intestinal epithelial cell metabolism and miRNAs. Small intestinal miRNA expression is significantly altered by high fat diet and excessive lipid exposure43,44. Furthermore, a study utilizing a mouse model of intestinal Dicer-1 knockout showed that disruption of intestinal miRNA expression results in changes to lipid absorption and accumulation45. Other studies have confirmed extensive regulation of lipid metabolism by intestinal miRNAs46. Although research exploring miRNA mediated regulation of intestinal metabolism in the context of alcohol and burn injury is lacking, our findings highlight intestinal epithelial cell metabolism as an important network impacted by miRNA changes after alcohol and burn injury. In accordance with the repressive nature of miRNAs, we found metabolic pathways (lipid biosynthesis, cellular respiration, energy derivation by oxidation of organic compounds, carbohydrate and protein metabolism) enriched among the miRNet derived validated gene targets of our downregulated DEMs (Fig. 2B). When we integrate gene expression data from RNA sequencing and extract the validated gene targets which also exhibit differential expression, the resulting network also exhibited significant association with regulation of metabolic processes (Table 1). This association with metabolism was further demonstrated in a separate integrated analysis which integrated both validated and predicted gene targets of DEMs with RNA sequencing data (Fig. 5, Supplementary Table ST2–ST5). Individual assessment of each DEM identified miR-674-3p and miR-185-5p as critical downregulated miRNAs with upregulated gene targets associated with lipid and protein metabolism (Supplementary Table ST4). Future studies investigating the mechanism by which these miRNAs influence IEC metabolism after alcohol and burn injury could provide new insights into their impact on gut barrier integrity.
A limitation of this study is the lack of ethanol alone or burn injury alone groups for comparison. However, several previous studies by our laboratory have established that alcohol or burn injury (~ 12.5%) alone do not have any significant effect on intestinal pathophysiology and show no changes in barrier function47,48,49. Furthermore, a recent study demonstrates that gene expression of microRNA biogenesis components Drosha and Ago2 are significantly downregulated in intestinal epithelial cells after combined insult but not alcohol or burn injury alone20. Although the contribution of burn and alcohol insult alone remains to be established, we feel that the findings included in this manuscript provide an adequate overview of global changes in miRNA expression and lay the foundation for future mechanistic studies underlying gut barrier dysfunction following alcohol and burn injury. We further recognize that this study is limited to male mice only and whether gender has any role in altered miRNA expression following alcohol and burn injury remains to be studied.
Altogether, our results demonstrate significant intestinal miRNA dysregulation following alcohol and burn injury. Furthermore, we revealed several miRNAs of interest, summarized in Fig. 6, which likely play an important role in decreasing gut barrier integrity by reducing intestinal proliferation (miR-98-3p and miR-381-3p), disrupting epithelial cell tight junctions (miR-29a-3p, miR-429-3p and miR-3535), and promoting intestinal apoptosis (Let-7d-5p and miR-130b-5p) and hypermetabolism (miR-674-3p and miR-185-5p). Overall, these finding open the door to future mechanistic studies that could increase our understanding of how miRNAs regulate and control gut barrier function and intestinal homeostasis.
Dysregulated miRNA expression promotes intestinal barrier disruption after alcohol and burn injury. Intestinal epithelial cells maintain the gut barrier via a variety of mechanisms that include the expression of tight junction proteins, strict regulation of cellular metabolism, and balancing cell death with cellular proliferation. Changes in these crucial functions results in disruption of the gut barrier following alcohol intoxication and burn injury, contributing to sepsis and multiple organ dysfunction. Our study indicates that dysregulated miRNA expression could significantly impact several pathways associated with intestinal barrier function after alcohol and burn injury. Here we particularly highlight upregulated miRNAs that were associated with reduced proliferation (miR-98-3p and miR-381-3p) and tight junctions (miR-29a-3p, miR-429-3p and miR3535), and downregulated miRNAs that were connected to the upregulation of apoptotic (Let-7d-5p and miR-130b-5p) and metabolic signaling (miR-674-3p and miR-185-5p).
Methods
Animals
10–12-week-old C57BL/6 male mice (23–25 g body weight) were obtained from Charles River Laboratories and maintained in animal housing facilities at Loyola University Chicago Health Sciences Division, Maywood, Illinois, USA.
Murine model of acute alcohol intoxication and burn injury
12 week old male C57/BL6 mice (22–26 g) were randomly assigned into two experimental groups: Sham injury + vehicle (water) (n = 6) or burn injury + ethanol (n = 7). On the day of injury, mice were gavaged with 400 µL of 25% ethanol in water (2.9 g/kg). Sham + vehicle animals were gavaged with 400 µL water. Four hours following the gavage, mice were given 1 mg/kg buprenorphine subcutaneously. Mice were anesthetized with a ketamine hydrochloride/xylazine cocktail (~ 80 mg/kg and ~ 1.2 mg/kg respectively) given by intraperitoneal injection. The dorsal surface of the mice was shaved before placing the mice in a prefabricated template exposing ~ 12.5% total body surface area calculated using Meeh’s formula as described by Walker and Mason(26). Burn group animals were immersed in ~ 85 °C water bath for ~ 7 s to induce a full-thickness scald burn injury. Sham animals were placed in a 37 °C water bath for 7 s. Following burn or sham injury, animals were dried gently and given 1.0 mL normal saline resuscitation by intraperitoneal injection. Animals were returned to their cages and allowed food and water ad libitum. All animal experiments were conducted in accordance with the guidelines set forth in the Animal Welfare Act and ARRIVE. In addition, they were approved by the Institutional Animal Care and Use Committee (IACUC) at Loyola University Health Sciences Division.
Small intestinal epithelial cell total RNA isolation
One day after injury, mice were euthanized, and the abdominal cavity was exposed via midline incision. ~ 8 cm of the distal small intestine was harvested and opened longitudinally and washed in cold PBS. Small intestines were then incubated in HBSS supplemented with 10 mMol/L HEPES, 50 μg/mL gentamicin, 100 U/mL penicillin, 100 μg/mL streptomycin, 5 mM EDTA and 1 mM DTT (pre-digestion solution) for 20 min at 37 °C with agitation and then vortexed to disrupt epithelial cells from the lamina propria. Epithelial cells were collected through a 100 µm strainer and the disruption process was repeated a second time50,51. Isolated epithelial cells were washed and then total RNA extracted using mirVana miRNA Isolation Kit (Invitrogen) according to manufacturer’s instructions. Total RNA concentration and purity were assessed using a Nanodrop spectrophotometer (ThermoScientific).
miRNA sequencing analysis
Isolated total RNA from small intestinal epithelial cells was submitted to Northwestern University Chicago’s NUSeq Core Facility for library preparation and miRNA sequencing (low quality RNA samples excluded and n = 5 per group chosen randomly). Analysis, including read clean up, genome mapping, and differential expression analysis, were performed at Loyola University by Dr. Michael Zilliox. Briefly, small RNA libraries were prepared using TruSeq Small RNA Library Preparation Kit according to manufacturer’s instructions. Resulting libraries were then sequenced at 100 million raw reads per sample using 75 base pair, single read sequencing via the NextSeq Illumina Platform. Raw sequencing reads were then cleaned using Cutadapt software to remove adaptor sequences and low quality reads52. The resulting clean reads were then mapped to the mouse genome, mm10. An annotation file from miRBase release 22.153,54, describing miRNA coordinates, and the sequencing alignment mappings were used as input for the Python package HTSeq to generate raw counts of miRNAs observed in the alignments55. Differential expression analysis was performed using the DESeq2 R package and Wald tests were conducted to determine significance56. Differentially expressed miRNAs were then identified using p-values both before and after adjustment using Benjamini and Hochberg’s approach to control for the false discovery rate (p value < 0.1 and padj < 0.1). Gene set enrichment analysis was performed using validated miRNA gene targets from the miRNet database21,22 and their built in software for Gene Ontology (GO) and KEGG pathway enrichment analysis57,58,59.
RNA sequencing analysis
Isolated total RNA from small intestinal epithelial cells was submitted to Novogene for library preparation and RNA sequencing analysis (low quality RNA samples excluded and n = 5 per group chosen randomly). Briefly, samples were enriched for mRNAs using oligo(dT) beads targeting the 3′ polyA tail. Enriched mRNAs were then fragmented randomly and non-stranded, non-directional library preparation performed using NEBNext kit according to manufacturer’s instructions. Resulting libraries were sequenced at 20 million raw reads per sample using 150 base pair, pair-ended sequencing via the Illumina Platform. Raw reads were obtained by CASAVA base recognition (Base Calling) and then filtered for adaptor contamination and low quality score. The resulting clean reads were then mapped to the mouse genome using STAR software60. FPKM (expected number of Fragments Per Kilobase of transcript sequence per Million base pairs sequenced) was used to quantify mRNA expression and then differential expression analysis was performed using the DESeq2 R package56. Differentially expressed genes were identified using p-values adjusted using Benjamini and Hochberg’s approach to control for the false discovery rate (padj < 0.05). Gene set enrichment analysis was performed using clusterProfiler software and the KEGG database57,58,61.
References
Association AB: Burn Incidence and Treatment in the United States: 2016. Burn Incidence Fact Sheet. https://ameriburn.org/who-we-are/media/burn-incidence-fact-sheet/
MacFie, J. et al. Gut origin of sepsis: A prospective study investigating associations between bacterial translocation, gastric microflora, and septic morbidity. Gut 45, 223–228. https://doi.org/10.1136/gut.45.2.223 (1999).
Chelakkot, C., Ghim, J. & Ryu, S. H. Mechanisms regulating intestinal barrier integrity and its pathological implications. Exp. Mol. Med. 50, 103. https://doi.org/10.1038/s12276-018-0126-x (2018).
He, W., Wang, Y., Wang, P. & Wang, F. Intestinal barrier dysfunction in severe burn injury. Burns Trauma 7, 24. https://doi.org/10.1186/s41038-019-0162-3 (2019).
Deitch, E. A. Intestinal permeability is increased in burn patients shortly after injury. Surgery 107, 411–416 (1990).
Smith, G. S., Branas, C. C. & Miller, T. R. Fatal nontraffic injuries involving alcohol: A metaanalysis. Ann. Emerg. Med. 33, 659–668. https://doi.org/10.1016/S0196-0644(99)70195-2 (1999).
Hadjizacharia, P. et al. Alcohol exposure and outcomes in trauma patients. Eur. J. Trauma Emerg. Surg. 37, 169–175. https://doi.org/10.1007/s00068-010-0038-5 (2011).
Silver, G. M. et al. Adverse clinical outcomes associated with elevated blood alcohol levels at the time of burn injury. J. Burn Care Res. 29, 784–789. https://doi.org/10.1097/BCR.0b013e31818481bc (2008).
Chen, M. M. et al. Intoxication by intraperitoneal injection or oral gavage equally potentiates postburn organ damage and inflammation. Mediat. Inflamm. 2013, 971481. https://doi.org/10.1155/2013/971481 (2013).
Shults, J. A. et al. Impaired respiratory function and heightened pulmonary inflammation in episodic binge ethanol intoxication and burn injury. Alcohol 49, 713–720. https://doi.org/10.1016/j.alcohol.2015.06.006 (2015).
Bartel, D. P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116, 281–297. https://doi.org/10.1016/s0092-8674(04)00045-5 (2004).
He, L. & Hannon, G. J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5, 522–531. https://doi.org/10.1038/nrg1379 (2004).
Friedman, R. C., Farh, K. K., Burge, C. B. & Bartel, D. P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105. https://doi.org/10.1101/gr.082701.108 (2009).
Du, T. & Zamore, P. D. Beginning to understand microRNA function. Cell Res. 17, 661–663. https://doi.org/10.1038/cr.2007.67 (2007).
McKenna, L. B. et al. MicroRNAs control intestinal epithelial differentiation, architecture, and barrier function. Gastroenterology 139, 1654–1664. https://doi.org/10.1053/j.gastro.2010.07.040 (2010).
Wang, J. Y. & Xiao, L. Posttranscriptional regulation of intestinal epithelial integrity by noncoding RNAs. Wiley Interdiscip. Rev. RNA. https://doi.org/10.1002/wrna.1399 (2017).
Cao, B. et al. Role of MiRNAs in inflammatory bowel disease. Dig. Dis. Sci. 62, 1426–1438. https://doi.org/10.1007/s10620-017-4567-1 (2017).
James, J. P. et al. MicroRNA biomarkers in IBD-differential diagnosis and prediction of colitis-associated cancer. Int. J. Mol. Sci. https://doi.org/10.3390/ijms21217893 (2020).
Tang, X. J., Wang, W. & Hann, S. S. Interactions among lncRNAs, miRNAs and mRNA in colorectal cancer. Biochimie 163, 58–72. https://doi.org/10.1016/j.biochi.2019.05.010 (2019).
Morris, N. L. et al. Dysregulation of microRNA biogenesis in the small intestine after ethanol and burn injury. Biochim. Biophys. Acta Mol. Basis Dis. 1863, 2645–2653. https://doi.org/10.1016/j.bbadis.2017.03.025 (2017).
Fan, Y. et al. miRNet—dissecting miRNA-target interactions and functional associations through network-based visual analysis. Nucleic Acids Res. 44, W135-141. https://doi.org/10.1093/nar/gkw288 (2016).
Chang, L., Zhou, G., Soufan, O. & Xia, J. miRNet 2.0: Network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 48, W244–W251. https://doi.org/10.1093/nar/gkaa467 (2020).
Raudvere, U. et al. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 47, W191–W198. https://doi.org/10.1093/nar/gkz369 (2019).
Chen, Y. & Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 48, D127–D131. https://doi.org/10.1093/nar/gkz757 (2020).
Agarwal, V., Bell, G. W., Nam, J. W. & Bartel, D. P. Predicting effective microRNA target sites in mammalian mRNAs. Elife https://doi.org/10.7554/eLife.05005 (2015).
Capaldo, C. T., Powell, D. N. & Kalman, D. Layered defense: How mucus and tight junctions seal the intestinal barrier. J. Mol. Med. (Berl.) 95, 927–934. https://doi.org/10.1007/s00109-017-1557-x (2017).
Choudhry, M. A. et al. Impaired intestinal immunity and barrier function: A cause for enhanced bacterial translocation in alcohol intoxication and burn injury. Alcohol 33, 199–208. https://doi.org/10.1016/j.alcohol.2004.05.004 (2004).
Li, X., Akhtar, S. & Choudhry, M. A. Alteration in intestine tight junction protein phosphorylation and apoptosis is associated with increase in IL-18 levels following alcohol intoxication and burn injury. Biochim. Biophys. Acta 1822, 196–203. https://doi.org/10.1016/j.bbadis.2011.09.019 (2012).
Williams, F. N., Herndon, D. N. & Jeschke, M. G. The hypermetabolic response to burn injury and interventions to modify this response. Clin. Plast. Surg. 36, 583–596. https://doi.org/10.1016/j.cps.2009.05.001 (2009).
Williams, F. N. & Herndon, D. N. Metabolic and endocrine considerations after burn injury. Clin. Plast. Surg. 44, 541–553. https://doi.org/10.1016/j.cps.2017.02.013 (2017).
Clayton, R. P., Herndon, D. N., Abate, N. & Porter, C. The effect of burn trauma on lipid and glucose metabolism: Implications for insulin sensitivity. J. Burn Care Res. 39, 713–723. https://doi.org/10.1093/jbcr/irx047 (2018).
Jeschke, M. G., Barrow, R. E. & Herndon, D. N. Extended hypermetabolic response of the liver in severely burned pediatric patients. Arch. Surg. 139, 641–647. https://doi.org/10.1001/archsurg.139.6.641 (2004).
Molina, P. E. et al. Alcohol’s burden on immunity following burn, hemorrhagic shock, or traumatic brain injury. Alcohol Res. 37, 263–278 (2015).
Lang, C. H., Derdak, Z. & Wands, J. R. Strain-dependent differences for suppression of insulin-stimulated glucose uptake in skeletal and cardiac muscle by ethanol. Alcohol Clin. Exp. Res. 38, 897–910. https://doi.org/10.1111/acer.12343 (2014).
Idrovo, J. P., Shults, J. A., Curtis, B. J., Chen, M. M. & Kovacs, E. J. Alcohol intoxication and the postburn gastrointestinal hormonal response. J. Burn Care Res. 40, 785–791. https://doi.org/10.1093/jbcr/irz083 (2019).
Brubaker, P. L. Glucagon-like peptide-2 and the regulation of intestinal growth and function. Compr. Physiol. 8, 1185–1210. https://doi.org/10.1002/cphy.c170055 (2018).
Ren, W. et al. Glucagon-like peptide-2 improve intestinal mucosal barrier function in aged rats. J. Nutr. Health Aging 22, 731–738. https://doi.org/10.1007/s12603-018-1022-8 (2018).
Basson, A. R. et al. Regulation of intestinal inflammation by dietary fats. Front. Immunol. 11, 604989. https://doi.org/10.3389/fimmu.2020.604989 (2020).
Stenman, L. K., Holma, R., Eggert, A. & Korpela, R. A novel mechanism for gut barrier dysfunction by dietary fat: Epithelial disruption by hydrophobic bile acids. Am. J. Physiol. Gastrointest. Liver Physiol. 304, G227-234. https://doi.org/10.1152/ajpgi.00267.2012 (2013).
Mroz, M. S., Lajczak, N. K., Goggins, B. J., Keely, S. & Keely, S. J. The bile acids, deoxycholic acid and ursodeoxycholic acid, regulate colonic epithelial wound healing. Am. J. Physiol. Gastrointest. Liver Physiol. 314, G378–G387. https://doi.org/10.1152/ajpgi.00435.2016 (2018).
Suzuki, T. Regulation of the intestinal barrier by nutrients: The role of tight junctions. Anim. Sci. J. 91, e13357. https://doi.org/10.1111/asj.13357 (2020).
Vienberg, S., Geiger, J., Madsen, S. & Dalgaard, L. T. MicroRNAs in metabolism. Acta Physiol. (Oxf.) 219, 346–361. https://doi.org/10.1111/apha.12681 (2017).
Sedgeman, L. R., Michell, D. L. & Vickers, K. C. Integrative roles of microRNAs in lipid metabolism and dyslipidemia. Curr. Opin. Lipidol. 30, 165–171. https://doi.org/10.1097/MOL.0000000000000603 (2019).
Gil-Zamorano, J. et al. Docosahexaenoic acid modulates the enterocyte Caco-2 cell expression of microRNAs involved in lipid metabolism. J. Nutr. 144, 575–585. https://doi.org/10.3945/jn.113.189050 (2014).
Gil-Zamorano, J. et al. Intestinal miRNAs regulated in response to dietary lipids. Sci. Rep. 10, 18921. https://doi.org/10.1038/s41598-020-75751-w (2020).
Ruiz-Roso, M. B. et al. Intestinal lipid metabolism genes regulated by miRNAs. Front. Genet. 11, 707. https://doi.org/10.3389/fgene.2020.00707 (2020).
Hammer, A. M. et al. The effects of alcohol intoxication and burn injury on the expression of Claudins and Mucins in the small and large intestines. Shock 45, 73–81. https://doi.org/10.1097/SHK.0000000000000483 (2016).
Li, X., Kovacs, E. J., Schwacha, M. G., Chaudry, I. H. & Choudhry, M. A. Acute alcohol intoxication increases interleukin-18-mediated neutrophil infiltration and lung inflammation following burn injury in rats. Am. J. Physiol. Lung Cell Mol. Physiol. 292, L1193-1201. https://doi.org/10.1152/ajplung.00408.2006 (2007).
Li, X., Akhtar, S., Kovacs, E. J., Gamelli, R. L. & Choudhry, M. A. Inflammatory response in multiple organs in a mouse model of acute alcohol intoxication and burn injury. J. Burn Care Res. 32, 489–497. https://doi.org/10.1097/BCR.0b013e3182223c9e (2011).
Hammer, A. M. et al. Interleukin-22 prevents microbial dysbiosis and promotes intestinal barrier regeneration following acute injury. Shock 48, 657–665. https://doi.org/10.1097/SHK.0000000000000900 (2017).
Weigmann, B. et al. Isolation and subsequent analysis of murine lamina propria mononuclear cells from colonic tissue. Nat. Protoc. 2, 2307–2311. https://doi.org/10.1038/nprot.2007.315 (2007).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal Bioinform. Action 17, 10–12 (2011).
Griffiths-Jones, S., Grocock, R. J., van Dongen, S., Bateman, A. & Enright, A. J. miRBase: MicroRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 34, D140-144. https://doi.org/10.1093/nar/gkj112 (2006).
Kozomara, A., Birgaoanu, M. & Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 47, D155–D162. https://doi.org/10.1093/nar/gky1141 (2019).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. https://doi.org/10.1093/bioinformatics/btu638 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. https://doi.org/10.1186/s13059-014-0550-8 (2014).
Ogata, H. et al. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 27, 29–34. https://doi.org/10.1093/nar/27.1.29 (1999).
Kanehisa, M., Goto, S., Kawashima, S. & Nakaya, A. The KEGG databases at GenomeNet. Nucleic Acids Res. 30, 42–46. https://doi.org/10.1093/nar/30.1.42 (2002).
Ashburner, M. et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29. https://doi.org/10.1038/75556 (2000).
Dobin, A. et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. https://doi.org/10.1093/bioinformatics/bts635 (2013).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287. https://doi.org/10.1089/omi.2011.0118 (2012).
Acknowledgements
The authors acknowledge the support from the National Institutes of Health R01 GM128242, T32 AA013527, and F30 DK123929.
Author information
Authors and Affiliations
Contributions
C.H. and M.C. conceptualized the study and designed experiments. C.H., M.L. and X.L. carried out the experiment. C.H. and M.Z. performed data analysis. C.H. wrote the manuscript with guidance from M.C.. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Herrnreiter, C.J., Li, X., Luck, M.E. et al. Integrated analysis of dysregulated microRNA and mRNA expression in intestinal epithelial cells following ethanol intoxication and burn injury. Sci Rep 11, 20213 (2021). https://doi.org/10.1038/s41598-021-99281-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-99281-1
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.