Abstract
CYP2C9, one of the most abundant hepatic cytochrome P450 enzymes, is involved in metabolism of 15–20% of clinically important drugs (warfarin, sulfonylureas, phenytoin, non-steroid anti-inflammatory drugs). To avoid adverse events and/or impaired drug-response, CYP2C9 pharmacogenetic testing is recommended. The impact of CYP2C9 polymorphic alleles (CYP2C9*2, CYP2C9*3) and phenoconverting non-genetic factors on CYP2C9 function and expression was investigated in liver tissues from Caucasian subjects (N = 164). The presence of CYP2C9*3 allele was associated with CYP2C9 functional impairment, and CYP2C9*2 influenced tolbutamide 4′-hydroxylase activity only in subjects with two polymorphic alleles, whereas the contribution of CYP2C8*3 was not confirmed. In addition to CYP2C9 genetic polymorphisms, non-genetic factors (co-medication with CYP2C9-specific inhibitors/inducers and non-specific factors including amoxicillin + clavulanic acid therapy or chronic alcohol consumption) contributed to the prediction of hepatic CYP2C9 activity; however, a CYP2C9 genotype–phenotype mismatch still existed in 32.6% of the subjects. Substantial variability in CYP2C9 mRNA levels, irrespective of CYP2C9 genotype, was demonstrated; however, CYP2C9 induction and non-specific non-genetic factors potentially resulting in liver injury appeared to modify CYP2C9 expression. In conclusion, complex implementation of CYP2C9 genotype and non-genetic factors for the most accurate estimation of hepatic CYP2C9 activity may improve efficiency and safety of medication with CYP2C9 substrate drugs in clinical practice.
Similar content being viewed by others
Introduction
Human CYP2C9 is one of the most abundant drug metabolizing cytochrome P450 (CYP) enzymes, being expressed in the liver at protein level similar to the CYP3A41. CYP2C9 is involved in the metabolism of approximately 15–20% of clinically used drugs, such as anticoagulants (S-acenocoumarol, S-warfarin), antiepileptics (phenytoin, valproate), non-steroid anti-inflammatory drugs (diclofenac, ibuprofen, flurbiprofen, celecoxib, valdecoxib), oral sulfonylurea antidiabetics (tolbutamide, glyburide), diuretics (torsemide, sulfinpyrazone), and antihypertensive angiotensin II receptor blockers (losartan, irbesartan)2,3,4. Many of these drugs, most prominently warfarin, acenocoumarol, sulfonylureas, valproate and phenytoin have a narrow therapeutic index; therefore, continuous monitoring of blood coagulation, serum glucose level and/or drug concentration is recommended. Substantial inter-individual variability in the metabolism of these drugs has been reported, which is mainly attributed to the genetic polymorphisms of CYP2C94,5. More than 60 CYP2C9 allelic variants have been identified, several of which are associated with changes, typically decreasing, in enzyme activity (https://www.pharmvar.org/gene/CYP2C9, access date: 19 May 2021). The polymorphic CYP2C9*2 and CYP2C9*3 alleles occur at the highest frequencies though with significant interethnic differences4,6. In Caucasian populations, CYP2C9*2 is common (10–17%), whereas it is relatively rare in Africans and Asians (2–4.6%), particularly in East Asian populations (0.03%). CYP2C9*3 allele is also frequent in Caucasian and Asian populations (6–7% and 2–11%, respectively), while it is less prevalent in African or African-American ethnicity groups (1%). Other CYP2C9 allelic variants associated with altered expression or enzyme function are rare (< 0.1%) or absent in Caucasian individuals; though some of them (CYP2C9*5, *6, *8 and *11) occur predominantly in populations with African descent4.
CYP2C9*2, one of the well-characterized CYP2C9 allele, is associated with an amino acid change (Arg144 \(\to\) Cys144) and decreased enzyme activity due to the 3608C > T transition in exon 3 (rs1799853). The Arg144Cys amino acid substitution has been demonstrated to affect the interaction of the enzyme with NADPH-cytochrome P450 oxidoreductase that can significantly impair the CYP2C9 catalytic function7. However, alterations in the CYP catalytic cycle have also been suggested to diminish metabolic activity of CYP2C9*28. CYP2C9*3 allele with a nucleotide change of 42614A > C in exon 7 (rs1057910) results in an amino acid substitution of Ile359 \(\to\) Leu359 in the active site of the enzyme that has been suggested to play an important role in substrate recognition or in formation of iron-oxene complex8,9. The Ile359Leu amino acid change leads to a significant decrease in catalytic activity due to an increase in Km values and a decrease in maximal enzyme activity (vmax) for most CYP2C9 substrates10,11. In addition, some single nucleotide polymorphisms (SNPs) in the promoter region of CYP2C9 have been identified, and these genetic variations were assumed to influence the constitutive expression or the pregnane X receptor mediated induction of CYP2C912. As a consequence of CYP2C9 polymorphisms, substantial reduction in oral clearance of many clinically important drugs, such as S-warfarin, fluvastatin, gliclazide or glimepiride, celecoxib, phenytoin or valproate, has been reported that requires dose adjustment to avoid serious adverse reactions4,5,11. In patients with loss-of-function CYP2C9*2 or CYP2C9*3 alleles, reduced ability to metabolize the anticoagulant S-warfarin and therefore increased risk of over-anticoagulation are well-established13,14. CYP2C9 genotype-guided dosing has been demonstrated to reduce bleeding complications during initiation of warfarin therapy13,15,16. In vitro studies with human liver microsomes and clinical studies involving patients with epilepsy have shown that carriers of CYP2C9*2 and/or CYP2C9*3 alleles resulted in decreased valproate metabolism compared to individuals with CYP2C9*1/*1 wild-type genotypes17,18,19. Although the association between CYP2C9 genotype and valproate blood concentrations was clearly demonstrated in children whose metabolic pathways of valproate other than CYP2C9-mediated routes are poorly developed, the impact of CYP2C9 genetic variability on valproate clearance was not significant in adult patients20.
Investigation of clinically relevant CYP2C9 polymorphisms with decreased enzyme function is important in the interpretation of altered efficacy and/or toxicity of CYP2C9 substrate drugs, particularly of those with narrow therapeutic ranges21. Applying CYP2C9 genotype testing in clinical practice may contribute to better understanding of phenotypic effect and thus to avoiding adverse events and/or impaired drug-response. Several clinical studies have been implemented to apply CYP2C9 genotype-based therapies; however, inter-individual variability in CYP2C9 activity can be partly attributed to genetic polymorphisms. Non-genetic factors (age, hormonal status, disease, co-medication, nutrition) can further modulate CYP2C9 expression and CYP2C9 activity resulting in phenoconversion6,22. Inhibition of CYP2C9 activity or transcriptional induction of CYP2C9 gene due to co-medication and other non-genetic factors can transiently switch into poor or extensive metabolizer phenotype. The CYP2C9 promoter region contains several nuclear receptor responsive elements (e.g. for pregnane X receptor, constitutive androstane receptor, glucocorticoid receptor) that are involved in transcriptional regulation of CYP2C9 expression23,24. Considering both the genetic and non-genetic variations may be required for an accurate estimation of CYP2C9-mediated drug metabolism. The present study attempted to investigate the impact of the two most common CYP2C9 alleles (CYP2C9*2 and CYP2C9*3) on CYP2C9 activity in liver tissue samples from adult organ donors belonging to Hungarian (Caucasian) population. The hepatic microsomal CYP2C9 activity was characterized by using tolbutamide as the CYP2C9-selective probe substrate. Since a minor role of CYP2C8 in tolbutamide 4′-hydroxylation has been suggested25, the impact of the clinically most relevant, loss-of-function CYP2C8*3 (rs11572080) on tolbutamide 4′-hydroxylase activity of tissue donors was also investigated. Furthermore, we aimed to identify some non-genetic factors (demographic parameters, co-medication) that can potentially modify the expression or the activity of CYP2C9. Incorporating these factors in prediction of CYP2C9 activity may improve CYP2C9 genotype-based therapy.
Material and methods
Human liver microsomes and RNA samples
Human liver tissues (N = 164) were obtained from organ transplant donors at the Department of Transplantation and Surgery, Semmelweis University (Budapest, Hungary). The present study was approved by the Hungarian Committee of Science and Research Ethics, Medical Research Council (125/PI/2011, 4799-0/2011EKU) (https://ett.aeek.hu/en/secretariat/ access date: 08 June, 2021). The study was performed in accordance with the relevant guidelines and regulations (Act CLIV of 1997 on Health, decree 23/2002 of the Minister of Health of Hungary and the declaration of Helsinki). Written informed consent for CYP testing of liver tissues was obtained from transplant recipients. Clinical histories of the tissue donors are shown in Table 1. Human livers were perfused with Euro-Collin’s solution (Fresenius AG, Bad Homburg vdH, Germany) and excised. For the isolation of microsomal fraction, the tissues were homogenized in 0.1 M Tris–HCl buffer (pH 7.4) containing 1 mM EDTA and 154 mM KCl, and differential centrifugation was performed as described by van der Hoeven and Coon26. Microsomal protein content was determined by the method of Lowry et al. with bovine serum albumin as the standard27. Total RNA was also extracted from approximately 50 mg of liver tissues homogenized in 1 ml of TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. The RNA was precipitated by using ethanol and stored in 0.1% diethylpyrocarbonate contained ultra pure water at − 80 °C for further analyses.
CYP2C9 enzyme assay
The method of Miners and Birkett was followed to determine tolbutamide 4′-hydroxylation activity selective for CYP2C928. The incubation mixture contained NADPH-generating system (1 mM NADP, 10 mM glucose 6-phosphate, 5 mM MgCl2 and 2 units/ml glucose 6-phosphate dehydrogenase), human liver microsomes (1 mg/ml) and tolbutamide (1 mM). After 20-min incubation, the reaction was terminated by ice-cold methanol and the incubation mixture was centrifuged for 10 min at 10,000 × g. High-performance liquid chromatographic analysis was performed according to published method28. CYP2C9 enzyme assay for each donor was performed in triplicate.
CYP2C genotyping
Genomic DNA was isolated from liver samples by Quick-DNA™ Universal Kit (Zymo Research, Irvine, CA). Hydrolysis SNP analysis for CYP2C9*2, CYP2C9*3 and CYP2C8*3 was performed by polymerase chain reaction (PCR) with TaqMan probes (Metabion, Planegg/Steinkirchen, Germany) as previously reported29. Real-time PCR was carried out with 30 ng of genomic DNA by using Luminaris Color Probe qPCR Master Mix (Thermo Fisher Scientific, Waltham, MA).
Analysis of CYP2C9 mRNA levels by quantitative real-time PCR
Total RNA (3 μg) was reverse-transcribed into single-stranded cDNA by using Maxima First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Wilmington, DE). The real-time PCR with human cDNA was performed by using KAPA Fast Probes Mastermix (KAPA Biosystems, Cape Town, South Africa) and TaqMan probes for CYP2C9 (BioSearch Technologies, Novato, CA). The quantity of the target RNA relative to that of the housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was determined. GAPDH expression is constant in all cells and independent from experimental conditions; therefore, its expression was set to 1, and CYP2C9 mRNA levels were normalized by GAPDH expression. The sequences of primers and probes used for the real-time PCR analyses of CYP2C9 and GAPDH expression were previously reported by Déri et al. (2020)30.
Statistical analysis
For liver tissue donors (N = 164), CYP2C9 and CYP2C8 genotypes (for CYP2C9*2, CYP2C9*3 and CYP2C8*3) as well as hepatic activities (N = 144) and/or mRNA expression (N = 109) of CYP2C9 were determined. Linkage disequilibrium between CYP2C SNPs was calculated using Haploview (v4.2; Broad Institute, Cambridge, MA)31. The frequency distribution of CYP2C9 activities were determined in 144 subjects, and three categories (low, intermediate, high) were distinguished for poor, intermediate and extensive metabolizers. The comparison of CYP2C9 enzyme activities or mRNA levels between various CYP2C9 genotype groups was performed by Kruskal–Wallis ANOVA followed by Dunn’s multiple comparisons test (GraphPad Instat v3.05; GraphPad Software, San Diego, CA). The frequencies of CYP2C9 activity reducing factors were compared in subjects carrying CYP2C9*1/*1, and the differences between high intermediate/extensive metabolizers and low intermediate/poor metabolizer subjects were calculated by Fisher’s exact test. A 2-tailed P-value < 0.05 was considered to be statistically significant.
Results
CYP2C9 genotypes of liver tissue donors
The two loss-of-function CYP2C9 alleles most common in Caucasian populations (CYP2C9*2: 3608C > T, rs1799853 and CYP2C9*3: 42614A > C, rs1057910) were identified in liver tissue donors (N = 164). The wild-type CYP2C9*1 allele was assigned in the absence of CYP2C9*2 and CYP2C9*3. The genetic linkage between CYP2C9*2 and CYP2C8*3 is well-characterized32; therefore, the loss-of-function CYP2C8*3 (2130G > A, rs11572080; 30411A > G, rs10509681) allele was also identified in the tissue donors. The relative allele frequencies of CYP2C9*2, CYP2C9*3 and CYP2C8*3 alleles were found to be similar to those in the Caucasian populations (Table 2)6,33. The majority (approximately two thirds) of organ donors carried CYP2C9*1/*1 genotype, possessing the potential for having functional CYP2C9 enzyme. More than one fourth of tissue donors were heterozygous; 24 subjects carried CYP2C9*1/*2 and 21 displayed CYP2C9*1/*3 genotype. Six subjects carried mutations (SNPs) associated with decreased CYP2C9 activity in both alleles (CYP2C9*2/*2 or CYP2C9*2/*3). CYP2C9*3/*3 genotype was not detected in the investigated population. The SNPs in CYP2C9*2 and CYP2C8*3 alleles were in significant linkage (D’ 0.87; LOD 18.8) in tissue donors all belonging to the Caucasian population, whereas other SNP pairs were in linkage disequilibrium. Most subjects with CYP2C9*1/*2 were heterozygous for CYP2C8*3 (with CYP2C8*1/*3 genotype, 19/24), whereas of those with CYP2C9*1/*3, none of the subjects carried CYP2C8*3. Of the two tissue donors with homozygous for CYP2C9*2, one was also homozygous for CYP2C8*3; however, the other carried CYP2C8*1/*3 genotype. Furthermore, of those subjects carrying the combination of both CYP2C9 alleles (CYP2C9*2/*3), two were heterozygous for CYP2C8*3 (CYP2C8*1/*3), and the other two were with homozygous wild genotype (CYP2C8*1/*1). Of 113 subjects having homozygous wild genotype (CYP2C9*1/*1), only two subjects were CYP2C8*1/*3 carrier.
Hepatic CYP2C9 activities
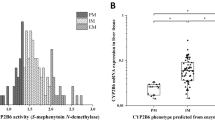
CYP2C9 enzyme activity was characterized in microsomal fractions from 144 human liver tissues. Tolbutamide was used as the probe substrate for CYP2C9, and the formation of 4′-hydroxy-tolbutamide was quantified based on “per mg microsomal protein per minute”. Variations of tolbutamide 4′-hydroxylation ranged from extremely low to rather high values displaying skewed distribution and more than two orders of magnitude differences between the lowest and the highest activities (9.47–1056 pmol*mg−1*min−1) (Fig. 1). On the basis of CYP2C9 activities, the liver tissue donors were classified into poor, intermediate and extensive metabolizer phenotype categories, and the cutoff values between the categories were 110 and 490 pmol*mg−1*min−1. In the intermediate metabolizer group, high and low intermediate metabolizer phenotypes were distinguished by the cutoff value of the median CYP2C9 activity (240 pmol*mg−1*min−1). The majority of liver tissues (N = 97) showed intermediate CYP2C9 activity, while 23 of the 144 were poor and 24 were classified as extensive metabolizers. No associations were found between CYP2C9 activities and the tissue donors’ demographic parameters, such as age and sex (P > 0.05, data not shown).
Impact of CYP2C9 genotype and non-genetic factors on hepatic CYP2C9 activities
For prediction of CYP2C9 activity and CYP2C9 phenotype from CYP2C9 genotyping data, the basic assignments of poor-intermediate-extensive metabolizer phenotypes for various CYP2C9 diplotypes were applied. Refinement of the genotype-to-phenotype classifications by Mostafa et al. and activity scoring recommended by Clinical Pharmacogenetic Implementation Consortium were also considered (Table 3)34,35. Subjects with two functional wild-type alleles were predicted to be normal metabolizers with extensive and high intermediate CYP2C9 activity. Those carrying CYP2C9*1/*2 were distinguished from those with CYP2C9*1/*3 displaying high intermediate and intermediate metabolizer phenotypes, respectively. While those subjects with two reduced-function alleles (CYP2C9*2/*2, CYP2C9*2/*3) were considered to be poor metabolizers (Table 3). However, the CYP2C9 genotype-based phenotype prediction was not always consistent with the categories based on hepatic tolbutamide 4′-hydroxylation activity. Therefore, in addition to CYP2C9 genotype, the impact of CYP2C8*3 allele and non-genetic phenoconverting factors (sex, medication, alcohol consumption) on CYP2C9 activity was investigated22,32,36,37. The impact of CYP2C9 inducers (dexamethasone, methylprednisolone, midazolam) and CYP2C9 inhibitors (amlodipine, tamoxifen) as well as of non-specific non-genetic factors, such as chronic alcohol consumption and amoxicillin + clavulanic acid therapy that can increase transcriptional expression of CYP2C9 or decrease CYP2C9 enzyme activities 3,37,38,39,40,41 was taken into account in CYP2C9 phenotype prediction (Table 3). The cohort of tissue donors were divided into four groups according to their CYP2C9 genotypes (CYP2C9*1/*1, CYP2C9*1/*2, CYP2C9*1/*3 and CYP2C9*2/*2 or CYP2C9*2/*3). Subjects carrying CYP2C9*2/*2 and CYP2C9*2/*3 were grouped, because of the low number of tissue donors, and because these genotypes were predicted to have similar effect on CYP2C9 activity.
Although in various CYP2C9 genotype groups, no gender-based differences in tolbutamide 4′-hydroxylation were demonstrated, male subjects carrying CYP2C9*1/*3 displayed significantly lower activities than those males with CYP2C9*1/*1 or CYP2C9*1/*2 genotypes (CYP2C9*1/*1, CYP2C9*1/*2, CYP2C9*1/*3 and CYP2C9*2/*2 or *2/*3: 303.7 ± 180.78, 279.1 ± 129.96, 126.1 ± 90.26 and 130.0 ± 59.02 pmol mg−1 min−1; Kruskal–Wallis ChiSq = 15.9, N = 76, P = 0.0012) (Fig. 2). In female tissue donors, however, similar differences between various genotype groups were not observed (CYP2C9*1/*1, CYP2C9*1/*2, CYP2C9*1/*3 and CYP2C9*2/*2 or *2/*3: 332.4 ± 251.99, 310.8 ± 202.79, 201.0 ± 175.71 and 135.7 ± 37.45 pmol mg−1 min−1; Kruskal–Wallis ChiSq = 5.01, N = 68, P > 0.05). Therefore, gender-independent evaluation of phenoconversion was applied for other non-genetic factors.
Most of the subjects with CYP2C9*1/*1 genotype predicting normal (high intermediate/extensive) CYP2C9 activity (Table 3) displayed intermediate (68.3%), extensive (19.8%) and poor (11.9%) tolbutamide 4′-hydroxylation (317.0 ± 216.19 pmol*mg−1*min−1) (Fig. 3A and Table 4). Of the 20 extensive metabolizers, 3 were treated with CYP2C9 inducers (dexamethasone, midazolam) that confirmed rather high CYP2C9 activity. Fourty three tissue donors, despite their homozygous wild genotype, displayed lower tolbutamide 4′-hydroxylase activity than predicted from the genotype, and were thus categorized as low intermediate and poor metabolizers. It was clearly recognized that the activity reducing non-genetic factors in the medical history of the CYP2C9*1/*1 carrier subjects were associated with low intermediate and poor CYP2C9 activities (14/43 in low intermediate/poor metabolizers vs 0/58 in high intermediate/extensive metabolizers, OR: 57.5, 95%CI: 3.31–998.61, P < 0.0001) (Fig. 3A). Fourteen of these low intermediate and poor metabolizer subjects carrying CYP2C9*1/*1 genotype were reported non-genetic factors in the medical history (amoxicillin + clavulanic acid therapy for 5, CYP2C9 inhibitor therapy for 3 and chronic alcohol consumption for 6) that explained the low tolbutamide 4′-hydroxylase activities. However, in the medical history of 29 tissue donors, there was no relevant information on the non-genetic factors responsible for low CYP2C9 activity, which means that the low tolbutamide 4′-hydroxylation activity of 28.7% of the CYP2C9*1/*1 carriers (29/101) could be explained by neither the polymorphic CYP2C9 alleles investigated nor by the non-genetic phenoconverting factors reported in the medical history, and there was still a gap between CYP2C9 genotype and phenotype. Significant differences in hepatic tolbutamide 4′-hydroxylation activity were observed between the subjects carrying CYP2C9*1/*1 or CYP2C9*1/*2 and CYP2C9*1/*3 (317.0 ± 216.19 or 299.1 ± 176.10 vs 155.3 ± 130.77 pmol*mg−1*min−1, Kruskal–Wallis ChiSq = 20.3, P = 0.0008). Although high intermediate metabolizer phenotype was predicted from CYP2C9*1/*2 genotype (Table 3), tolbutamide 4′-hydroxylation activity ranged from intermediate to extensive metabolism (Table 4). Of the 19 subjects with CYP2C9*1/*2 genotype, 4 tissue donors (2 extensive and 2 intermediate metabolizers) received CYP2C9 inducer drugs, whereas the medical history of 10 with low intermediate tolbutamide 4′-hydroxylation did not indicate any non-genetic factors that potentially decreased CYP2C9 activity (Fig. 3A). The subjects carrying CYP2C9*1/*3 (N = 18) were predicted to be intermediate metabolizers (Table 3); the CYP2C9 activities, however, extended wide range from poor to extensive metabolism (Table 4). The one extensive metabolizer subject with CYP2C9*1/*3 genotype was on CYP2C9 inducer therapy that clearly explained the relatively high activity. For one further CYP2C9*1/*3 carrier subject, amoxicillin + clavulanic acid therapy was reported in the medical history that might have resulted in low CYP2C9 activity. However, for the other 7 heterozygous subjects with CYP2C9*1/*3, there was no relevant information available that could explain the low CYP2C9 activities. For 45.9% of the tissue donors with one wild type and one polymorphic alleles (CYP2C9*1/*2 and CYP2C9*1/*3) (17/37), neither the CYP2C9 genotype nor the non-genetic factors reported in the medical history explained the lower CYP2C9 activity than predicted from the genotype. The tissue donors with two reduced function alleles (CYP2C9*2/*2 and CYP2C9*2/*3) predicting poor metabolizer phenotype (Table 3) displayed poor-intermediate CYP2C9 activities that significantly differed from the activity of the CYP2C9*1/*1 genotype group (317.0 ± 216.19 vs 131.9 ± 48.78 pmol*mg−1*min−1, P = 0.0395). For 3 of them, CYP2C9 inducer therapy explained the elevated activities, whereas for 1 subject (16.6%), CYP2C9 genotype and non-genetic phenoconverting factors could not confirm the intermediate activity. In conclusion, the CYP2C9 genotype-predicted tolbutamide 4′-hydroxylation activity (Table 3) was false in approximately half of the tissue donor subjects (68/144), and non-genetic phenoconverting factors improved the activity prediction merely to 67.4% (false in 47 of 144, 32.6%). However, the CYP2C9 genotype–phenotype mismatch still existed in 32.6% of the subjects.
Hepatic CYP2C9 activity (tolbutamide 4′-hydroxylation) (A) and CYP2C9 expression (B) in subjects carrying various CYP2C9 genotypes. CYP2C8*3 carriers and non-genetic factors (CYP2C9 inducer and inhibitor therapy, amoxicillin + clavulanic acid treatment, chronic alcohol consumption) found in clinical reports of the tissue donors are indicated. The median CYP2C9 activity (dotted line) is for the cutoff value between high and low intermediate metabolizers. PM poor metabolizer, IM intermediate metabolizer, EM extensive metabolizer, low low expression, intermediate intermediate expression, high high expression. *P < 0.05; **P < 0.001.
Although it is known that CYP2C8 is able to metabolise several of CYP2C9 substrates to some extent, it is not clear whether CYP2C8 polymorphism can contribute to the poor metabolism of tolbutamide; therefore, the effect of CYP2C8*3 on 4′-hydroxylation activity was also considered. The majority of liver tissue donors with CYP2C9*1/*2 genotype (17/19) carried the CYP2C8*3 allele, only 2 subjects with the lowest CYP2C9 activities of the CYP2C9*1/*2 group carried homozygous wild CYP2C8*1/*1 genotype. Furthermore, on the basis of tolbutamide 4′-hydroxylation activity, those subjects carrying CYP2C8*3 allele in CYP2C9*1/*1 genotype group were categorized to be high intermediate metabolizer. Of the subjects with two reduced-function alleles (CYP2C9*2/*2 and CYP2C9*2/*3), 4 carried CYP2C8*3 allele; however, they were phenotypic intermediate and poor metabolizers. From these results, CYP2C8*3 appeared to have no impact on hepatic tolbutamide 4′-hydroxylation.
Association between CYP2C9 genotype and hepatic CYP2C9 expression
In the present work, it was also investigated whether CYP2C9 genotype was associated with hepatic CYP2C9 mRNA levels in 109 tissue donors (Fig. 3B and Table 4). In the evaluation, we considered non-genetic factors in the medical history of liver tissue donors that may affect CYP2C9 expression (CYP2C9 inducer therapy, chronic alcohol consumption, amoxicillin + clavulanic acid therapy). Most of the 109 subjects (75.2%) carried CYP2C9*1/*1 genotype. Of these homozygous wild CYP2C9*1/*1 genotype subjects, 17% expressed CYP2C9 mRNA at low, 61% at intermediate and 22% at high levels. The hepatic CYP2C9 expression of the heterozygous subjects carrying either CYP2C9*1/*2 or CYP2C9*1/*3, and of those with two polymorphic alleles (CYP2C9*2/*2 and CYP2C9*2/*3) did not differ from each other or from those of CYP2C9*1/*1 subjects (CYP2C9*1/*1, CYP2C9*1/*2, CYP2C9*1/*3 and CYP2C9*2/*2 or *2/*3: 0.628 ± 0.451, 0.657 ± 0.448, 0.651 ± 0.551 and 0.243 ± 0.098; Kruskal–Wallis ChiSq = 3.31, N = 109, P > 0.05). The non-specific non-genetic factors, such as chronic alcohol consumption and amoxicillin + clavulanic acid therapy were often demonstrated to be the culprit of the low CYP2C9 expression independently from CYP2C9 genotype (for 12 subjects with CYP2C9*1/*1 genotype, for 1 with CYP2C9*1/*2, for 1 with CYP2C9*1/*3 genotype and for 1 with CYP2C9*2/*3 genotype) (Fig. 3B). Furthermore, CYP2C9 inducer therapy substantially increased CYP2C9 expression often evoking high CYP2C9 mRNA levels (for 3 subjects with CYP2C9*1/*1 genotype, for 2 with CYP2C9*1/*2 and for 2 with CYP2C9*1/*3 genotype). As a consequence of CYP2C9 inducer therapy, intermediate expression was observed in 2 additional subjects with CYP2C9*1/*2 genotype and in 2 subjects with two loss-of-function CYP2C9 alleles (CYP2C9*2/*2 and CYP2C9*2/*3).
Discussion
CYP2C9 is in the focus of pharmacogenetic studies for genotype-based drug therapy, because it is one of the most abundant hepatic CYP enzymes and catalyses the metabolism of many clinically important drugs, particularly of those with narrow therapeutic concentration range4,5,35. The association between the clearance of the anticoagulant S-warfarin and CYP2C9 polymorphism was the most intensively investigated, and to avoid bleeding complications, genotype-guided dosing was recommended13,14,15,42. Although no significant difference in in vitro losartan metabolism was found between CYP2C9*1/*2 and CYP2C9*1/*1 genotypes, a substantial decrease was detected in hepatic microsomes from subjects with CYP2C9*1/*3, CYP2C9*2/*2, and CYP2C9*3/*3 genotypes comparing to those with the homozygous wild-type genotype43. Similarly, no or negligible reduction of flurbiprofen metabolism was observed in healthy volunteers carrying CYP2C9*1/*2, whereas the metabolic rates of flurbiprofen were markedly decreased in subjects with CYP2C9*1/*3 or with two polymorphic alleles (CYP2C9*2/*2, CYP2C9*2/*3, CYP2C9*3/*3) comparing to those carrying two wild-type alleles44. Genetic variability of CYP2C9, however, can partly explain the substantial inter-individual variations in pharmacokinetics of CYP2C9 substrates. Due to phenoconversion, non-genetic factors can significantly modify CYP2C9 expression and/or enzyme activity predicted from the genotype36,45. The aim of the present work was to elucidate the impact of CYP2C9 polymorphisms common in Caucasian populations on the activity and expression of CYP2C9 in liver tissues from 164 Hungarian organ donors. CYP2C9 enzyme activity was characterized by tolbutamide 4′-hydroxylation in microsomal fractions isolated from the liver tissues. Since a minor role of CYP2C8 enzyme in the hydroxylation of tolbutamide was suggested25, the contribution of the most common CYP2C8*3 allele to the reduction of tolbutamide 4′-hydroxylation was also investigated.
The allele frequencies for CYP2C9*2, CYP2C9*3 and CYP2C8*3 in the 164 tissue donors were similar to the allele frequency data previously reported in Caucasian populations4,5,6,33. The significant linkage between the CYP2C9*2 and CYP2C8*3 alleles (D′ = 0.87) demonstrated in the liver tissue donors was concordant with previous findings32,46,47. Since several CYP2C9 substrates (montelukast, fluoxetine, ibuprofen, rosiglitazone, tolbutamide, zopiclone) are also metabolized to some extent by CYP2C848, reduced activity of CYP2C8*3 has been assumed to contribute to the low metabolic rates of CYP2C9 substrates in CYP2C9*2 carriers. A substantial decrease in ibuprofen clearance and as a consequence, increased risk of gastrointestinal bleeding have been reported in heterozygous CYP2C9*1/*2 subjects when CYP2C9*2 was present in combination with CYP2C8*346,49,50. In vitro studies on diclofenac metabolism identified CYP2C9 as the major catalyst with minor contribution of CYP2C851; however, the association between diclofenac clearance and genetic polymorphisms of CYP2C9 and CYP2C8 appear to be controversial52,53. The impact of CYP2C9 genetic polymorphisms on tolbutamide clearance has clearly been demonstrated54; however, no data available for the CYP2C8 polymorphic alleles. Although both CYP2C9*2 and CYP2C9*3 alleles are associated with decreased enzyme activity55,56, in heterozygous subjects carrying one wild-type and one polymorphic alleles (CYP2C9*1/*2 or CYP2C9*1/*3), the role of the CYP2C9*2 in modification of tolbutamide 4′-hydroxylase activity was found to be negligible in the liver tissues, whereas CYP2C9*3 was associated with significant reduction of CYP2C9 activity comparing to those with CYP2C9*1/*1 genotype. Furthermore, tolbutamide 4′-hydroxylase activity of the liver tissue donors with two loss-of-function alleles (CYP2C9*2/*2, CYP2C9*2/*3) was lower than in those with CYP2C9*1/*1. Our data are consistent with those of Kirchheiner et al.54 and Jetter et al.57, who demonstrated tolbutamide clearance in CYP2C9*1/*2 carriers to be indistinguishable from the subjects with two wild-type alleles, whereas the oral plasma clearance was significantly reduced in CYP2C9*1/*3 heterozygotes and in subjects with two polymorphic alleles comparing with those carrying CYP2C9*1/*1. In the present work, we have demonstrated for the first time that the presence of the loss-of-function CYP2C8*3 had no impact on tolbutamide 4′-hydroxylation in CYP2C9*1/*2 carriers. Even more, CYP2C8*3 in two of the CYP2C9*1/*1 carriers did not result in significant reduction of tolbutamide 4′-hydroxylase activity, but both subjects were high intermediate metabolizers.
In subjects with homozygous wild-type genotype (CYP2C9*1/*1), hepatic CYP2C9 mRNA expression has been reported to correlate with CYP2C9 activity29. However, the polymorphic CYP2C9*2 and CYP2C9*3 alleles seemed to have no impact on CYP2C9 expression. No significant differences in CYP2C9 mRNA levels were found between the subjects with CYP2C9*1/*1 genotype and those carrying one or two polymorphic CYP2C9 alleles. The C > T transition at 3608 position in CYP2C9*2 allele and the nucleotide change of 42614A > C in CYP2C9*3 are well-described to result in amino acid changes that significantly impair CYP2C9 activity; however, these SNPs seem to have no impact on CYP2C9 mRNA expression4. Our findings confirmed that the CYP2C9*2 and CYP2C9*3 polymorphic alleles did not influence the hepatic CYP2C9 mRNA expression. It also means that in those carrying polymorphic CYP2C9 alleles, CYP2C9 expression does not inform about hepatic CYP2C9 activity, whereas CYP2C9 mRNA levels correlate with CYP2C9 activity merely in those subjects with CYP2C9*1/*1 genotype, in line with previous findings29.
Because of the risk of serious adverse reactions, Clinical Pharmacogenetic Implementation Consortium has recommended CYP2C9 genotype-based activity scoring to clinicians when prescribing CYP2C9 substrate drugs, such as non-steroidal anti-inflammatory agents, anticoagulant warfarin or anticonvulsant phenytoin35,58,59; however, internal and environmental non-genetic factors have also been suggested to consider. Due to phenoconversion, the concomitant treatments with CYP2C9 inhibitors and inducers have been proposed to improve CYP2C9 phenotype prediction. As a consequence of CYP2C9 inhibitor therapy, an individual with CYP2C9*1/*1 genotype can transiently become low intermediate or poor metabolizer, while the poor-metabolizer status of subjects with two polymorphic alleles has been assumed to be not affected by inhibitors34. In liver tissue donors carrying CYP2C9*1/*1 genotype, medication with CYP2C9 inhibitors (amlodipine, tamoxifen) was associated with low tolbutamide 4′-hydroxylation activity; furthermore, amoxicillin + clavulanic acid therapy and chronic alcohol consumption resulted in a decrease in CYP2C9 activity and mRNA levels. The racemic mixture of the anti-hypertensive amlodipine is used for therapeutic purposes; however, vasodilation is ascribed only to its S-enantiomer. Amlodipine has been reported to inhibit CYP2C9 activity in a stereoselective manner, and R-enantiomer was proved to be more potent CYP2C9 inhibitor than S-amlodipine41. The risk of drug interactions with CYP2C9 substrates has also been predicted during co-administration of the selective estrogen receptor modulator tamoxifen60. Tamoxifen and its anti-estrogenic hydroxylated metabolites (4-hydroxy-tamoxifen, endoxifen and norendoxifen) potently inhibited the activities of CYP2C enzymes. These findings were confirmed by the reduced tolbutamide 4′-hydroxylation activities in hepatic microsomes of tissue donors treated with amlodipine or tamoxifen. The pathomechanism of chronic alcohol consumption induced liver disease has long been studied, and there is a large body of evidence indicating that impaired drug metabolism is related to severe liver disease61,62,63. Amoxicillin, the widely used antibiotic is often applied in combination with clavulanic acid. Hepatotoxic effect of this combination is generally mild; however, amoxicillin + clavulanic acid therapy rarely leads to drug-induced liver injury or severe acute liver failure, for which liver transplantation is the only life-saving intervention64,65. Although both chronic alcohol consumption and amoxicillin + clavulanic acid therapy have been reported to exert liver injury, information about their CYP2C9 inhibitory potential is hardly available. In contrast to amlodipine and tamoxifen that have the capability to inhibit CYP2C9 function, chronic alcohol consumption and amoxicillin + clavulanic acid therapy evoking liver injury were likely to have non-specific impact on CYP enzyme function rather than direct CYP2C9 inhibitory properties66,67.
CYP2C9 inducers have also been reported to modify CYP2C9 metabolic activity; therefore, concomitant treatment with CYP2C9 inducer drugs is recommended to take into account during phenotype prediction34. As a consequence of inducer therapy such as rifampicin, dexamethasone, carbamazepine or phenobarbitone, higher CYP2C9 activity is expected in patients with one or two wild-type CYP2C9 alleles than predicted from the genotype, whereas CYP2C9 function is assumed to be unchanged in those subjects with two polymorphic alleles34. The promoter region of the CYP2C9 gene contains a number of nuclear receptor binding sites through which the gene can be transcriptionally induced. The synthetic glucocorticoid dexamethasone or the corticosteroid methylprednisolone are known to activate glucocorticoid receptor and pregnane X receptor, and to increase CYP2C9 transcription, whereas midazolam has been reported to act as a pregnane X receptor activator and to induce the expression of CYP2C93,38,39,40,68,69. The liver tissue donors known to receive dexamethasone, methylprednisolone or midazolam displayed high CYP2C9 mRNA expression and activity. In those subjects carrying one or two wild-type alleles, the high tolbutamide 4′-hydroxylation activity and CYP2C9 mRNA level were associated with CYP2C9 inducer drug therapy, resulting in extensive or high intermediate metabolizer phenotypes. However, in those with two polymorphic alleles (CYP2C9*2/*2 or CYP2C9*2/*3), the effect of CYP2C9 inducer therapy on increasing CYP2C9 expression and function was observed, which did not confirm the phenoconversion prediction by Mostafa et al.34. Since both CYP2C9*2 and CYP2C9*3 display some residual activity, it is reasonable to assume that the presence of CYP2C9 inducers ameliorated the poor function of CYP2C9 predicted from genotype. In vitro studies with various CYP2C9 allelic variants demonstrated that the inducibility of CYP2C9*2 and CYP2C9*3 by rifampicin was similar to that of CYP2C9*112. As a result of rifampin treatment (pregnane X receptor activator), an increase in tolbutamide clearance (generally twofold) was reported in healthy volunteers with various CYP2C9 genotypes, and the CYP2C9 inducibility by rifampin was observed in all genotype groups, even in CYP2C9*2/*2, CYP2C9*3/*3 or CYP2C9*2/*3 carriers70.
Some limitations of the present study should be considered. First, CYP2C9*3/*3 genotype was not detected in the present population, although the prevalence of this genotype was reported to be 0–5.7% in Caucasian populations5,32,71,72,73. Definite conclusion nevertheless could be drawn from CYP2C9*1/*3 genotype regarding the impact of CYP2C9*3 allele on the enzyme activity and the expression of CYP2C9. Second, we did not assess CYP2C9 alleles other than CYP2C9*2 and CYP2C9*3; however, the prevalence of other clinically relevant CYP2C9 alleles in Caucasian populations are extremely low74. Third, one may assume that the medical history of some subjects was incompletely reported, and some relevant non-genetic factors that can decrease or increase CYP2C9 activity were not included.
In conclusion, the impact of CYP2C9 polymorphic alleles (CYP2C9*2, CYP2C9*3) and non-genetic factors on CYP2C9 function and mRNA expression was demonstrated in human liver tissues. The role of the CYP2C9*3 allele in functional impairment was clearly confirmed, whereas the influence of CYP2C9*2 allele on hepatic tolbutamide 4′-hydroxylation activity was evident in those subjects carrying two polymorphic alleles. Furthermore, the contribution of CYP2C8*3 to tolbutamide 4′-hydroxylase activity was not confirmed. Although CYP2C9 genotype was found to be a major factor in CYP2C9 function, non-genetic factors such as co-medication with CYP2C9 inhibitors and inducers as well as non-specific factors including amoxicillin + clavulanic acid therapy and chronic alcohol consumption significantly altered the CYP2C9 phenotype predicted from genotype. In more than two thirds of the liver tissue donors, the combined effect of the CYP2C9 genotype and non-genetic factors was found to correspond to CYP2C9 function. It should be emphasized that non-genetic factors affected tolbutamide 4′-hydroxylation activity of CYP2C9 both in CYP2C9*1/*1 carriers and in those subjects with one or two polymorphic alleles. In contrast, substantial variability in hepatic CYP2C9 mRNA levels, irrespective of the CYP2C9 genotype, was demonstrated; however, CYP2C9 induction and non-specific non-genetic factors potentially resulting in liver injury appeared to contribute to CYP2C9 expression. As such these results supported the complex implementation of CYP2C9 genotype and non-genetic factors for the most accurate estimation of hepatic CYP2C9 enzyme activity that can improve efficiency and safety of medication with CYP2C9 substrate drugs in clinical practice. However, a CYP2C9 genotype–phenotype mismatch still existed in 32.6% of the subjects.
Abbreviations
- CYP:
-
Cytochrome P450
- GAPDH:
-
Glyceraldehyde 3-phosphate dehydrogenase
- PCR:
-
Polymerase chain reaction
- SNP:
-
Single nucleotide polymorphism
References
Ohtsuki, S. et al. Simultaneous absolute protein quantification of transporters, cytochromes P450, and UDP-glucuronosyltransferases as a novel approach for the characterization of individual human liver: Comparison with mRNA levels and activities. Drug Metab. Dispos. 40, 83–92 (2012).
Miners, J. O. & Birkett, D. J. Cytochrome P4502C9: An enzyme of major importance in human drug metabolism. Br. J. Clin. Pharmacol. 45, 525–538 (1998).
Zhou, S.-F., Zhou, Z.-W., Yang, L.-P. & Cai, J.-P. Substrates, inducers, inhibitors and structure-activity relationships of human cytochrome P450 2C9 and implications in drug development. Curr. Med. Chem. 16, 3480–3675 (2009).
Daly, A. K., Rettie, A. E., Fowler, D. M. & Miners, J. O. Pharmacogenomics of CYP2C9: Functional and clinical considerations. J. Pers. Med. 8, 1–31 (2018).
Zhou, S.-F., Zhou, Z.-W. & Huang, M. Polymorphisms of human cytochrome P450 2C9 and the functional relevance. Toxicology 278, 165–188 (2010).
Zanger, U. M. & Schwab, M. Cytochrome P450 enzymes in drug metabolism: Regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 138, 103–141 (2013).
Crespi, C. L. & Miller, V. P. The R144C change in the CYP2C9*2 allele alters interaction of the cytochrome P450 with NADPH: Cytochrome P450 oxidoreductase. Pharmacogenetics 7, 203–210 (1997).
Wei, L., Locuson, C. W. & Tracy, T. S. Polymorphic variants of CYP2C9: mechanisms involved in reduced catalytic activity. Mol. Pharmacol. 72, 1280–1288 (2007).
Sano, E. et al. Mechanism of the decrease in catalytic activity of human cytochrome P450 2C9 polymorphic variants investigated by computational analysis. J. Comput. Chem. 31, 2746–2758 (2010).
Takanashi, K. et al. CYP2C9 Ile359 and Leu359 variants: Enzyme kinetic study with seven substrates. Pharmacogenetics 10, 95–104 (2000).
Kirchheiner, J., Tsahuridu, M., Jabrane, W., Roots, I. & Brockmöller, J. The CYP2C9 polymorphism: From enzyme kinetics to clinical dose recommendations. Per. Med. 1, 63–84 (2004).
Kramer, M. A., Rettie, A. E., Rieder, M. J., Cabacungan, E. T. & Hines, R. N. Novel CYP2C9 promoter variants and assessment of their impact on gene expression. Mol. Pharmacol. 73, 1751–1760 (2008).
Higashi, M. K. et al. Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. JAMA 287, 1690–1698 (2002).
Flora, D. R., Rettie, A. E., Brundage, R. C. & Tracy, T. S. CYP2C9 genotype-gependent warfarin pharmacokinetics: Impact of CYP2C9 genotype on R- and S-warfarin and their oxidative metabolites. J. Clin. Pharmacol. 57, 382–393 (2017).
Margaglione, M. et al. Genetic modulation of oral anticoagulation with warfarin. Thromb. Haemost. 84, 775–778 (2000).
Zhang, J. et al. Effect of gene-based warfarin dosing on anticoagulation control and clinical events in a real-world setting. Front. Pharmacol. 10, 1527 (2020).
Ho, P. C., Abbott, F. S., Zanger, U. M. & Chang, T. K. H. Influence of CYP2C9 genotypes on the formation of a hepatotoxic metabolite of valproic acid in human liver microsomes. Pharmacogenom. J. 3, 335–342 (2003).
Tóth, K. et al. Phenoconversion of CYP2C9 in epilepsy limits the predictive value of CYP2C9 genotype in optimizing valproate therapy. Per. Med. 12, 199–207 (2015).
Tan, L. et al. The influence of cytochrome oxidase CYP2A6, CYP2B6, and CYP2C9 polymorphisms on the plasma concentrations of valproic acid in epileptic patients. Clin. Neurol. Neurosurg. 112, 320–323 (2010).
Monostory, K. et al. Relevance of CYP2C9 function in valproate therapy. Curr. Neuropharmacol. 17, 99–106 (2019).
Pratt, V. M. et al. Recommendations for clinical CYP2C9 genotyping allele selection: A joint recommendation of the Association for Molecular Pathology and College of American Pathologists. J. Mol. Diagn. 21, 746–755 (2019).
Shah, R. R. & Smith, R. L. Addressing phenoconversion: The Achilles’ heel of personalized medicine. Br. J. Clin. Pharmacol. 79, 222–240 (2015).
Gerbal-Chaloin, S. et al. Transcriptional regulation of CYP2C9 gene. Role of glucocorticoid receptor and constitutive androstane receptor. J. Biol. Chem. 277, 209–217 (2002).
Monostory, K. & Pascussi, J.-M. Regulation of drug-metabolizing human cytochrome P450s. Acta Chim. Slov. 55, 20–37 (2008).
Komatsu, K. et al. Prediction of in vivo drug-drug interactions between tolbutamide and various sulfonamides in humans based on in vitro experiments. Drug Metab. Dispos. 28, 475–481 (2000).
van der Hoeven, T. A. & Coon, M. J. Preparation and properties of partially purified cytochrome P-450 and reduced nicotinamide adenine dinucleotide phosphate-cytochrome P-450 reductase from rabbit liver microsomes. J. Biol. Chem. 249, 6302–6310 (1974).
Lowry, O. H., Rosebrough, N. J., Farr, A. L. & Randall, R. J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193, 265–275 (1951).
Miners, J. O. & Birkett, D. J. Use of tolbutamide as a substrate probe for human hepatic cytochrome P450 2C9. Methods Enzymol. 272, 139–145 (1996).
Temesvári, M. et al. Estimation of drug-metabolizing capacity by cytochrome P450 genotyping and expression. J. Pharmacol. Exp. Ther. 341, 294–305 (2012).
Déri, M. T. et al. End-stage renal disease reduces the expression of drug-metabolizing cytochrome P450s. Pharmacol. Rep. 72, 1695–1705 (2020).
Barrett, J. C., Fry, B., Maller, J. & Daly, M. J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265 (2005).
Yasar, Ü. et al. Linkage between the CYP2C8 and CYP2C9 genetic polymorphisms. Biochem. Biophys. Res. Commun. 299, 25–28 (2002).
Xie, H. G., Prasad, H. C., Kim, R. B. & Stein, C. M. CYP2C9 allelic variants: Ethnic distribution and functional significance. Adv. Drug Deliv. Rev. 54, 1257–1270 (2002).
Mostafa, S., Kirkpatrick, C. M. J., Byron, K. & Sheffield, L. An analysis of allele, genotype and phenotype frequencies, actionable pharmacogenomic (PGx) variants and phenoconversion in 5408 Australian patients genotyped for CYP2D6, CYP2C19, CYP2C9 and VKORC1 genes. J. Neural Transm. 126, 5–18 (2019).
Theken, K. N. et al. Clinical Pharmacogenetics Implementation Consortium guideline (CPIC) for CYP2C9 and nonsteroidal anti-inflammatory drugs. Clin. Pharmacol. Ther. 108, 191–200 (2020).
Shah, R. R. et al. CYP450 genotype and pharmacogenetic association studies: A critical appraisal. Pharmacogenomics 17, 259–275 (2016).
Klomp, S. D., Manson, M. L., Guchelaar, H.-J. & Swen, J. J. Phenoconversion of cytochrome P450 metabolism: A systematic review. J. Clin. Med. 9, 2890 (2020).
Monostory, K. & Dvorak, Z. Steroid regulation of drug-metabolizing cytochromes P450. Curr. Drug Metab. 12, 154–172 (2011).
’t Hoen, P. A. C., Bijsterbosch, M. K., van Berkel, T. J., Vermeulen, N. P. E. & Commandeur, J. N. M. Midazolam is a phenobarbital-like cytochrome P450 inducer in rats. J. Pharmacol. Exp. Ther. 299, 921–927 (2001).
Vrzal, R., Kubesova, K., Pavek, P. & Dvorak, Z. Benzodiazepines medazepam and midazolam are activators of pregnane X receptor and weak inducers of CYP3A4: Investigation in primary cultures of human hepatocytes and hepatocarcinoma cell lines. Toxicol. Lett. 193, 183–138 (2010).
Krasulova, K., Holas, O. & Anzenbacher, P. Influence of amlodipine enantiomers on human microsomal cytochromes P450: Stereoselective time-dependent inhibition of CYP3A enzyme activity. Molecules 22, 1–14 (2017).
Scordo, M. G. et al. Influence of CYP2C9 and CYP2C19 genetic polymorphisms on warfarin maintenance dose and metabolic clearance. Clin. Pharmacol. Ther. 72, 702–710 (2002).
Yasar, Ü. et al. Role of CYP2C9 polymorphism in losartan oxidation. Drug Metab. Dispos. 29, 1051–1056 (2001).
Vogl, S., Lutz, R. W., Schönfelder, G. & Lutz, W. K. CYP2C9 genotype vs. metabolic phenotype for individual drug dosing—A correlation analysis using flurbiprofen as probe drug. PLoS ONE 10, e0120403 (2015).
Shah, R. R. Genotype-guided warfarin therapy: Still of only questionable value two decades on. J. Clin. Pharm. Ther. 45, 547–560 (2020).
García-Martín, E., Martínez, C., Tabarés, B., Frías, J. & Agúndez, J. A. G. Interindividual variability in ibuprofen pharmacokinetics is related to interaction of cytochrome P450 2C8 and 2C9 amino acid polymorphisms. Clin. Pharmacol. Ther. 76, 119–127 (2004).
Dreisbach, A. W. et al. The prevalence of CYP2C8, 2C9, 2J2, and soluble epoxide hydrolase polymorphisms in African Americans with hypertension. Am. J. Hypertens. 18, 1276–1281 (2005).
Backman, J. T., Filppula, A. M., Niemi, M. & Neuvonen, P. J. Role of cytochrome P450 2C8 in drug metabolism and interactions. Pharmacol. Rev. 68, 168–241 (2016).
Karaźniewicz-Łada, M., Luczak, M. & Główka, F. Pharmacokinetic studies of enantiomers of ibuprofen and its chiral metabolites in humans with different variants of genes coding CYP2C8 and CYP2C9 isoenzymes. Xenobiotica 39, 476–485 (2009).
Blanco, G. et al. Interaction of CYP2C8 and CYP2C9 genotypes modifies the risk for nonsteroidal anti-inflammatory drugs-related acute gastrointestinal bleeding. Pharmacogenet. Genom. 18, 37–43 (2008).
Bort, R. et al. Hepatic metabolism of diclofenac: Role of human CYP in the minor oxidative pathways. Biochem. Pharmacol. 58, 787–796 (1999).
Kirchheiner, J. et al. Pharmacokinetics of diclofenac and inhibition of cyclooxygenases 1 and 2: No relationship to the CYP2C9 genetic polymorphism in humans. Br. J. Clin. Pharmacol. 55, 51–61 (2003).
Dorado, P. et al. Relationship between CYP2C8 genotypes and diclofenac 5-hydroxylation in healthy Spanish volunteers. Eur. J. Clin. Pharmacol. 64, 967–970 (2008).
Kirchheiner, J. et al. Impact of CYP2C9 and CYP2C19 polymorphisms on tolbutamide kinetics and the insulin and glucose response in healthy volunteers. Pharmacogenetics 12, 101–109 (2002).
Niinuma, Y. et al. Functional characterization of 32 CYP2C9 allelic variants. Pharmacogenom. J. 14, 107–114 (2014).
Du, H. et al. Functional characterization of human CYP2C9 allelic variants in COS-7 cells. Front. Pharmacol. 7, 98 (2016).
Jetter, A. et al. Cytochrome P450 2C9 phenotyping using low-dose tolbutamide. Eur. J. Clin. Pharmacol. 60, 165–171 (2004).
Karnes, J. H. et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2C9 and HLA-B genotypes and phenytoin dosing: 2020 update. Clin. Pharmacol. Ther. 109, 302–309 (2020).
Johnson, J. A. et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for pharmacogenetics-guided warfarin dosing: 2017 update. Clin. Pharmacol. Ther. 102, 397–404 (2017).
Johänning, J. et al. The formation of estrogen-like tamoxifen metabolites and their influence on enzyme activity and gene expression of ADME genes. Arch. Toxicol. 92, 1099–1112 (2018).
Ceni, E., Mello, T. & Galli, A. Pathogenesis of alcoholic liver disease: Role of oxidative metabolism. World J. Gastroenterol. 20, 17756–17772 (2014).
Lown, K. et al. The erythromycin breath test selectively measures P450IIIA in patients with severe liver disease. Clin. Pharmacol. Ther. 51, 229–238 (1992).
Seitz, H. K. & Mueller, S. Alcoholic liver disease. In Clinical Hepatology: Principles and Practice of Hepatobiliary Diseases (ed. Dancygier, H.) 1111–1151 (Springer-Verlag, 2010).
Fontana, R. J., Shakil, A. O., Greenson, J. K., Boyd, I. & Lee, W. M. Acute liver failure due to amoxicillin and amoxicillin/clavulanate. Dig. Dis. Sci. 50, 1785–1790 (2005).
Gresser, U. Amoxicillin-clavulanic acid therapy may be associated with severe side effects—Review of the literature. Eur. J. Med. Res. 6, 139–149 (2001).
Zhang, Q. et al. Amoxicillin/clavulanic acid-warfarin drug interaction: A randomized controlled trial. Br. J. Clin. Pharmacol. 71, 232–236 (2011).
Niwa, T. et al. Effect of penicillin-based antibiotics, amoxicillin, ampicillin, and piperacillin, on drug-metabolizing activities of human hepatic cytochromes P450. J. Toxicol. Sci. 41, 143–146 (2016).
Pascussi, J.-M., Drocourt, L., Fabre, J.-M., Maurel, P. & Vilarem, M.-J. Dexamethasone induces pregnane X receptor and retinoid X receptor-α expression in human hepatocytes: Synergistic increase of CYP3A4 induction by pregnane X receptor activators. Mol. Pharmacol. 58, 361–372 (2000).
Pascussi, J.-M., Gerbal-Chaloin, S., Fabre, J.-M., Maurel, P. & Vilarem, M.-J. Dexamethasone enhances constitutive androstane receptor expression in human hepatocytes: Consequences on cytochrome P450 gene regulation. Mol. Pharmacol. 58, 1441–1450 (2000).
Vormfelde, S. V. et al. Relative impact of genotype and enzyme induction on the metabolic capacity of CYP2C9 in healthy volunteers. Clin. Pharmacol. Ther. 86, 54–61 (2009).
Dai, D. et al. Polymorphisms in human CYP2C8 decrease metabolism of the anticancer drug paclitaxel and arachidonic acid. Pharmacogenetics 11, 597–607 (2001).
Takahashi, H. & Echizen, H. Pharmacogenetics of CYP2C9 and interindividual variability in anticoagulant response to warfarin. Pharmacogenom. J. 3, 202–214 (2003).
Scordo, M. G., Caputi, A. P., D’Arrigo, C., Fava, G. & Spina, E. Allele and genotype frequencies of CYP2C9, CYP2C19 and CYP2D6 in an Italian population. Pharmacol. Res. 50, 195–200 (2004).
Zhou, Y., Ingelman-Sundberg, M. & Lauschke, V. M. Worldwide distribution of cytochrome P450 alleles: A meta-analysis of population-scale sequencing projects. Clin. Pharmacol. Ther. 102, 688–700 (2017).
Acknowledgements
This work was supported by the Grant of VEKOP-2.3.3-15-2017-00014.
Author information
Authors and Affiliations
Contributions
Study conception and design: K.Monostory, F.F.; CYP genotyping, measurement of enzyme activity and expression: F.F., K.Mangó, M.D., E.I., A.M.; data analysis and interpretation: F.F., K.Monostory; manuscript drafting: F.F., K.Monostory. All authors reviewed the manuscript and approved the final version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Fekete, F., Mangó, K., Déri, M. et al. Impact of genetic and non-genetic factors on hepatic CYP2C9 expression and activity in Hungarian subjects. Sci Rep 11, 17081 (2021). https://doi.org/10.1038/s41598-021-96590-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-96590-3
This article is cited by
-
Global distribution of functionally important CYP2C9 alleles and their inferred metabolic consequences
Human Genomics (2023)
-
Role of Cytochrome P450 2C9 in COVID-19 Treatment: Current Status and Future Directions
European Journal of Drug Metabolism and Pharmacokinetics (2023)
-
Phenytoin Metabolic Ratio, a Marker of CYP2C9 Activity, is Superior to the CYP2C9 Genotype as a Predictor of (S)-Warfarin Clearance
Clinical Pharmacokinetics (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.