Abstract
Prior knowledge of allele frequencies of cytochrome P450 polymorphisms in a population is crucial for the revision and optimization of existing medication choices and doses. In the current study, the frequency of the CYP2C9*2, CYP2C9*3, CYP2C19*2, CYP2C19*3, CYP2C19*6, CYP2C19*17, and CYP3A4 (rs4646437) alleles in a Thai population across different regions of Thailand was examined. Tests for polymorphisms of CYP2C9 and CYP3A4 were performed using TaqMan SNP genotyping assay and CYP2C19 was performed using two different methods; TaqMan SNP genotyping assay and Luminex x Tag V3. The blood samples were collected from 1205 unrelated healthy individuals across different regions within Thailand. Polymorphisms of CYP2C9 and CYP2C19 were transformed into phenotypes, which included normal metabolizer (NM), intermediate metabolizer (IM), poor metabolizer (PM), and rapid metabolizers (RM). The CYP2C9 allele frequencies among the Thai population were 0.08% and 5.27% for the CYP2C9*2 and CYP2C9*3 alleles, respectively. The CYP2C19 allele frequencies among the Thai population were 25.60%, 2.50%, 0.10%, and 1.80% for the CYP2C19*2, CYP2C19*3, CYP2C19*6, and CYP2C19*17 alleles, respectively. The allele frequency of the CYP3A4 (rs4646437) variant allele was 28.50% in the Thai population. The frequency of the CYP2C9*3 allele was significantly lower among the Northern Thai population (P < 0.001). The frequency of the CYP2C19*17 allele was significantly higher in the Southern Thai population (P < 0.001). Our results may provide an understanding of the ethnic differences in drug responses and support for the utilization of pharmacogenomics testing in clinical practice.
Similar content being viewed by others
Introduction
Genetic variations exist across different human populations and are often associated with the variation of drug response between populations1. Pharmacogenomics is the study of genetic variants that influence drug effects, typically through alterations in pharmacokinetics and pharmacodynamics2. Genetic polymorphisms in phase-1 drug-metabolizing enzymes, such as cytochrome P450 oxidases (CYPs), can alter the pharmacokinetic properties of the administered drugs, their metabolites, or both at the target site, resulting in variability in drug responses3. The genetic variability in CYP enzymes results in an enzyme with increased, normal, decreased, or no enzyme activity3. CYP enzymes consist of 57 functional members, which are classified into 18 families and 43 subfamilies based on sequence similarity. CYP enzymes metabolize the majority of the common clinically prescribed drugs4. Polymorphisms in CYP genes categorize the population as poor metabolizer (PM), intermediate metabolizer (IM), normal metabolizer (NM), rapid metabolizer (RM) and ultrarapid metabolizer (UM)5. Genetic polymorphisms within CYP2C9, CYP2C19, and CYP3A4 significantly affect most clinically used drugs, and the prediction of phenotypes by detecting polymorphisms of these CYP genes is useful in drug therapy.
The CYP2C9*2 (R144C) and CYP2C9*3 (I359L) alleles are the clinically relevant defective variants associated with decreased enzyme activity and impaired drug metabolism phenotypes. CYP2C9 metabolizes approximately 25% of clinically-administered drugs including phenytoin, warfarin, glipizide, tolbutamide and non-steroidal anti-inflammatory drugs (NSAIDs)2,3. The frequency of CYP2C9*2 is 12.68% in European, 4.60% in South Asian, 2.35% in African, and < 1% in East Asian ancestry. The frequency of CYP2C9*3 is 11.31% in South Asian, 6.88% in European, 3.38% in East Asian, and 1.26% in African ancestry6.
Among the CYP2C19 polymorphisms, CYP2C19*2, CYP2C19*3, CYP2C19*6, and CYP2C19*17 are the common variants responsible for interindividual differences in the pharmacokinetics and response to CYP2C19 substrates7. CYP2C19 is involved in the metabolism of tricyclic antidepressants, selective serotonin reuptake inhibitors, voriconazole, and clopidogrel8. The allelic frequencies of CYP2C19*2 and CYP2C19*3 responsible for the majority of PM phenotypes in the metabolism of CYP2C19 substrate drugs are higher in Asian populations9.
CYP3A4 is responsible for the metabolism of ∼50% of drugs used therapeutically such as clarithromycin, erythromycin, diltiazem, itraconazole, ketoconazole, ritonavir. There is considerable interindividual variability in CYP3A4 activity10. CYP3A4 rs4646437 polymorphism was related to the risk of hypertension in the Chinese population11. Assessment of inter-individual differences in the allele and genotype frequencies in the Thai population is important for the better outcome of pharmacotherapy. In this study, we have analyzed the frequency of specific gene polymorphisms of CYP2C9, CYP2C19, and CYP3A4 (Table 1) in the Thai population. We also compared the the real-time polymerase chain reaction technique (real-time PCR) (ViiA7) and Luminex bead-based multiplex assays for the detection of CYP2C19 variants.
Methods
Subjects
For the allele frequency of CYP2C9, CYP2C19, and CYP3A4 polymorphisms, a total of 1,205 blood specimens of unrelated healthy donors were obtained from the Thai National Health Examination Survey. The samples were selected from five regions of Thailand, including Northern, Northeastern, Central, Southern, and Bangkok. For the comparison between the real-time PCR technique (ViiA7) and Luminex bead-based multiplex assays for the detection CYP2C19 variants, voriconazole-treated patients (N = 180) were recruited between 2012 to 2015 from the Division of Infectious Disease, Department of Medicine, Faculty of Medicine, Ramathibodi Hospital, Mahidol University, Thailand. This study was approved by the ethical committee of the Ramathibodi Hospital, Faculty of Medicine, Mahidol University (MURA2013/292/SP6) and conducted in accordance with the Declaration of Helsinki. The study protocol was clearly explained to all participants and/or their legal guardians, and informed consent was obtained before the study.
Genomic DNA extraction and genotyping
Genomic DNA was extracted from EDTA blood sample using a MagNA Pure LC DNA Isolation Kit I (Roche, Mannheim, Germany) and quantified using NanoDrop ND-1000 Spectrophotometer (Thermo Fisher Scientific, DE, USA). TaqMan SNP Genotyping Assays of CYP2C9*2, CYP2C9*3, CYP2C19*2, CYP2C19*3, CYP2C19*6, CYP2C19*17, and CYP3A4 (rs4646437) were performed on the ViiA 7 real-time PCR System (Applied Biosystems, Foster City, California), according to the manufacturer's instructions. CYP2C19 genotyping using Luminex xTAG v3 was performed on the Luminex 100/200 System (Luminex Molecular Diagnostics Inc., Austin, Texas).
Statistical analysis
Statistical analyses were performed using the SPSS software package (SPSS version 18.0 for Windows, SPSS Inc., Chicago, IL, https://www.ibm.com/products/spss-statistics ). The genotype distributions were evaluated for Hardy–Weinberg equilibrium by using Fisher’s exact test. Allele frequencies in the Thai population among the different regions of Thailand were compared using the χ2-test. P-value < 0.05 was considered significance. Cohen's Kappa (κ) was used to calculate the agreement between the Luminex xTAG v3 and TaqMan SNP genotyping methods.
Results
Genotype and allele frequencies of polymorphisms of CYP2C9, CYP2C19, and CYP3A4 in Thai
The genotype and allele frequencies for all the polymorphisms screened are shown in Table 2. All the genotypes were in Hardy–Weinberg equilibrium (P > 0.05). The sample size for the analysis was 1,205. The major allele was defined as the most commonly occurring allele in the population. The population allele frequencies were calculated from genotype numbers.
In our sample of Thai unrelated healthy population, two individuals were heterozygous for the CYP2C9*2 variant. CYP2C9*2 variant allele frequency was 0.08%. Carriers of the CYP2C9*3 variant allele were found in 125 individuals; two of these individuals were homozygous for the CYP2C9*3 variant while 123 individuals were heterozygous for the CYP2C9*3 variant. CYP2C9*3 variant allele frequency was 5.27%.
Regarding CYP2C19 variants, the homozygous CYP2C19*2 variant was found in 64 individuals, while 489 individuals were heterozygous for the CYP2C19*2 variant. The allele frequency of the CYP2C19*2 variant was 25.6%. Fifty-eight individuals carried the CYP2C19*3 variant allele; one individual was homozygous for the CYP2C19*3 variant while 57 individuals were heterozygous for the CYP2C19*3 variant. CYP2C19*3 variant allele frequency was 2.5%. The heterozygous CYP2C19*6 variant was found in two individuals. CYP2C19*6 variant allele frequency was 0.1%. Forty-three individuals were heterozygous for the CYP2C19*17 variant, and the CYP2C19*17 variant allele frequency was 1.8%.
Carriers of the CYP3A4 (rs4646437) variant allele were found in 595 individuals in this study population; 89 of these individuals were homozygous for the rs4646437 variant while 506 individuals were heterozygous for the rs4646437 variant. The allele frequency of the rs4646437 variant allele was 28.5%.
Frequency distribution of polymorphisms of CYP2C9, CYP2C19, and CYP3A4 in different regions of Thailand
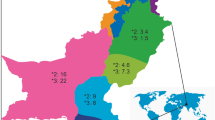
The distribution of CYP2C9, CYP2C19, and CYP3A4 alleles in different regions of Thailand are shown in Table 3 and Fig. 1. The frequency of the CYP2C9*3 allele was significantly lower among the Northern Thai population (P < 0.001). The frequency of the CYP2C19*17 allele was significantly higher in the Southern Thai population (P < 0.001). Overall, the prevalence of CYP2C19 alleles was significantly different among the different regions of Thailand (P < 0.001). The frequency of the CYP3A4 rs4646437 allele in different regions of Thailand was found to be comparable.
The distribution of CYP2C9, CYP2C19, and CYP3A4 alleles in Thai population. The figure was created using Microsoft Excel 365 (https://www.office.com).
Frequencies of the CYP2C9 and CYP2C19 genotypes and predicted phenotypes in a sample of the Thai population
The CYP2C9 and CYP2C19 genotype and phenotype frequencies are summarized in Table 4. In our sample of the Thai population, 89.46% possessed a wild-type CYP2C9 genotype while 10.54% possessed a mutated CYP2C9 genotype. There were 1078 subjects (89.46%) classified as CYP2C9 NM (*1/*1), 125 subjects (10.37%) classified as CYP2C9 IM (*1/*2 and *1/*3), and 2 subjects (0.17%) classified as CYP2C9 PM (*3/*3).
Regarding CYP2C19, 48.22% possessed wild type CYP2C19 genotype while 51.78% possessed a mutated CYP2C19 genotype. There were 581 subjects (48.22%) classified as CYP2C19 NM (*1/*1), 518 subjects (42.98%) classified as CYP2C19 IM (*1/*2, *1/*3, *1/*6, and *2/*17), 80 subjects (6.64%) classified as CYP2C19 PM (*2/*2, *2/*3, and *3/*3), and 26 subjects (2.16%) classified as CYP2C19 RM (*1*17).
Analysis of CYP2C19 variants accordance between Luminex xTAG and TaqMan real-time PCR
We compared the allele and genotype frequencies pattern of CYP2C19 variants between Luminex xTAG and TaqMan real-time PCR platforms. CYP2C19 variants genotyped by TaqMan real-time PCR included *2, *3, and *17. CYP2C19 variants genotyped by Luminex xTAG included *2, *3, *4, *5, *6, *7, *8, *9, *10, and *17. The presence of the wild type allele, CYP2C19*1, was inferred from the absence of other CYP2C19 variants. Detailed comparisons of alleles and genotypes frequencies produced by the Luminex xTAG and TaqMan real-time PCR platforms are shown in Supplementary Tables 1 and 2. The accordant rate for allele and genotype frequencies was 98.89% (178/180) between the two platforms. The Kappa value was 0.931 indicating an almost perfect agreement between the two platforms.
Discussion
The interethnic variability in the drug metabolism capacity and pharmacokinetics between populations is the impact of the differences in the allele distribution of pharmacogenes14. Drug metabolizing enzymes CYP2C9, CYP2C19, and CYP3A4 are polymorphic and cause substantial interpersonal differences in drug and metabolite exposure15. To date, numerous studies have analyzed the frequencies of the polymorphisms in CYP2C9, CYP2C19, and CYP3A4 in the Thai population; yet the available frequency data have been derived from a small sample of the Thai population. In this study, we report the frequency of specific gene polymorphisms of CYP2C9, CYP2C19, and CYP3A4 in 1,205 Thai subjects. Furthermore, this works are able to represent the majority allele frequencies of Thai people. Since the samples were recruited to cover all the regions in Thailand.
Regarding CYP2C9, the frequencies of the allelic variants CYP2C9*2 and CYP2C9*3 in the present study were consistent with the figures reported for East Asians in PharmGKB (https://www.pharmgkb.org/variant/PA166153972, accessed September 06, 2020). The frequency of CYP2C9*3 was slightly higher in our study compared to what has been reported in PharmGKB for East Asians (5.27% vs 3.37%). About differences in the frequency of alleles across different regions of Thailand, the frequency of the CYP2C9*3 allele was significantly lower among the Northern Thai population (Table 3). The CYP2C9*2 allele was observed with a frequency of 0.08% in our study. By contrast, the CYP2C9*2 allele frequency was zero in previous reports that were examined in Thais16,17. Pratt et al. reported that CYP2C9*2 and CYP2C9*3 have decreased function and are the most well-characterized variant CYP2C9 alleles compared with other alleles18. Both CYP2C9*2 and CYP2C9*3 alleles have been reported at higher frequencies among the Caucasian population19. Concerning the metabolic predicted phenotypes, the frequencies of CYP2C9 IM and PM were 10.37% and 0.17%, respectively. Interestingly, these frequencies were more consistent with the Han Chinese population (8.23% IM and 0.16% PM)20. The Clinical Pharmacogenetics Implementation Consortium (CPIC) has published recommendations for pharmacogenetic testing on the CYP2C9 gene for dosing of phenytoin, (NSAIDs), and warfarin21,22,23.
In our research, we determined and compared the frequency of four pharmacologically relevant CYP2C19 variants among Thai populations. The frequencies of CYP2C19*2, CYP2C19*3, CYP2C19*6, and CYP2C19*17 were 25.6%, 2.5%, 0.1%, and 1.8% respectively. We observed the significant variability in the distribution of CYP2C19 variants across different regions of Thailand. The CYP2C19*2, CYP2C19*3, and CYP2C19*6 alleles are the no-function alleles and characterized as PM, while the CYP2C19*17 allele is associated with high CYP2C19 activity and characterized as UM24,25,26,27. The frequency of the CYP2C19*2 allele in our study is less than the frequency in Asian populations (~ 30%) but higher than the allele frequency of 18% in Africans and Europeans26. The prevalence of the CYP2C19*3 allele presented in our study is lower than that of Han Chinese (7%), Koreans (12%), Japanese (11%), and Vietnamese (14%)28. The distribution of CYP2C19*2 and CYP2C19*3 alleles in Northeastern Thailand are consistent with the prior study of Tassaneeyakul et al. conducted in 774 Thais29. We observed a very low frequency of the CYP2C19*6 allele in our study. The prevalence of the CYP2C19*6 allele is very low, and sometimes absent in general populations30. The CYP2C19*17 allele, associated with accelerated metabolism, is prevalent at 18.2% in African-American, 6.2% in Asian, 15.8% in Caucasian, 15.2% in Hispanic, and 19.8% in Ashkenazi Jewish populations30. The frequency of the NM, IM, PM, and UM phenotypes was 48.22%, 42.98%, 6.64%, and 2.16% respectively. Notably, a similar trend was observed for the IM among the Thais and Han Chinese in previous studies20,31. The CYP2C19 metabolic phenotypes are uniformly distributed across African-American, Asian, Caucasian, Hispanic, and Ashkenazi Jewish populations30. Currently, there are dosing guidelines from the CPIC for CYP2C19 genotyping and the personalization of clopidogrel, tricyclic antidepressants, selective serotonin reuptake inhibitors, voriconazole, and proton pump inhibitor therapy32,33,34,35,36.
The CYP3A4 enzyme is the most abundant metabolic enzyme in the human liver, encoded by the CYP3A4 gene37. A lot of studies have reported the effects of CYP3A4 rs4646437 on drug metabolism and outcomes of drug therapy38,39,40,41. The frequency of CYP3A4 rs4646437 in our study was 28.5% in the Thai population and 29.9% in the northeastern Thai population, which is slightly higher from the findings in another study conducted in the northeastern Thai population42. According to the 1000 Genomes project, the CYP3A4 rs4646437 is highly prevalent among African, East Asian, and South Asian populations, but not so frequent among the Europeans (https://www.pharmgkb.org/variant/PA166157391).
In addition to the allele, genotype, and phenotype frequencies in our study subjects, we also compared the Luminex xTAG v3 and TaqMan SNP genotyping methods for the detection of CYP2C19 variants (Supplementary Table 3). Because of the lower turnaround time (~ 2.15 h), simple workflow, and lesser amount of DNA needed for PCR in TaqMan assay, we conclude that TaqMan SNP genotyping assay is better than Luminex xTAG v3 system for the detection of CYP2C19 variants in Thai population.
Conclusions
In conclusion, our findings confirm the interregional differences in the CYP2C9, CYP2C19, and CYP3A4 allele and genotype frequencies among Thais. Our results further support the need to identify individuals who have altered pharmacokinetics for CYP2C9, CYP2C19, or CYP3A4 substrates so that prescribers could personalize appropriate dosages for optimal drug responses. TaqMan SNP genotyping assay is better than the Luminex xTAG v3 system in the genotyping of CYP2C19 variants in the Thai population.
References
Bachtiar, M. & Lee, C. G. L. Genetics of population differences in drug response. Curr. Genet. Med. Rep. 1, 162–170 (2013).
Relling, M. V. & Evans, W. E. Pharmacogenomics in the clinic. Nature 526(7573), 343–350 (2015).
Ahmed, S. et al. Pharmacogenomics of drug metabolizing enzymes and transporters: Relevance to precision medicine. Genomics Proteomics Bioinform. 14(5), 298–313 (2016).
Preissner, S.C. et al. Polymorphic cytochrome P450 enzymes (CYPs) and their role in personalized therapy. PLoS One 8(12), e82562 (2013).
Dorji, P. W., Tshering, G. & Na-Bangchang, K. CYP2C9, CYP2C19, CYP2D6 and CYP3A5 polymorphisms in South-East and East Asian populations: A systematic review. J. Clin. Pharm. Ther. 44(4), 508–524 (2019).
Daly, A.K. et al. Pharmacogenomics of CYP2C9: Functional and clinical considerations. J. Pers. Med. 8(1) (2017).
Lee, S. J. Clinical application of CYP2C19 pharmacogenetics toward more personalized medicine. Front. Genet. 3, 318 (2012).
Petrovic, J., Pesic, V. & Lauschke, V. M. Frequencies of clinically important CYP2C19 and CYP2D6 alleles are graded across Europe. Eur. J. Hum. Genet. 28(1), 88–94 (2020).
Zhu, W. Y. et al. Association of CYP2C19 polymorphisms with the clinical efficacy of clopidogrel therapy in patients undergoing carotid artery stenting in Asia. Sci. Rep. 6, 25478 (2016).
Zhou, X. Y. et al. Enzymatic activities of CYP3A4 allelic variants on quinine 3-hydroxylation in vitro. Front. Pharmacol. 10, 591 (2019).
Wang, J. et al. Association between CYP3A4 gene rs4646437 polymorphism and the risk of hypertension in Chinese population: A case-control study. Biosci. Rep. 39(4) (2019).
Ingelman-Sundberg, M. The Human Cytochrome P450 (CYP) Allele Nomenclature Database
Schirmer, M. et al. Sex-dependent genetic markers of CYP3A4 expression and activity in human liver microsomes. Pharmacogenomics 8(5), 443–453 (2007).
van Dyk, M. et al. Assessment of inter-racial variability in CYP3A4 activity and inducibility among healthy adult males of Caucasian and South Asian ancestries. Eur. J. Clin. Pharmacol. 74(7), 913–920 (2018).
Bishop, J. R. Pharmacogenetics. Handb. Clin. Neurol. 147, 59–73 (2018).
Maneechay, W. et al. Genotype distributions of CYP2C9 and VKORC1 in southern Thais and their association with warfarin maintenance dose in patients with cardiac surgery. Asian Biomed. 11(1), 81–87 (2017).
Sangviroon, A. et al. Pharmacokinetic and pharmacodynamic variation associated with VKORC1 and CYP2C9 polymorphisms in Thai patients taking warfarin. Drug Metab. Pharmacokinet. 25(6), 531–538 (2010).
Pratt, V. M. et al. Recommendations for clinical CYP2C9 genotyping allele selection: A joint recommendation of the Association for Molecular Pathology and College of American Pathologists. J. Mol. Diagn. 21(5), 746–755 (2019).
Cavallari, L.H., Momary, K.M. Pharmacogenetics in Cardiovascular Diseases. Pharmacogenomics: Challenges and Opportunities in Therapeutic Implementation, 2nd edn (eds. Lam, Y.W.F., Scott, S.A.). (Academic Press, 2019).
He, L. et al. Genetic and phenotypic frequency distribution of CYP2C9, CYP2C19 and CYP2D6 in over 3200 Han Chinese. Clin. Exp. Pharmacol. Physiol. 47(10), 1659–1663 (2020).
Johnson, J. A. et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for pharmacogenetics-guided warfarin dosing: 2017 update. Clin. Pharmacol. Ther. 102(3), 397–404 (2017).
Karnes, J.H. et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2C9 and HLA-B genotypes and phenytoin dosing: 2020 update. Clin. Pharmacol. Ther. (2020).
Theken, K. N. et al. Clinical Pharmacogenetics Implementation Consortium Guideline (CPIC) for CYP2C9 and nonsteroidal anti-inflammatory drugs. Clin. Pharmacol. Ther. 108(2), 191–200 (2020).
Ibeanu, G. C. et al. Identification of new human CYP2C19 alleles (CYP2C19*6 and CYP2C19*2B) in a Caucasian poor metabolizer of mephenytoin. J. Pharmacol. Exp. Ther. 286(3), 1490–1495 (1998).
Ionova, Y. et al. CYP2C19 allele frequencies in over 2.2 million direct-to-consumer genetics research participants and the potential implication for prescriptions in a large health system. Clin. Transl. Sci. (2020).
Jarrar, M. et al. Cytochrome allelic variants and clopidogrel metabolism in cardiovascular diseases therapy. Mol. Biol. Rep. 43(6), 473–484 (2016).
Takahashi, M. et al. Functional characterization of 21 CYP2C19 allelic variants for clopidogrel 2-oxidation. Pharmacogenomics J. 15(1), 26–32 (2015).
Zhong, Z. et al. Analysis of CYP2C19 genetic polymorphism in a large ethnic Hakka Population in Southern China. Med. Sci. Monit. 23, 6186–6192 (2017).
Tassaneeyakul, W. et al. CYP2C19 genetic polymorphism in Thai, Burmese and Karen populations. Drug Metab. Pharmacokinet. 21(4), 286–290 (2006).
Martis, S. et al. Multi-ethnic distribution of clinically relevant CYP2C genotypes and haplotypes. Pharmacogenomics J. 13(4), 369–377 (2013).
Sukasem, C. et al. CYP2C19 polymorphisms in the Thai population and the clinical response to clopidogrel in patients with atherothrombotic-risk factors. Pharmgenomics Pers. Med. 6, 85–91 (2013).
Scott, S. A. et al. Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin. Pharmacol. Ther. 94(3), 317–323 (2013).
Hicks, J. K. et al. Clinical Pharmacogenetics Implementation Consortium guideline for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants. Clin. Pharmacol. Ther. 93(5), 402–408 (2013).
Hicks, J. K. et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin. Pharmacol. Ther. 98(2), 127–134 (2015).
Moriyama, B. et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for CYP2C19 and voriconazole therapy. Clin. Pharmacol. Ther. 102(1), 45–51 (2017).
Lima, J.J. et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2C19 and proton pump inhibitor dosing. Clin. Pharmacol. Ther. (2020).
Wang, L. et al. Effects of CYP3A4 polymorphisms on drug addiction risk among the Chinese Han population. Front. Public Health 7, 315 (2019).
Diekstra, M. H. et al. Sunitinib-induced hypertension in CYP3A4 rs4646437 A-allele carriers with metastatic renal cell carcinoma. Pharmacogenomics J. 17(1), 42–46 (2017).
Liu, J. et al. Donor and recipient P450 gene polymorphisms influence individual pharmacological effects of tacrolimus in Chinese liver transplantation patients. Int. Immunopharmacol. 57, 18–24 (2018).
He, H. R. et al. Effects of CYP3A4 polymorphisms on the plasma concentration of voriconazole. Eur. J. Clin. Microbiol. Infect. Dis. 34(4), 811–819 (2015).
Sharaki, O. et al. Impact of CYP3A4 and MDR1 gene (G2677T) polymorphisms on dose requirement of the cyclosporine in renal transplant Egyptian recipients. Mol. Biol. Rep. 42(1), 105–117 (2015).
Areesinpitak, T., et al. Prevalence of CYP2C19, CYP3A4 and FMO3 genetic polymorphisms in healthy northeastern Thai volunteers. ScienceAsia 46 (2020).
Acknowledgements
This study was supported by grants from the (1) Faculty of Medicine, Ramathibodi Hospital, Mahidol University, (2) The Health System Research Institute under Genomics Thailand Strategic Fund and (3) The International Research Network-The Thailand Research Fund (IRN60W003).
Author information
Authors and Affiliations
Contributions
R.S. wrote the manuscript; S.C., A.P., and C.S. designed the research; N.K., T.J., S.P., J.R., and N.J. collected the samples and data; R.S., N.K., and P.J. performed the research; R.S. and N.N. analyzed the data; R.S., S.C. and C.S. revised the critical revision of the article; A.P. and C.S. approved the final version to be published.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sukprasong, R., Chuwongwattana, S., Koomdee, N. et al. Allele frequencies of single nucleotide polymorphisms of clinically important drug-metabolizing enzymes CYP2C9, CYP2C19, and CYP3A4 in a Thai population. Sci Rep 11, 12343 (2021). https://doi.org/10.1038/s41598-021-90969-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-90969-y
This article is cited by
-
CYP2C19 loss-of-function is associated with increased risk of hypertension in a Hakka population: a case-control study
BMC Cardiovascular Disorders (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.