Abstract
As an approach to the exploration of cold-active enzymes, in this study, we isolated a cold-active protease produced by psychrotrophic bacteria from glacial soils of Thajwas Glacier, Himalayas. The isolated strain BO1, identified as Bacillus pumilus, grew well within a temperature range of 4–30 °C. After its qualitative and quantitative screening, the cold-active protease (Apr-BO1) was purified. The Apr-BO1 had a molecular mass of 38 kDa and showed maximum (37.02 U/mg) specific activity at 20 °C, with casein as substrate. It was stable and active between the temperature range of 5–35 °C and pH 6.0–12.0, with an optimum temperature of 20 °C at pH 9.0. The Apr-BO1 had low Km value of 1.0 mg/ml and Vmax 10.0 µmol/ml/min. Moreover, it displayed better tolerance to organic solvents, surfactants, metal ions and reducing agents than most alkaline proteases. The results exhibited that it effectively removed the stains even in a cold wash and could be considered a decent detergent additive. Furthermore, through protein modelling, the structure of this protease was generated from template, subtilisin E of Bacillus subtilis (PDB ID: 3WHI), and different methods checked its quality. For the first time, this study reported the protein sequence for psychrotrophic Apr-BO1 and brought forth its novelty among other cold-active proteases.
Similar content being viewed by others
Introduction
Psychrophilic and psychrotrophic bacteria inhabit almost all permanently cold habitats, from polar to non-polar glaciers and deep sea1,2. The prime significance of these bacteria stems from their ability to survive and retain functionality in cold temperatures and grow optimally even at warm temperatures3. These bacteria offer an immense natural resource of cold-active enzymes that function at low temperatures (10–20 °C). Indeed, these bacteria have been targeted for their industrial and biotechnological potential, for they provide energy-saving and economically beneficial alternatives to the enzymes from mesophilic homologues at low temperature4,5. These enzymes have evolved with increased conformational flexibility stemming from weakened non-covalent interactions, like salt bridges, hydrophobic interactions, aromatic-aromatic interactions and hydrogen bonding6. Among various enzymes, alkaline proteases that are usually active at different pH and temperature ranges7,8 are ranked high in commercial applications. Capable of producing large amounts of extracellular serine proteases, Bacilli are the chief source of almost all major serine alkaline proteases used in detergents9. While the contribution of bacterial alkaline proteases to total protease market share stands at 60%10,11,12, the data related to cold-active alkaline proteases from psychrotrophs is essentially missing4.
Nonetheless, a handful of previous studies have successfully isolated and characterized the small number of alkaline cold-active proteases from Bacillus sp. and demonstrated their application in detergent industries4,13,14. Though numerous cold-active proteases have been isolated from different cold environments, only a few are alkaline and appropriate as detergent additives15. In addition, crystal structures of numerous alkaline serine proteases from Bacilli have been identified16,17,18,19,20. The commercially important detergent proteases like subtilisin BPNʹ18, Carlsberg21, Savinase and Esperase19,22, are stable at higher pH (≥ 9) and temperature (25–40 °C), but the majority of the other proteases are comparatively unstable. Notably, in bleach-based detergents, these proteases do not withstand the oxidizing agents like hydrogen peroxide (H2O2), surfactants like Tween-80 and sodium dodecyl sulfate (SDS)23.
Despite advancement, the dearth of novel alkaline cold-active proteases characterized by being active at low temperatures, stable in the presence of metal ions, surfactants, organic solvents and oxidizing agents still exists. The main aim of the present study was to purify and characterize serine alkaline protease from a psychrotrophic bacterium that can catalyze reactions at low temperature and be used as an industrial additive. The cultural conditions like media, temperature, pH, and incubation time were modified to match psychrotrophic bacterium and obtain maximum yield at conditions suitable for the strain. This study successfully characterized and purified cold-active serine alkaline protease from psychrotrophic Bacillus pumilus BO1 isolated from western Himalayan glacial soil.
Results
Bacterial isolation and screening
The strain BO1 was selected among other isolated strains for further study after it displayed the highest proteolytic activity during primary screening. The isolated BO1 strain was able to grow at temperatures between 4 and 30 °C with the optimum growth at 20 °C.
Identification and qualitative screening of BO1 strain
The colony morphology and physiological characteristics of BO1 strain were round with undulate margin, flat elevation, and spore-forming rod-shaped Gram-positive aerobe. The amplified 16S rRNA gene sequence showed a 100–98% sequence identity to Bacillus pumilus when blasted against the nucleotide sequence database at NCBI. The 16S rRNA gene sequence of BO1 strain was submitted in NCBI GenBank under Accession number MN094861. The 16S rRNA phylogenetic tree (Supplementary Fig. S1) of psychrotrophic Bacillus pumilus BO1 showed significant similarity with other highly-homologous Bacillus pumilus species by sharing the same branch in the phylogenetic tree. The BO1 strain showed highest protease activity (zone of hydrolysis) on casein (Supplementary Fig. S2) out of three different media used (Fig. 1a).
(a) Qualitative screening of BO1 strain for its proteolytic activity on three different substrates. (b) Substrate efficacy of purified Apr-BO1. (c) Effect of pH on the activity and stability of purified cold-active Apr-BO1. (d) Effect of temperature on the activity and stability of purified Apr-BO1.
Purification and molecular mass determination of cold-active protease
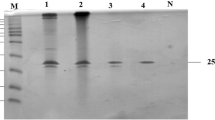
The protease (Apr-BO1) achieved a 4.43-fold increase in its purification with 80% ammonium sulphate saturation, followed by dialysis, which increased the purification by 17.63-folds. 92.55-fold purification was achieved with Sephadex G-50 superfine (Sigma, USA) gel filtration column. The purification profile of cold-active protease is given in Table 1. Further, the single-band appearance on SDS-PAGE indicated that purified Apr-BO1 had a molecular mass of 38 kDa (Fig. 2a).
(a) SDS-PAGE of purified Apr-BO1 (lane P). Lane M, molecular protein marker. (b) Lineweaver–Burk plot of purified cold-active Apr-BO1. The kinetic parameters (Km and Vmax) were studied by varying casein concentration from 0.02 to 0.22 g%. The original image (a) was cropped; the full-length image is presented as Supplementary Fig. S6b along with SDS-PAGE analysis of crude extracts (Supplementary Fig. S6a).
Effect of pH and temperature on activity and stability of the purified enzyme
The enzyme showed an increase in its activity from pH 6.0, reaching its optimum activity (100%) at pH 9.0, after which the activity started declining with an increase in pH up to 12.0 (Fig. 1c). Compared to protease activity at pH 9.0, it showed 42.13, 62.83 and 98.20% activity at pH 6.0, 7.0, and 8.0, respectively. Further, when the pH of the reaction mixture was increased towards alkalinity, Apr-BO1 displayed the relative activity of 92.8, 73.23 and 48.03% at pH 10.0, 11.0, and 12.0, respectively. As a result, Apr-BO1 was classified as an alkaline protease. On the other hand, the pH-dependent enzyme stability revealed that Apr-BO1 was stable over the pH range 6.0–12.0 (Fig. 1c), with only a 7.33–3.35% reduction in its activity at pH 7.0 and 8.0, respectively (concerning activity at pH 9.0). When incubated at pH 12.0, however, there was a 28.0% reduction in its activity.
The temperature-dependent enzyme activity showed an increase in relative activity with an increase in temperature from 5 to 20 °C, with optimum activity being observed at 20 °C. Further, increase in incubation temperature above 20 °C sharply decreased the enzyme activity, with approximately 71.0% activity being retained at 35 °C concerning 100% relative activity at 20 °C. While as the enzyme thermostability varied from 86.0 to 99.0% at 5–20 °C, respectively, which reveals that Apr-BO1 is stable at low temperatures. The enzyme showed decreased stability above 20 °C and retained average stability of 91.0, 80.0 and 67.0% (with respect to optimum stability at 20 °C) at 25 °C, 30 °C and 35 °C, respectively (Fig. 1d).
Effect of metal ions, surfactants, organic solvents, inhibitors and oxidizing agents on the activity of purified cold-active protease
The results of the metal ion effect on the activity of protease are given in Table 2. As it is evident, Fe2+ showed an increase in the relative activity of the enzyme by 13.0% concerning control, which was better than Cu2+ (9%) at a lower concentration (1.0 mM). Further, Cu2+ and Zn2+ also increased the enzyme activity up to 5.0 mM while as the activity decreased at 10.0 mM concentration of Cu2+ and Zn2+. However, Fe2+ did not show any significant effect on enzyme activity at higher concentration. Mn2+ showed a gradual increase with increasing concentration and exhibited a 16.02% increase in enzyme activity at 10.0 mM concentration. Ca2+ also increased the enzyme activity by 12.5, 19.0 and 23% at 1.0 mM, 5.0 mM and 10.0 mM, respectively. On the other hand, metal ions like Hg2+, Co2+ and Cd2+ decreased the enzyme activity with increasing concentration (Table 2). Besides, the protease showed enough activity in the presence of surfactants, oxidizing agents (Table 3) excluding, SDS which decreased the enzyme activity by 16.74%, 28.6% at 0.5% and 1.0% concentrations, respectively. Cold-active protease also showed good tolerance towards the oxidizing agent (H2O2). The inhibitor EDTA showed a 32.0% decrease at 1.0 mM concentration, while a sharp decline of 98.0% in enzyme activity was observed at 10.0 mM EDTA concentration. The organic solvents, xylene (100%), did not affect the enzyme activity, while toluene (95.0%) and benzene (86.0%) had a moderate effect on it. In addition, the enzyme showed good stability with all the organic solvents tested (Table 4).
Substrate efficacy
The protease, Apr-BO1, showed relative activity towards casein, skim milk, bovine serum albumin (BSA) and gelatin to a diverse extent. When standardized to casein (100%), the activity of enzyme towards these substrates was 77.91% for skim milk, 6.4% for BSA, and 3.34% for gelatin (Fig. 1b).
Enzyme kinetics (Km and Vmax) of purified Apr-BO1 towards casein
The rate of reaction exhibited a sharp increase with the increase in casein concentration up to 0.15 g%, after which the reaction rate stabilized. Apr-BO1 displayed Km of 1.0 mg/ml (4.16 × 10–5 M) and Vmax of 10.0 µmol/ml/min against casein, showing its high affinity and efficient catalytic role (Fig. 2b).
Protease gene identification and sequence analysis
Comparing it with the NCBI database, the nucleotide sequence of amplified protease gene (Supplementary Fig. S3) showed high similarity (96%) with serine alkaline proteases of Bacillus pumilus. The protease gene was provided GenBank accession no. MT178236. The inferred protein sequence of Apr-BO1 showed high similarity to serine peptidase S8A subfamily from Bacillus species (Fig. 3). As per the MEROPS database (http://merops.sanger.ac.uk/) phylogenetic classification and sequence analysis, it was concluded that Apr-BO1 belongs to S8A subfamily of serine alkaline protease. Protein sequence alignment in Fig. 4 contains sequence alignment of Apr-BO1 and other homologous subtilisins (S8A subfamily) like, Pro-TK-SP from Thermococcus kodakarensis (GenBank No. WP011250626), alkaline protease from Alkalihalobacillus halodurans (GenBank No. WP010897028), alkaline protease from Alkalihalobacillus clausii (GenBank No. WP094423791), alkaline protease from Bacillaceae (GenBank No. WP095239263), subtilisin Carlsberg from Bacillus licheniformis (GenBank No. P00781), ProMP27 from Bacillus pumilus (GenBank No. KX431582), serine protease from Bacillus subtilis (GenBank No. WP013351733), AprE from Bacillus pumilus (GenBank No. P07518), AprE from Bacillus (GenBank No. WP003233171), subtilisin amylosacchariticus from Bacillus subtilis subsp. amylosacchariticus (GenBank No. P00783) and subtilisin J from Geobacillus stearothermophilus (GenBank No. P29142). Besides, it shows the conserved and conservatively substituted regions of the Apr-BO1 protein sequence. The Apr-BO1 consisted of 346 amino acids with theoretical isoelectric point (pI) of 6.5.
Analysis of amino acid sequences by multiple alignments using homologous proteases. The residues highlighted by the black background are strictly conserved, and those boxed are conservatively substituted. The figure was produced using ESPript 3.0 program88.
Protein modelling
The refined model was developed from template subtilisin E of Bacillus subtilis (PDB ID: 3WHI). This model (Fig. 5a) qualified the stereochemical quality check, as shown by Ramachandran plot of the refined model where 90.6% of the residues occurred in most favored regions, and 8.1% of the residues were found in additional allowed regions (Supplementary Fig. S4). Further, the model passed Verify 3D analysis with high success as indicated by the favourable averaged 3D-1D score (Supplementary Fig. S5a). Moreover, the results of ProSA-web were highly encouraging, and the z-score of this model (− 8.81) was found to be well within the reference range, indicating the absence of errors (Supplementary Fig. S5b). Additionally, the root means square deviation value of model and template (3WHI) was 0.372 Å again stamping model quality (Fig. 5b). Our InterPro investigation revealed two domains of Apr-BO1, and further analysis by ScanProsite showed subtilase region (91–346). ScanProsite tool revealed aspartate 118, histidine 150 and serine 307 as the catalytic triad residues (Fig. 6).
(a) UCSF Chimera rendered a refined model of alkaline cold-active protease (Apr-BO1). Model generation was done by GalaxyTBM and among the five predicted models the best one was refined by GalaxyRefine. (b) Superimposed structures of Apr-BO1 model (yellow colour) and template (PDB ID: 3WHI) in hot pink. This ID corresponds to subtilisin E of Bacillus subtilis. Root mean square deviation (RMSD) was calculated between template and model using the Chimera tool, and it was observed that RMSD between 323 pruned atom pairs is 0.372 Å. RMSD is a quantitative estimate of similarity between two protein structures. Lower the value of model-template RMSD better is the model quality.
Analysis of domains and active site region of Apr-BO1. This protease showed two domains after InterPro Scan. (a) These domains are inhibitor 19 (ranging from amino acid residue 21–84) shown in purple and peptidase S8 domain (from 109 positions to 345th residue) represented in blue. (b) The subtilase ranges from 91 to 346 amino acids, and it is known that the S8 family possess a catalytic triad composed of Aspartate/Histidine/Serine, unlike the S53 family, which have glutamate instead of histidine as a part of this triad. ScanProsite tool revealed aspartate 118, histidine 150 and serine 307 as the catalytic triad residues. (c) This triad functions in a charge relay mechanism crucial for transforming substrate to products.
Industrial application
Wash analysis and detergent stability
Bloodstain was removed in the combination of Apr-BO1 and commercially available detergent (Tide and Surf excel). Comparing the stain removal efficiency of Apr-BO1 with detergents showed better results compared to Surf excel and almost equal stain removal efficiency compared to Tide (Fig. 7a). While as detergent stability test revealed good stability with two out of four commercial detergents (Fig. 7c). Stability of 74% and 67% was observed with Ghari and Wheel, respectively, while only 16.63% stability was observed with Tide and 10.0% with Surf excel.
(a) Application of purified Apr-BO1 and commercial detergents (Tide and surf Excel) on blood-stained cloth. The treatments given include: (i) untreated cloth, control; (ii) Apr-BO1 only; (iii) Detergent (Tide only); (iv) Detergent (Surf Excel only); (v) Detergent (Tide) + Apr-BO1; (vi) Detergent (Surf Excel) + Apr-BO1. (b) Visual observation of feather fibril detachment when Apr-BO1 enzyme treatment was performed for 8 h at 20 °C. The dehaired feather is shown in the right (h,j), whereas control (without treatment) is shown in the left (g,i). (c) Detergent stability of purified Apr-BO1.
Feather hydrolysis
The detachment of fibril from the feather branch (barb) was visibly observed (Fig. 7b) after the chicken feather was incubated for 8 h in the enzyme (100 µg/ml) at 20 °C. The results showed that it could prove effective in industrial dehairing processes at low temperatures.
Discussion
Given that very little is known regarding the microbial communities of Himalayan glacial soils, the present study reporting the potential proteolytic psychrotrophic microbial strain from Thajwas glacier is presumably the first report from northwestern Kashmir Himalaya. We were able to isolate a psychrotrophic Bacillus pumilus BO1 from these soils capable of producing Apr-BO1, a cold-active serine alkaline protease. Previously, few cold-active enzymes have been characterized from psychrophilic and psychrotrophic Bacillus species for their industrial uses4,24. However, there is still a demand for a cold-active enzyme that can show tolerance to low temperature, alkaline pH, surfactants, oxidizing agents and organic solvents. As per the literature, ours is the first cold-active serine protease with such properties. Apr-BO1 retained its stability and activity under alkaline conditions over a broad array of pH (7.0–11.0), exhibiting its optimal pH at 9.0. This enzyme also retained its stability and activity at low temperatures (5–20 °C) in the presence of several organic solvents. Further, it retained sufficient activity in the presence of several surfactants, oxidizing agents and metal ions than earlier reported cold-active alkaline proteases (CAAP) from Bacillus25,26, Stenotrophomonas27,28 and Pseudoalteromonas29.
Apart from these properties, a high yield of the enzyme at low temperature is deemed highly significant for industrial applications30. The CAAP isolated from psychrophilic and psychrotrophic Bacillus species has either low yield per hour of cultivation time25,26 or are active at temperatures above 20 °C14. Apr-BO1, on the other hand, was found to have a high yield at a low temperature (20 °C), making it the first CAAP from psychrotrophic Bacillus pumilus BO1. The purification results are also higher (Table 1) than what has been reported earlier for CAAP isolated from Bacillus25,26. The molecular weight (38 kDa) of Apr-BO1 is comparable to what has been reported so far, such as serine alkaline proteases isolated from Bacillus pumilus CBS31, Bacillus sp. RKY332 and Bacillus subtilis WLCP125.
The tolerance to organic solvents is crucial for the enzyme to withstand harsh industrial processes8. In agreement with Shah et al.33 mentioning that CAAP isolated from Bacillus cereus AK1871 retained maximum activity with organic solvents like benzene, toluene and xylene, Apr-BO1 for the present study also retained its maximum activity in their presence (Table 4). This tolerance suggests its potential application in peptide synthesis. Besides, the Apr-BO1 retained 85.0 and 71.0% (Table 3) activity on treatment for 30 min with 0.5 and 1.0% SDS, respectively. Previous studies on the stability of S8 subtilisins with SDS showed almost similar observations in Bacillus sp. JB-99 protease34, alkaline proteases from Bacillus clausii I-5235, Bacillus subtilis RD736, and novel Bacillus caseinilyticus37. Inhibition observed in Apr-BO1 enzyme for the present study was not as sturdy as reported for various serine alkaline proteases from Bacillus pumilus AI38, Bacillus sp. ZJ150239, Bacillus safensis S40640 and Bacillus invictae AHI41, adds to its novelty.
Serine proteases are generally inactivated by H2O2 due to the oxidization of methionine18,42,43.
However, Apr-BO1 remained stable in the presence of H2O2, and its activity was marginally increased by 3.5% and 5.8%, respectively, at 0.5% and 1.0% (v/v) H2O2 (Table 3). Thus it appears comparable with other novel serine alkaline proteases like SBcas3.3 from E. coli44, Aprx-SK37 from Virgibacillus sp. SK3743, Apr-PCC from Idiomarina sp. C9-142 and oxidase stable protease from Bacillus cereus SV145. It retained its activity for 30 min that presents its unique nature to withstand H2O2 incubation for a longer duration than others. Contrary to earlier reports that Hg2+ and Cd2+ exert an inhibitory effect on proteases as Hg2+ reacts with thiol group residues, histidine and tryptophan42,46, the results of the present study showed that Hg2+ and Cd2+ had an impact on enzyme activity but did not completely inhibit it (Table 2). For other metal ions like Fe2+ and Cu2+, no influence on the enzyme activity was recorded. These ions slightly increased the enzyme activity at lower concentrations (Table 2). Similarly, Apr-BO1 showed increased activity with increasing Mn2+ concentration, which has also been observed in alkaline serine proteases from Bacillus pumilus TMS5547, Bacillus acquimaris VITP448, Bacillus circulans L.49, Bacillus strain SH150 and proteases from Bacillus laterosporusn51. Ca2+ also showed the stimulating effect on the enzyme activity at 1.0 mM to 10.0 mM concentration. Alkaline protease from Virgibacillus pantothenticus52, a serine protease from novel Bacillus lehensis53 and alkaline protease from Bacillus altitudinis W354 also reported similar stimulating effects. S8 proteases have previously been activated by Ca2+ and inhibited by EDTA. According to previous research on subtilisin, Ca2+ plays an important structural role in the enzyme, and the effect of Ca2+ on enzyme activity may be due to the formation of a stable active conformation54. Subtilisin has two binding sites, one strong and one weak, in most cases21. As a result, the Apr-BO1 structure may have Ca2+ binding sites.
Moreover, in comparison to previous reports suggesting proteases from Bacilli had low Km between 0.57 and 4.0 mg/ml at higher temperatures (45–60 °C)41,55,56,57, the Apr-BO1 had a low Km (1.0 mg/ml) and Vmax (10.0 µmol/ml/min) at low temperature (20°), which indicates that it has high substrate affinity and efficient catalytic activity. The sequence of Apr-BO1 and other similar proteases showed low similarity after comparing with the database available at UniProt (https://www.uniprot.org/blast/). The highest identity of 96.0% was observed with Bacillus pumilus SARF-032, displaying the novel protease nature of Apr-BO1. The Ramachandran Plot of the refined model showed that 90.6% residues occurred in most favored regions while as 8.1% of the residues occurred in additional allowed regions (Supplementary Fig. S4). This is significant, and good quality model possesses above 90% residues in the most favored regions58,59. The quality check through ProSA further confirmed the model accuracy as its z-score was well within the confines of z-scores of experimental structures possessing similar size60,61. Our study showed that Asp 118/His 150/Ser 307 constitutes this protease’s catalytic triad. These results parallel with the findings where it has been demonstrated that S8 proteases have Asp/His/Ser as a catalytic triad, unlike S53, which have glutamate instead of histidine (Fig. 6). Serine proteases use a charge relay mechanism for catalysis, and these catalytic triad residues play a key role in this mechanism62. Besides, the predicted pI of the 11 subtilisin-like serine proteases and their molecular mass in comparison to the predicted pI (6.5) and molecular mass of Apr-BO1 showed that the molecular mass of Apr-BO1 was almost the same as that of other proteases except few. At the same time, two proteases (WP010897028 and P07518) had pI similar to Apr-BO1 (see Supplementary Table S1). Previous studies show that pI of most of the S8 proteases varies between 4 and 954,63.
Apr-BO1 efficiently removed bloodstain from the cloth without the assistance of any detergent (Fig. 7a). This compares well with previous results showing similar observations in mesophilic Bacillus pumilus8 and psychrophilic Bacillus subtilis25. The stain removal efficiency of Apr-BO1 to that of previously reported alkaline proteases28,64,65,66 shows that Apr-BO1 has higher efficiency at low concentrations (100 µg/ml) and low temperatures (20 °C), which makes it an appropriate detergent additive. Further, it also demonstrated good detergent stability, which reveals that it can be used in detergents and other industrial processes to carry out operations at lower energy expenditures. The Apr-BO1 also showed promising results in detachment of chicken feather after 8 h incubation at 20 °C in 4U of the enzyme. Similar results were obtained after 8 h incubation of chicken feather in 4U of enzyme along with 0.1% beta-mercaptoethanol (βME) at 45 °C and after 24 h incubation without βME. When compared with the previously isolated keratinolytic serine protease from thermophilic Bacillus subtilis67. The present study is the first report of such a kind from psychrotrophic bacteria. This enzyme activity could provide an energy-saving alternative to enzymes from thermophilic bacteria where the temperature of the substrate is raised so that the enzyme could function effectively4.
Novel, environmentally friendly enzymes are in high demand for industrial processes to reduce rising industrial costs and pollution. CAAP are emerging as essential enzymes with high commercial value in the dehairing and detergent industries due to their high substrate affinity at lower temperatures. The results from present study show that Apr-BO1 exhibits a high yielding capacity over a short duration (48 h), with a low Km value. It indicates that it exhibits a high affinity towards the substrate at low temperature, highlighting its novel behaviour. In addition, Apr-BO1 was active and stable over a wide pH and temperature range and retained its maximum activity with various metal ions, surfactant and organic solvents. This unparallel catalytic nature makes it suitable for industrial purposes. The modelled and certified structure of this protease may provide opportunities for further studies on this enzyme.
Material and methods
Sample collection and culture conditions
Soil samples were collected from Thajwas Glacier (34° 21′ 53.7′′ N latitude and 75° 21′ 03.6′′ E longitude) Kashmir, India at an altitude of 3900 m (a.m.s.l) in sterile poly-bags, plastic vials and transported to the laboratory68. A sixfold dilution of the soil sample was prepared, and the isolation of bacterial strain was done according to Zhang et al.69. Further, the initial proteolytic screening of the isolated bacterial strains was carried out on skim milk agar70.
Identification of psychrotrophic bacterial strain BO1
The cell morphology of BO1 strain was studied by Gram’s stain under a light microscope (Olympus IX71, Japan). The genomic DNA was extracted using the bacterial genomic DNA isolation kit GenElute (Sigma-Aldrich, USA), and for molecular identification, 16S rRNA gene amplification was done by PCR69. The 16S rRNA gene sequence was matched with the NCBI database using BLASTn program. The alignment of nucleotide sequences of BO1 strain and that of similar species was done by ClustalW. The phylogenetic tree for the 16S rRNA gene was constructed by the Neighbor-Joining method in MEGA 7.071.
Protease production
Qualitative screening
Qualitative screening of BO1 strain for protease production was done on three different media (agar 1.5% w/v, peptone 0.5%, NaCl 0.5% w/v, beef extract 0.3%) containing skim milk, casein and gelatin (1% w/v) as protein source separately. The media plates were incubated at 20 °C for 48 h, followed by submerging the bacterial colonies in mercuric chloride reagent and observing the zone of hydrolysis65.
Quantitative screening
The culture media contained: peptone (0.75% w/v), casein (1% w/v), MgSO4·7H2O (0.5% w/v), KH2PO4 (0.5% w/v), glucose (0.5% w/v) and FeSO4·7H2O (0.01% w/v)65 and pH was adjusted at 9.0 using sterilized Tris–HCl buffer. The overnight grown bacterial culture was inoculated in the ratio of 5:100 into protease production media, and media flasks were incubated for 48 h in a shaking incubator at 20 °C, 150 rpm. The media was centrifuged at 16,128×g for 10 min at 4 °C to collect the supernatant containing crude extracellular enzyme, and the purification of the enzyme was carried out.
Purification and characterization of alkaline protease
The protein aggregate and non-protein polymers were separated from the supernatant by adding ammonium sulphate to 40% saturation. Ammonium salt was further added up to 80% saturation to recollected supernatant. Centrifugation for both saturations was carried out for 10 min at 16,128×g and 4 °C72. 50 mM Tris–HCl buffer (pH 9.0) was utilized to dissolve the protein precipitate, and dialysis was carried out to remove ammonium sulphate residue from protein precipitate using a dialysis bag (Sigma Aldrich, USA). The resultant dialysate was loaded onto the Sephadex G-50 superfine column (Sigma-Aldrich, USA). Collected pooled fractions were saturated up to 60% with pre-chilled acetone (added slowly with gentle mixing) and kept at 4 °C for 4 h to precipitate73.
Electrophoresis
The SDS-PAGE74 was used to determine the molecular mass of the purified Apr-BO1, with 5% stacking gel and 12% resolving gel. The protein bands were stained by Coomassie Brilliant Blue R-250 and a standard protein marker (Thermo scientific PageRuler) was used as a reference.
Protein determination and protease assay
Protein content (mg/ml) was determined following Lowry et al.75 using bovine serum albumin (BSA) as a reference after each stage of the enzyme purification process. While as the proteolytic activity was measured by a modification of Zhou et al.42. The reaction mixture containing 0.6% (w/v) casein was dissolved in 200 µl 0.05 M Tris–HCl buffer (pH 9.0) and 100 µl enzyme. The reaction mixture was incubated at 20 °C for 30 min, and the reaction was stopped by adding 300 µl of 10% (w/v) trichloroacetic acid (TCA) while for control (blank) TCA was added just before incubation. Centrifugation of the reaction mixture was done for 10 min at 16,128×g and 4 °C, and 500 µl of supernatant was pipetted out into a new tube to which 2 ml of Na2CO3 (0.4 M) and 250 µl of Folin–Ciocalteu reagent (1 N) were added. Incubation of the mixture was done for 25 min at 30 °C and absorbance was measured at 660 nm. Protease activity (1U) was defined as the amount of enzyme (Apr-BO1) needed to release 1.0 µmol of tyrosine for every ml for each minute under standard assay conditions. The amount of tyrosine released was determined by the tyrosine standard curve.
Effect of pH and temperature on the protease activity and stability
The optimal pH of Apr-BO1 was assayed at 20 °C in 0.1 M Phosphate buffer (pH 6.0–7.0), 0.1 M Tris–HCl (pH 8.0–9.0) and 0.1 M Glycine–NaOH (pH 10.0–12.0) containing 0.6% casein (w/v). The effect of pH on enzyme stability was assayed by pre-incubating enzyme in the said buffers for 1 h at 20 °C. The % residual activities (i.e., sample activity after incubation/activity of sample before incubation × 100) were then measured.
The temperature was assayed between 5 and 40 °C with a gradient of 5 °C for 30 min in 0.1 M Tris–HCl buffer (pH 9.0) containing 0.6% casein (w/v). The temperature stability was determined by assessing enzyme residual activity by pre-incubating the enzyme at different temperatures (5–40 °C) with the gradient of 5 °C for 1 h in 0.1 M Tris–HCl buffer, pH 9.0.
Effect of metal ions and other reagents on the protease activity
The effect of various metal ions (Hg2+, Co2+, Fe2+, Cd2+, Cu2+, Mg2+, Mn2+, Ca2+ and Zn2+) on the enzyme catalytic activity was studied by assaying the relative activity with the addition of 1.0 mM, 5.0 mM and 10.0 mM concentration of metal ions. Similarly, the effect of ethylenediaminetetraacetic acid (EDTA) on enzyme activity was analyzed by the addition of 1.0 mM, 5.0 mM and 10.0 mM concentration. In addition, the effect of organic solvents (benzene, toluene and xylene), surfactants (Triton-x-100, SDS and Tween-80) and oxidizing agent (H2O2) on relative activity of enzyme was analyzed by adding, 0.5% and 1.0% (v/v; w/v for SDS) concentrations of organic solvents, surfactants and oxidizing agent to 500 µl of Tris–HCl buffer (pH 9.0) containing 0.6% casein (w/v). The stability of enzyme against organic solvents (benzene, toluene and xylene) was assessed by pre-incubating 100 µl of the enzyme at 20 °C in each of these solvents for 30 min and assayed accordingly.
Substrate efficacy
In order to check the substrate efficacy of enzyme, a concentration of 1.0% of four substrates (casein, gelatin, BSA and skim milk) was assayed at 20 °C and pH 9.0.
Enzyme kinetics of cold-active Apr-BO1
Enzyme kinetic parameters of Apr-BO1 were calculated at 0.02–0.22 g% concentrations of casein at pH 9.0 and temperature 20 °C for 30 min. Lineweaver- Burk plot76 was used to calculate the Km and Vmax of the cold-active protease against casein.
Amplification of alkaline protease gene
The conserved protein gene (apr) encoding the Apr-BO1 was amplified using a pair of primers (AprF: ATGTGCGTGAAAAAGAAAAATGTG and AprR: TTAGTTAGAAGCTGCTTGAACGTT) designed at NCBI’s Primer Blast tool. Genomic DNA already extracted for the identification of BO1 strain was now used again as a template for the amplification of alkaline protease gene by PCR with similar PCR conditions used for the 16S rRNA gene. The translated amino acid sequence of the Apr-BO1 gene was analyzed in the NCBI BLASTp program, and the sequences were aligned using Clustal Omega. Phylogenetic tree for protein sequence was constructed using MEGA 7.0 with Neighbor-Joining method71. The theoretical isoelectric point (pI) of Apr-BO1 was calculated at https://web.expasy.org/cgi-bin/compute_pi/pi_tool.
Protein modelling and refinement
GalaxyTBM was used for producing the protein structure from a given amino acid sequence. It first generates the structure of reliable core region through a template-based approach, after which the unreliable local regions are recognized and re-modelled through ab initio method77,78. Among five predicted models, the best one was chosen for refinement. GalaxyRefine—another service of GalaxyWEB was used to improve the quality of model generated79. The refined model was rendered with the help of UCSF Chimera (1.10.2) software80. For generating this model, the template used was subtilisin E of Bacillus subtilis (PDB ID: 3WHI)81. Root mean square deviation (RMSD) was estimated by superimposing the model on a template using MatchMaker of Chimera software. Other valuable information regarding this protease, including several domains and catalytic triad, was investigated using the pandemically popular tools InterPro and ScanProsite82,83.
Model validation through different approaches
The stereochemical quality of the model was checked by PROCHECK, and the model compatibility with its sequence was evaluated by Verify 3D59,84,85. The errors in the refined model were examined using Protein Structure Analysis (ProSA)-web86.
Industrial application
Detergent stability and wash analysis
The stability of cold-active enzyme with four commercially available detergents viz; Surf Excel, Wheel (Hindustan Unilever Limited), Tide (Procter & Gamble) and Ghari (RSPL Limited) was assayed by incubating 100 µl of enzyme in 200 µl of 7.0 mg/ml detergents solution. The detergent solution was incubated at 100 °C for 1 h to deactivate the already present proteases in the detergents before adding the enzyme.
To check the stain removal efficiency of the enzyme Furhan et al.25 was followed with modification. White cotton cloths measuring 4 cm × 4 cm were stained with blood and dried. The stained pieces of cloths were given following treatments: (a) 400 µl of purified enzyme (100 µg/ml) + 20 ml of tap water + blood-stained cloth; (b) 400 µl of purified enzyme + 20 ml of detergent solution (0.07% w/v) + blood-stained cloth; (c) 0.07% (w/v) detergent solution + blood-stained cloth and (d) bloodstained cloth + 20 ml of tap water was used for blank control. The treatments were given in triplicates and incubated at 20 °C for 30 min. The cloth was then rinsed with tap water, dried and evaluated for stain removal efficiency.
Feather fibril detachment
Hydrolysis and detachment of feather fibril by Apr-BO1 was carried87 with few modifications. The chicken feathers were acquired from a local slaughterhouse and washed first with tap water, rinsed with distilled water and sterilized by autoclaving. The feathers were left overnight to dry at room temperature and incubated in 2 ml of the purified enzyme with 100 µg/ml concentrations for 8 h at 20 °C, washed and dried.
Ethical statement
The authors in the current study carried out no animal or human studies. The authors further confirm that this work is original and has not been published, nor is it currently under consideration for publication elsewhere.
References
Rafiq, M. et al. Recovery of metallo-tolerant and antibiotic resistant psychrophilic bacteria from Siachen glacier, Pakistan. PLoS ONE 12, e0178180 (2017).
Kumar, S., Suyal, D. C., Yadav, A., Shouche, Y. & Goel, R. Microbial diversity and soil physiochemical characteristic of higher altitude. PLoS ONE 14, 1–15 (2019).
Margesin, R. Effect of temperature on growth parameters of psychrophilic bacteria and yeasts. Extremophiles 13, 257–262 (2009).
Al-Ghanayem, A. A. & Joseph, B. Current prospective in using cold-active enzymes as eco-friendly detergent additive. Appl. Microbiol. Biotechnol. 104, 2871–2882 (2020).
Perfumo, A., Freiherr von Sass, G. J., Nordmann, E. L., Budisa, N. & Wagner, D. Discovery and characterization of a new cold-active protease from an extremophilic bacterium via comparative genome analysis and in vitro expression. Front. Microbiol. 11, 881 (2020).
Bhatia, R. K. et al. Psychrophiles: A source of cold-adapted enzymes for energy efficient biotechnological industrial processes. J. Environ. Chem. Eng. 9(1), 104607 (2020).
Sharma, K. M., Kumar, R., Panwar, S. & Kumar, A. Microbial alkaline proteases: Optimization of production parameters and their properties. J. Genet. Eng. Biotechnol. 15, 115–126 (2017).
Jayakumar, R., Jayashree, S., Annapurna, B. & Seshadri, S. Characterization of thermostable serine alkaline protease from an alkaliphilic strain Bacillus pumilus MCAS8 and its applications. Appl. Biochem. Biotechnol. 168, 1849–1866 (2012).
Gupta, R., Beg, Q. & Lorenz, P. Bacterial alkaline proteases: Molecular approaches and industrial applications. Appl. Microbiol. Biotechnol. 59, 15–32 (2002).
Sarethy, I. P. et al. Alkaliphilic bacteria: Applications in industrial biotechnology. J. Ind. Microbiol. Biotechnol. 38(7), 769–790 (2011).
Thomas, N. N., Archana, V., Shibina, S. & Edwin, B. T. Isolation and characterization of a protease from Bacillus sp.. Mater. Today Proc. 41, 685–691 (2020).
Razzaq, A. et al. Microbial proteases applications. Front. Bioeng. Biotechnol. 7, 110 (2019).
Liu, Y. et al. Improvement of cold adaptation of Bacillus alcalophilus alkaline protease by directed evolution. J. Mol. Catal. B Enzym. 106, 117–123 (2014).
Yang, J., Li, J., Mai, Z., Tian, X. & Zhang, S. Purification, characterization, and gene cloning of a cold-adapted thermolysin-like protease from Halobacillus sp. SCSIO 20089. J. Biosci. Bioeng. 115, 628–632 (2013).
Kuddus, M. & Ramteke, P. W. Recent developments in production and biotechnological applications of cold-active microbial proteases. Crit. Rev. Microbiol. 38, 330–338 (2012).
Dauter, Z., Betzel, C., Genov, N., Pipon, N. & Wilson, K. S. Complex between the subtilisin from a mesophilic bacterium and the Leech inhibitor eglin-C. Acta Crystallogr. Sect. B 47, 707–730 (1991).
Eschenburg, S. et al. Crystal structure of subtilisin DY, a random mutant of subtilisin Carlsberg. Eur. J. Biochem. 257, 309–318 (1998).
Bott, R. et al. The three-dimensional structure of Bacillus amyloliquefaciens subtilisin at 1.8 A and an analysis of the structural consequences of peroxide inactivation. J. Biol. Chem. 263, 7895–7906 (1988).
Betzel, C. et al. Crystal structure of the alkaline proteinase SavinaseTM from Bacillus lentus at 1.4 Å resolution. J. Mol. Biol. 223, 427–445 (1992).
Almog, O. et al. The 0.93 Å crystal structure of sphericase: A calcium-loaded serine protease from Bacillus sphaericus. J. Mol. Biol. 332, 1071–1082 (2003).
Neidhart, D. J. & Petsko, G. A. The refined crystal structure of subtilisin Carlsberg at 2.5 Å resolution. Protein Eng. Des. Sel. 2, 271–276 (1988).
Outtrup, H., Dambmann, C., Christiansen, M. & Aaslyng, D. A. Bacillus sp. JP 395, Method of making and detergent composition. US patent Number, 5466594 (1995).
Maurer, K. H. Detergent proteases. Curr. Opin. Biotechnol. 15, 330–334 (2004).
Furhan, J. & Sharma, S. Cold-active alkophilic proteases from various microbial sources: Benefits and applications. Int. J. Acad. Res. Dev. 3, 340–345 (2018).
Furhan, J., Awasthi, P. & Sharma, S. Biocatalysis and agricultural biotechnology biochemical characterization and homology modelling of cold-active alkophilic protease from Northwestern Himalayas and its application in detergent industry. Biocatal. Agric. Biotechnol. 17, 726–735 (2019).
Joshi, G. K., Kumar, S. & Sharma, V. Production of moderately halotolerant, SDS stable alkaline protease from Bacillus cereus MTCC 6840 isolated from lake Nainital, Uttaranchal State, India. Braz. J. Microbiol. 38, 773–779 (2007).
Saba, I. et al. Purification and characterization of a cold active alkaline protease from Stenotrophomonas sp., isolated from Kashmir, India. World J. Microbiol. Biotechnol. 28, 1071–1079 (2012).
Kuddus, M. & Ramteke, P. W. Cold-active extracellular alkaline protease from an alkaliphilic Stenotrophomonas maltophilia: Production of enzyme and its industrial applications. Can. J. Microbiol. 55, 1294–1301 (2009).
Vázquez, S. C., Hernández, E. & Mac-Cormack, W. P. Extracellular proteases from the Antarctic marine Pseudoalteromonas sp. P96–47 strain. Rev. Argent. Microbiol. 40, 63–71 (2008).
Singh, R., Kumar, M., Mittal, A. & Mehta, P. K. Microbial enzymes: Industrial progress in 21st century. 3 Biotech 6, 174 (2016).
Jaouadi, B., Ellouz-Chaabouni, S., Rhimi, M. & Bejar, S. Biochemical and molecular characterization of a detergent-stable serine alkaline protease from Bacillus pumilus CBS with high catalytic efficiency. Biochimie 90, 1291–1305 (2008).
Reddy, L. V. A., Wee, Y. J. & Ryu, H. W. Purification and characterization of an organic solvent and detergent-tolerant novel protease produced by Bacillus sp. RKY3. J. Chem. Technol. Biotechnol. 83, 1526–1533 (2008).
Shah, K., Mody, K., Keshri, J. & Jha, B. Purification and characterization of a solvent, detergent and oxidizing agent tolerant protease from Bacillus cereus isolated from the Gulf of Khambhat. J. Mol. Catal. B Enzym. 67, 85–91 (2010).
Johnvesly, B. & Naik, G. R. Studies on production of thermostable alkaline protease from thermophilic and alkaliphilic Bacillus sp. JB-99 in a chemically defined medium. Process Biochem. 37, 139–144 (2001).
Joo, H.-S., Kumar, C. G., Park, G.-C., Paik, S. R. & Chang, C.-S. Oxidant and SDS-stable alkaline protease from Bacillus clausii I-52: Production and some properties. J. Appl. Microbiol. 95, 267–272 (2003).
Suberu, Y., Akande, I., Samuel, T., Lawal, A. & Olaniran, A. Cloning, expression, purification and characterization of serine alkaline protease from Bacillus subtilis RD7. Biocatal. Agric. Biotechnol. 20, 101264 (2019).
Mothe, T. & Sultanpuram, V. R. Production, purification and characterization of a thermotolerant alkaline serine protease from a novel species Bacillus caseinilyticus. 3 Biotech 6, 1–10 (2016).
Fakhfakh-Zouari, N., Hmidet, N., Haddar, A., Kanoun, S. & Nasri, M. A novel serine metallokeratinase from a newly isolated Bacillus pumilus A1 grown on chicken feather meal: Biochemical and molecular characterization. Appl. Biochem. Biotechnol. 162, 329–344 (2010).
Yu, P., Huang, X., Ren, Q. & Wang, X. Purification and characterization of a H2O2-tolerant alkaline protease from Bacillus sp. ZJ1502, a newly isolated strain from fermented bean curd. Food Chem. 274, 510–517 (2019).
Mhamdi, S. et al. Evaluation of the biotechnological potential of a novel purified protease BS1 from Bacillus safensis S406 on the chitin extraction and detergent formulation. Int. J. Biol. Macromol. 104, 739–747 (2017).
Hammami, A., Bayoudh, A. & Abdelhedi, O. Response-surface methodology for the production and the purification of a new H2O2-tolerant alkaline protease from Bacillus invictae AH1 strain. Biotechnol. Prog. 36, 1–16 (2020).
Zhou, C. et al. A novel alkaline protease from alkaliphilic Idiomarina sp. C9–1 with potential application for eco-friendly enzymatic dehairing in the leather industry. Sci. Rep. 8, 1–18 (2018).
Phrommao, E., Yongsawatdigul, J., Rodtong, S. & Yamabhai, M. A novel subtilase with NaCl-activated and oxidant-stable activity from Virgibacillus sp. SK37. BMC Biotechnol. 11, 65 (2011).
Biver, S., Portetelle, D. & Vandenbol, M. Characterization of a new oxidant-stable serine protease isolated by functional metagenomics. Springerplus 2, 1–10 (2013).
Manni, L. et al. An oxidant- and solvent-stable protease produced by Bacillus cereus SV1: Application in the deproteinization of shrimp wastes and as a laundry detergent additive. Appl. Biochem. Biotechnol. 160, 2308–2321 (2010).
Wang, B., Wu, W. & Liu, X. Purification and characterization of a neutral serine protease with nematicidal activity from Hirsutella rhossiliensis. Mycopathologia 163, 169–176 (2007).
Ibrahim, S. K., Muniyandi, J. & Pandian, S. K. Purification and characterization of manganese-dependent alkaline serine protease from Bacillus pumilus TMS55. J. Microbiol. Biotechnol. 21, 20–27 (2011).
Chittoor, J. T., Balaji, L. & Jayaraman, G. Optimization of parameters that affect the activity of the alkaline protease from halotolerant bacterium, Bacillus acquimaris VITP4, by the application of response surface methodology and evaluation of the storage stability of the enzyme. Iran. J. Biotechnol. 14, 23–32 (2016).
Sharaf, E. F. & Al-Fadel, K. Green recycling of chicken feather and sheep wool using the partially purified alkaline protease from Bacillus circulans L.. Nat. Sci. 5, 25–29 (2013).
Ningthoujam, D. S. & Kshetri, P. A thermostable alkaline protease from a moderately halo-alkalithermotolerant Bacillus subtilis strain SH1. Aust. J. Basic Appl. Sci. 4, 5126–5134 (2010).
Usharani, B. & Muthuraj, M. Production and characterization of protease enzyme from Bacillus laterosporus. African J. Microbiol. Res. 4, 1057–1063 (2010).
Gupta, A., Joseph, B., Mani, A. & Thomas, G. Biosynthesis and properties of an extracellular thermostable serine alkaline protease from Virgibacillus pantothenticus. World J. Microbiol. Biotechnol. 24, 237–243 (2008).
Joshi, S. & Satyanarayana, T. Characteristics and applications of a recombinant alkaline serine protease from a novel bacterium Bacillus lehensis. Bioresour. Technol. 131, 76–85 (2013).
Yang, S. et al. Mining of alkaline proteases from Bacillus altitudinis W3 for desensitization of milk proteins: Their heterologous expression, purification, and characterization. Int. J. Biol. Macromol. 153, 1220–1230 (2020).
Jain, D., Pancha, I., Mishra, S. K., Shrivastav, A. & Mishra, S. Purification and characterization of haloalkaline thermoactive, solvent stable and SDS-induced protease from Bacillus sp.: A potential additive for laundry detergents. Bioresour. Technol. 115, 228–236 (2012).
Bhatt, H. B. & Singh, S. P. Cloning, expression, and structural elucidation of a biotechnologically potential alkaline serine protease from a newly isolated haloalkaliphilic Bacillus lehensis JO-26. Front. Microbiol. 11, 941 (2020).
Gupta, A. et al. One-step purification and characterization of an alkaline protease from haloalkaliphilic Bacillus sp.. J. Chromatogr. A 1075, 103–108 (2005).
Al-Khayyat, M. Z. S. & Al-Dabbagh, A. G. A. In silico prediction and docking of tertiary structure of LuxI, an inducer synthase of Vibrio fischeri. Rep. Biochem. Mol. Biol. 4, 66–75 (2016).
Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993).
Ji, X., Wang, W., Zheng, Y., Hao, J. & Sun, M. Homology modeling and molecular dynamics simulation studies of a marine alkaline protease. Bioinform. Biol. Insights 6, 255–263 (2012).
Ia, S., Hrm, R. & Ah, A. Homology modeling and virtual screening studies of antigen MLAA-42 protein: Identification of novel drug candidates against leukemia—An in silico approach. Comput. Math. Methods Med. 2020, 8196147 (2020).
Di Cera, E. Serine proteases. IUBMB Life 61, 510–515 (2009).
Rao, M. B., Tanksale, A. M., Ghatge, M. S. & Deshpande, V. V. Molecular and biotechnological aspects of microbial proteases. Microbiol. Mol. Biol. Rev. 62, 597–635 (1998).
Patil, U., Mokashe, N. & Chaudhari, A. Detergent-compatible, organic solvent-tolerant alkaline protease from Bacillus circulans MTCC 7942: Purification and characterization. Prep. Biochem. Biotechnol. 46, 56–64 (2016).
Baweja, M., Tiwari, R., Singh, P. K., Nain, L. & Shukla, P. An alkaline protease from Bacillus pumilus MP 27: Functional analysis of its binding model toward its applications as detergent additive. Front. Microbiol. 7, 1195 (2016).
Nassar, F. R., Abdelhafez, A. A., El-Tayeb, T. S. & Abu-Hussein, S. H. Purification, characterization and applications of proteases produced by Bacillus amyloliquefaciens 35s isolated from soil of the Nile Delta of Egypt. Br. Microbiol. Res. J. 6, 286–302 (2015).
Pillai, P. & Archana, G. Hide depilation and feather disintegration studies with keratinolytic serine protease from a novel Bacillus subtilis isolate. Appl. Microbiol. Biotechnol. 78, 643–650 (2008).
Srinivas, T. N. R. et al. Comparison of bacterial diversity in proglacial soil from Kafni Glacier, Himalayan Mountain ranges, India, with the bacterial diversity of other glaciers in the world. Extremophiles 15, 673–690 (2011).
Zhang, D.-C., Brouchkov, A., Griva, G., Schinner, F. & Margesin, R. Isolation and characterization of bacteria from ancient siberian permafrost sediment. Biology (Basel) 2, 85–106 (2013).
Baghel, V. S. et al. Psychrotrophic proteolytic bacteria from cold environment of Gangotri glacier, Western Himalaya, India. Enzyme Microb. Technol. 36, 654–659 (2005).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016).
Green, A. A. & Hughes, W. L. Protein fractionation on the basis of solubility in aqueous solutions of salts and organic solvents. Methods Enzymol. 1, 67–90 (1955).
Sana, B., Ghosh, D., Saha, M. & Mukherjee, J. Purification and characterization of a salt, solvent, detergent and bleach tolerant protease from a new gamma-Proteobacterium isolated from the marine environment of the Sundarbans. Process Biochem. 41, 208–215 (2006).
Laemmli, U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 (1970).
Lowry, O. H., Rosebrough, N. J., Farr, A. L. & Randall, R. J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193, 265–275 (1951).
Lineweaver, H. & Burk, D. The determination of enzyme dissociation constants. J. Am. Chem. Soc. 56, 658–666 (1934).
Ko, J., Park, H. & Seok, C. GalaxyTBM: Template-based modeling by building a reliable core and refining unreliable local regions. BMC Bioinform. 13, 198 (2012).
Mushtaq, H. et al. Biochemical characterization and functional analysis of heat stable high potential protease of Bacillus amyloliquefaciens strain HM48 from soils of Dachigam National Park in Kashmir Himalaya. Biomolecules 11, 117 (2021).
Heo, L., Park, H. & Seok, C. GalaxyRefine: Protein structure refinement driven by side-chain repacking. Nucleic Acids Res. 41, 384–388 (2013).
Pettersen, E. F. et al. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Uehara, R., Angkawidjaja, C., Koga, Y. & Kanaya, S. Formation of the high-affinity calcium binding site in pro-subtilisin e with the insertion sequence IS1 of Pro-Tk-subtilisin. Biochemistry 52, 9080–9088 (2013).
Hunter, S. et al. InterPro: The integrative protein signature database. Nucleic Acids Res. 37, 211–215 (2009).
Sigrist, C. J. A. et al. New and continuing developments at PROSITE. Nucleic Acids Res. 41, 344–347 (2013).
Bowie, J. U., Lüthy, R. & Eisenberg, D. A method to identify protein sequences that fold into a known three-dimensional stucture. Science 253, 164–170 (1991).
Lüthy, R., Bowie, J. U. & Eisenberg, D. Assessment of protein models with three-dimensional profiles. Nature 356, 83–85 (1992).
Wiederstein, M. & Sippl, M. J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 35, 407–410 (2007).
Bockle, B., Galunsky, B. & Muller, R. Characterization of a keratinolytic serine proteinase from Streptomyces pactum DSM 40530. Appl. Environ. Microbiol. 61, 3705–3710 (1995).
Robert, X. & Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, 320–324 (2014).
Author information
Authors and Affiliations
Contributions
S.F. framed the work design, collected, compiled, interpreted the data, conducted experiments, wrote the manuscript and designed the figures. R.N. designed the research work, evaluated the data and did the revision of manuscript. S.G. designed the protein 3D structure, evaluated the data, did the revision of manuscript and arranged the figures. B.G. did the critical revision of the manuscript, calculated the enzyme kinetics. Both R.N. and B.G. helped in designing the experiments and supervised the overall work. All the authors helped in writing the manuscript and provided important feedbacks.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Farooq, S., Nazir, R., Ganai, S.A. et al. Isolation and characterization of a new cold-active protease from psychrotrophic bacteria of Western Himalayan glacial soil. Sci Rep 11, 12768 (2021). https://doi.org/10.1038/s41598-021-92197-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-92197-w
This article is cited by
-
Investigating potential protease activity of psychrotrophic bacteria from a municipal landfill for solid waste management
Biomass Conversion and Biorefinery (2024)
-
Microbial pathogen profiling and water quality assessment of Jammu Himalayan springs
Biologia (2023)
-
Psychrophilic and psychrotrophic bacterial diversity of Himalayan Thajwas glacial soil, India
Biologia (2022)
-
Molecular characterization of lipase from a psychrotrophic bacterium Pseudomonas sp. CRBC14
Current Genetics (2022)
-
Metagenomic views on taxonomic and functional profiles of the Himalayan Tsomgo cold lake and unveiling its deterzome potential
Current Genetics (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.