Abstract
Paulownia catalpifolia is an important, fast-growing timber species known for its high density, color and texture. However, few transcriptomic and genetic studies have been conducted in P. catalpifolia. In this study, single-molecule real-time sequencing technology was applied to obtain the full-length transcriptome of P. catalpifolia leaves treated with varying degrees of drought stress. The sequencing data were then used to search for microsatellites, or simple sequence repeats (SSRs). A total of 28.83 Gb data were generated, 25,969 high-quality (HQ) transcripts with an average length of 1624 bp were acquired after removing the redundant reads, and 25,602 HQ transcripts (98.59%) were annotated using public databases. Among the HQ transcripts, 16,722 intact coding sequences, 149 long non-coding RNAs and 179 alternative splicing events were predicted, respectively. A total of 7367 SSR loci were distributed throughout 6293 HQ transcripts, of which 763 complex SSRs and 6604 complete SSRs. The SSR appearance frequency was 28.37%, and the average distribution distance was 5.59 kb. Among the 6604 complete SSR loci, 1–3 nucleotide repeats were dominant, occupying 97.85% of the total SSR loci, of which mono-, di- and tri-nucleotide repeats were 44.68%, 33.86% and 19.31%, respectively. We detected 112 repeat motifs, of which A/T (42.64%), AG/CT (12.22%), GA/TC (9.63%), GAA/TTC (1.57%) and CCA/TGG (1.54%) were most common in mono-, di- and tri-nucleotide repeats, respectively. The length of the repeat SSR motifs was 10–88 bp, and 4997 (75.67%) were ≤ 20 bp. This study provides a novel full-length transcriptome reference for P. catalpifolia and will facilitate the identification of germplasm resources and breeding of new drought-resistant P. catalpifolia varieties.
Similar content being viewed by others
Introduction
Paulownia, one of the most important fast-growing timber species around the world, is native to China and widely grown in subtropical and warm temperate regions, which have acted an important part of timber supply, ecological environmental construction, soil improvement and so forth1. Paulownia catalpifolia is a typical and important species of Genus Paulownia in northern China, it exhibits some drought resistance and is renowned for its high density, good color, and beautiful texture. Recently, droughts and water shortages have seriously affected P. catalpifolia growth, causing mass deaths in some P. catalpifolia plantations. Therefore, high-quality and drought-resistant P. catalpifolia varieties are urgently needed. Conventional plant breeding methods, such as cross-breeding and selection breeding, have yielded little success in improving the traits of plants; this is due to genetic resistance, reproductive isolation and long generation cycles, among other factors2,3. Previous studies on P. catalpifolia have focused on the chemical composition of its fruits and seeds, as well as on tissue culture4,5,6; however, molecular studies of P. catalpifolia are lacking.
Microsatellites, also known as simple sequence repeats (SSRs), are DNA sequences consisting of continuously repeating motifs, which are composed of 1–6 bases7,8. The type and number of repeat motifs differ among SSRs, resulting in polymorphisms at each SSR locus. SSR molecular markers are widely distributed throughout plant genomes9 and are characterized by codominance, high polymorphism and good repeatability. SSR loci are conserved within and among genera10. Depending on their origin, SSR markers can be categorized as genomic SSRs or expressed sequence tag (EST) SSRs. EST-SSR markers are easier to obtain for a large number of plants that have no reference genome, although the polymorphism of EST-SSR is lower than that of genomic SSR markers. As functional molecular markers, EST-SSRs are more conserved, better universality, lower cost and more interspecific transferability11,12. Moreover, EST-SSR polymorphisms may be directly related to gene function13 and can be used for researches of other related species14,15. In recent years, EST-SSR markers have been developed and applied in various tree species, including Eucalyptus globulus16, Euphrates Poplar17, the rubber tree18, Robinia pseudoacacia19, Fraxinus velutina20, and Pinus koraiensis21.
Single-molecule real-time (SMRT) sequencing technology (Pacific Biosciences), also known as third generation sequencing technology, can efficiently and accurately obtain high-quality (HQ), long and intact transcripts containing 5′- and 3′-untranslated regions and polyadenosine tails without assembly22,23. SMRT sequencing can be used to accurately identify features such as fusion genes, gene families, long non-coding RNAs (lncRNAs) and alternative splicing (AS) events24,25. SMRT sequencing technology is a reliable method for obtaining full-length transcripts that can be used to study the transcriptomes of non-model plants which lack reference genomes, such as Paulownia and Chinese catalpa. SMRT sequencing technology has been successfully applied to full-length transcriptome sequencing studies in animals, plants and insects26,27,28. Furthermore, full-length transcriptome sequences obtained using SMRT sequencing contain numerous EST SSRs29,30, which can be used for genetic analyses of the sequenced species and their related species, as well as for studies of conservation biology and molecular assisted breeding23,31,32. To the best of our knowledge, no full-length transcriptome sequence of P. catalpifolia has been reported.
In this study, we performed a full-length transcriptomic analysis of mixed P. catalpifolia leaves treated with varying degrees of drought stress using SMRT sequencing. We then performed function annotation analyses using publicly available databases and used various bioinformatics software to predict AS, lncRNAs and SSRs and to further analyze SSRs characteristics deeply. In the absence of Paulownia reference genome, the full-length transcriptome sequence acquired in our study not only can be used as a reference sequence for transcriptome sequencing, but also will support further genetic analyses in Paulownia species. In addition, the SSRs predicted in our study will facilitate the development of drought-resistant SSR markers, the discovery of drought-resistant genes and the study of the genetic relationships between P. catalpifolia and other related species.
Results
SMRT sequencing of the full-length transcriptome
We acquired full-length transcriptomic of P. catalpifolia using SMRT sequencing technology and obtained 28.83 Gb sequencing data. After removing the adapter sequences, approximately 454,554 polymerase reads remained, which then formed 19,052,345 subreads with an average read length of 1470 bp. After self-correction and merging, the subreads formed 405,034 circular consensus sequences (CCSs) (Fig. 1a) with an average length of 1693 bp, and 349,745 full-length non-chimeric sequences (FLNCs) (Fig. 1b). A total of 30,953 transcripts were obtained after clustering and removal of redundant sequences using the PacBio SMRT LINK Cluster tool, and 30,928 HQ transcripts with ≥ 99% accuracy and a full-length read support ≥ 2 were sequenced (Fig. 1c). The length range of the HQ transcripts was 362–7922 bp, the N50 was 1768 bp, and the mean transcript length was 1618 bp. Of the HQ transcripts, 10.47% and 86.07% were 362–900 bp and 1000–3000 bp in length, respectively. Long-length HQ transcripts (> 3000 bp) constituted 3.46% of the total HQ transcripts. After error correction and removal of all 100% identical sequences, 25,969 HQ transcripts remained, its individual transcript length ranging from 362 to 7922 bp, the average length of 1624 bp, and N50 of 1781 bp, which were used in subsequent analyses.
Functional annotation of the full-length transcriptome sequences
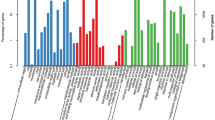
The functional annotation of the HQ transcripts was then performed. Of the 25,969 transcripts analyzed, 367 could not be functionally assigned by any of the databases used (Table 1). A total of 25,591 (98.54%) HQ transcripts were annotated using the NCBI non-redundant protein database and exhibited homology with known proteins of various species, including Sesamum indicum (75.38%), Erythranthe guttata (12.87%) and Dorcoceras hygrometricum (1.71%) (Fig. 2). The HQ transcripts were then searched against the gene ontology (GO) database to analyze their functions; 18,501 (71.24%) of the HQ transcripts were categorized into 50 GO group, which were divided into three broad classes: biological processes (37,536 HQ transcripts, 38.38%), cellular components (38,888, 39.76%) and molecular functions (21,377, 21.86%) (Fig. 3a). Following searches against the eukaryotic orthologous groups (KOG) database, the HQ transcripts were clustered into 26 KOG terms (Fig. 3b). Furthermore, 13,829 HQ transcripts were identified in the Kyoto encyclopedia of genes and genomes (KEGG) database and grouped into 129 KEGG pathways, which were divided into five broad categories: cellular processes (779 HQ transcripts, 5.63%), environmental information processing (523, 3.78%), genetic information processing (3207, 23.19%), metabolism (8962, 64.81%) and organismal systems (358, 2.59%) (Fig. 3c). Using Swiss-Prot, 22,606 (87.05%) HQ transcripts were annotated.
Identification of long non-coding RNAs, coding sequences and alternative splicing
The long non-coding RNAs (lncRNAs) are not translated into protein and its length are more than 200 nucleotides. LncRNAs are vital for regulating the neighboring gene expression33. A total of 149 common lncRNAs were identified in P. catalpifolia Using four methods (CPC2, CPAT, PLEK and CNCI) (Fig. 4a). TransDecoder software was used to predict 24,982 coding sequences (CDSs), of which 16,722 were intact. The lengths of the amino acids encoded by the intact CDSs were in the range of 100–1840, with the number of amino acids decreasing as the length increased except 100–300 (Fig. 4b). Alternative splicing (AS) is one of crucial biological phenomenons, and it is helpful to produce different mature transcripts using the same RNA sequence34. AS is highly correlated with biological function and a major source of proteomic diversity. A total of 179 AS events were predicted without reference to genomic information in our research.
Identification and characteristic analysis of SSRs
SSR loci were identified within the P. catalpifolia full-length transcriptome using MISA microsatellite software. A total of 7367 SSRs were identified, including 763 complex SSRs and 6604 complete SSRs. The total number of HQ transcripts containing SSRs was 6293, of which 747 contained ≥ 2 SSRs. SSRs occurred at a frequency of 24.23% (100% × total number of HQ transcripts containing SSRs/total number of HQ transcripts examined). The average distribution distance was 5.59 kb and the SSR appearance frequency was 28.37% (100% × total number of SSRs identified / total number of HQ transcripts examined) (Table 2).
The number of complete SSRs was 6604 in total and accounted for 89.64% of the total SSR loci, which included 2951 mononucleotide (44.68%), 2236 dinucleotide (33.86%), 1275 trinucleotide (19.31%), 50 tetranucleotide (0.76%), 24 pentanucleotide (0.36%) and 68 hexanucleotide SSRs (1.03%) (Fig. 5). The complete SSR lengths ranged from 10 to 88 bp, with a mean of 15.99 bp. The number of repeat SSR motifs ranged from 5 to 44, with a mean of 10.03. We found that SSRs with 6 motif repeats were the most common and accounted for 13.64% (901) of all SSRs, followed by SSRs with 10 repeats (897, 13.58%), 5 repeats (834, 12.63%) and 11 repeats (757, 11.46%), respectively. Furthermore, 4997 SSRs had motif repeat numbers ≤ 12, accounting for 75.67% of all SSR loci identified (Table 3).
A total of 112 repeat motifs were identified among the complete SSRs, of which there were 2 mononucleotides, 8 dinucleotides, 30 trinucleotides, 24 tetranucleotides, 12 pentanucleotides and 36 hexanucleotides, respectively (Table 3). Although SSR repeat types from mononucleotide to hexanucleotide all existed and they were also abundant, their occurrence frequency was quite different. The proportion of mononucleotide repeats dominated by A/T type was the highest (2951, 44.68%), and then dinucleotide repeats dominated by AG/CT and GA/TC (2236, 33.86%), trinucleotide repeats dominated by GAA/TTC and CCA/TGG (1275, 19.31%) and hexanucleotide repeats (68, 1.03%). Tetranucleotide and pentanucleotide repeat motifs exhibited relatively low frequencies, accounting for 0.76% and 0.36% of the total motif types, respectively. The statistical analysis of all SSR loci showed that the 5 repeat motif types with the highest occurrence frequency were in order as follows: A/T (2816, 42.64%), AG/CT (807, 12.22%), GA/TC (636, 9.63%), AC/GT (234, 3.54%) and CA/TG (215, 3.26%) (Table 3).
In P. catalpifolia, A/T was the most common mononucleotide repeat motif, accounting for 95.43% (2816) of all mononucleotide repeats, while C/G represented only 4.57% (135) (Fig. 6a). Of the dinucleotide repeats, AG/CT motif was the most frequent (807, 36.09%), followed by GA/TC (636, 28.44%), AC/GT (234, 10.47%) and CA/TG (215, 9.62%). The fewest dinucleotide motifs were GC/GC and CG/CG, each representing 1.79% (4) of the total dinucleotide repeats (Fig. 6b). There were 30 trinucleotide motifs present, of which GAA/TTC and CCA/TGG were the most frequent, accounting for 8.16% (104) and 8.00% (102) of the trinucleotide motifs, respectively, followed by AGA/TCT (82, 6.43%), CAG/CTG (81, 6.35%), AAG/CTT (78, 6.12%), CCG/CGG (78, 6.12%) and TCA/TGA (68, 5.33%). The fewest trinucleotide motifs were ACG/CGT (6, 0.47%), ACA/TGT (5, 0.39%) and GTA/TAC (2, 0.16%) (Table 3). Of the 24 tetranucleotide repeat motifs, CCCT/AGGG was the most frequent (8, 16%), followed by TGAA/TTCA (4, 8%) and GATT/AATC (4, 8%). The number of TTTG/CAAA, AAAT/ATTT, GAAA/TTTC, CTTT/AAAG and GGAG/CTCC all had 3 and accounted for 6%, 3 tetranucleotide motifs all with the number of 2 and another 13 repeat motif all with the number of 1. Within the 12 pentanucleotide repeat motifs, CCACC/GGTGG was the most frequent (9, 37.50%), followed by TCCTC/GAGGA (4, 16.67%) and CTTTT/AAAAG (2, 8.33%); the number of remaining 9 repeat motifs all were 1. Of the 36 hexanucleotide repeat motifs, AAGAGA/TCTCTT was the most frequent (8, 11.76%), followed by TTTCTT/AAGAAA (6, 8.82%). The number of 6 repeat motif types were all 3, 8 each were 2 and the remaining 20 each were 1 (Table 3).
Discussion
The lack of reference genome has impeded basic genetic research in P. catalpifolia and its related species. However, SMRT sequencing technology can generate full-length transcript sequences without a reference genome35,36,37 and has been widely used to predict and validate gene models related to some unique traits in species38. In this study, we used the SMRT technique to perform full-length transcriptome sequencing in P. catalpifolia using PacBio RS II platform. In total, 28.83 Gb sequencing data were obtained including 349,745 full-length non-chimeric sequence reads, which was similar to the number of FLNC reads in Rhododendron lapponicum39. After subjecting the reads to clustering, error correction and redundant sequence removal, a total of 25,969 HQ transcripts were finally obtained. Very-long-read sequences were generated using the SMRT sequencing technology, and one read is considered a full-length transcript under normal circumstances40. The HQ transcripts generated using SMRT sequencing were longer in length than those generated using an Illumina system. In this study, the average length of the HQ transcripts in P. catalpifolia was 1624 bp, while the mean unigene length was 945 bp in tung tree41, 683 bp in Pueraria lobata42 and 690 bp in Eucommia ulmoides43, each of which were sequenced using an Illumina system. In addition, we found that HQ transcripts > 1000 bp in length accounted for 84.04% of all HQ transcripts in our research, which was much higher than that in P. australis (40.09%)44 and P. tomentosa (42.16%)45 using Illumina sequencing technique. Our results demonstrated that SMRT sequencing is a reliable and efficient method to obtain full-length transcript sequences in species without an annotated reference genome.
We annotated 25,602 HQ P. catalpifolia transcripts using five public databases. The annotated HQ transcripts accounted for 98.59% of all HQ transcripts, a similar rate to those of transcriptomics studies in R. lapponicum39 and Medicago sativa46. The 367 HQ transcripts with no predicted functions are likely to be species-specific or unknown genes in P. catalpifolia. GO classification of the HQ transcripts indicated that the majority were associated with the GO terms metabolic processes, binding, catalytic activity, cellular processes, cell and cell part. HQ transcript annotation using KOG indicated that a large number of transcripts were involved in posttranslational modifications, protein turnover, chaperones, translation, and ribosomal structure and biogenesis. A total of 13,829 HQ transcripts were assigned to specific KEGG pathways, such as carbohydrate metabolism, energy metabolism, translation, folding, sorting and degradation. We also found that many HQ transcripts exhibited multiple molecular functions and participated in diverse biological pathways. Our study provides a wealth of genetic information for molecular research into the growth and development of P. catalpifolia leaves, particularly in response to drought stress.
In recent years, SSR molecular markers have been widely used for genetic map construction, genetic diversity analyses and functional gene mining. However, the traditional methods of SSR primer development are time-consuming, complex and costly, thus hindering their development seriously. While the SSR primers developed on the basis of transcriptome sequencing data information are economical, efficient, and abundant, which has gradually become one of important methods. Furthermore, SSR molecular markers are rapidly being developed alongside recent advancements in transcriptome sequencing technology47,48. In our study, a total of 7367 SSR loci were detected from 25,969 HQ transcripts, including 763 complex SSRs and 6604 complete SSRs. The frequency of the SSRs was 28.37%, and the average distribution distance was 5.59 kb. Among the 6604 complete SSRs, the most abundant and frequent mononucleotide, dinucleotide and trinucleotide motifs were A/T, AG/CT and GAA/TTC, respectively; studies examining SSRs in Hevea brasiliensis49, Chinese cabbage50 and R. lapponicum39 produced similar results. A/T was the most abundant mononucleotide motif (2816, 95.43%), which was consistent with a study performed by Lagercrantz et al51. AG/CT (807, 36.09%) and GA/TC (636, 28.44%) were the most abundant dinucleotide motifs, and CT repeats usually existed in transcriptional regions that might take part in antisense transcription and have an effect on gene regulation39,52. There were differences in SSR abundance of different plant species in diverse researches, and repeat number of 6, 10, 5, 11, and 12 occupied 59.30% of the total complete SSR loci in our study. The SSR markers that we have developed in this work will facilitate mining for drought resistance genes, breeding drought resistant varieties, genetic diversity analyses and genetic map construction in P. catalpifolia. Of course, the SSRs found in this study were predicted theoretically and should be verified experimentally before further using.
Materials and methods
Plant materials and RNA extraction
P. catalpifolia seedlings were planted in separate pots at Mengzhou Forest Farm at the Paulownia Research and Development Center of State Administration of Forestry and Grasslands (Jiaozuo, Henan, China, 112° 42′ 58″ E, 34° 51′ 38″ N). The third and fourth fully expanded functional leaves from the top of the stem were collected at 0, 8 and 16 days after drought stress, respectively. The leaves were immediately frozen in liquid nitrogen and stored at − 80 °C until the experiment23. The Paulownia catalpifolia used in this study were identified by Paulownia Research and Development Center of State Administration of Forestry and Grassland, and the collection and use of Paulownia catalpifolia samples in our experiment comply with the guidelines of Paulownia Research and Development Center of State Administration of Forestry and Grassland. Total RNAs extraction were performed using the EZ-10 DNAaway RNA mini-prep kit (Sangon Biotech Co., Shanghai, China) following the manufacturer’s instructions. The total RNAs of three samples above were mixed equally according to the method of Diao53 to form the sample S for transcriptome sequencing. The degrees of RNA degradation and contamination were evaluated using 1% agarose gels39. The RNA purity and concentration were checked using the NanoPhotometer spectrophotometer (Implen, CA, USA) and Qubit RNA Assay Kit (Life Technologies, CA, USA), respectively22. RNA integrity was analyzed using an Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA)22. The resulting high-quality RNA was used for full-length transcriptome sequencing.
cDNA library construction and SMRT sequencing of the full-length transcriptome
Full-length cDNA was synthesised from 1.0 μg purified mRNA using the SMARTer PCR cDNA Synthesis Kit (Clontech, USA) according to the manufacturer’s protocol, its size were selected using the BluePippin Size-Selection System (Sage Science, USA) and then PCR amplified again. The cDNA library was constructed after repairing the ends, connecting dumbbell-shaped SMRT adapters, performing exonuclease digestions and conducting a secondary screening using BluePippin. After the cDNA library had passed quality control using the Qubit 2.0 and Agilent 2100, full-length transcriptome sequencing of P. catalpifolia was performed using the PacBio RS II platform, based on the target data volume23.
Quality control and functional annotation of the full-length transcriptome
The raw SMRT data were pre-processed using the SMRT Pipe analysis workflow within the PacBio SMRT Analysis software suite. Examination of the polyadenosine signal and 5′ and 3′ adaptors, as well as error correction, were performed following the methods similar to the one described54. Full-length SMRT transcripts were identified, and non-redundant HQ transcripts were acquired using CD-HIT-EST software55. Clustering and removal of redundant sequences were performed using the PacBio SMRT LINK Cluster tool, and all HQ transcripts were aligned to nucleotide and protein databases using BLASTX54. The databases used in this study were NCBI non-redundant, gene ontology (GO), eukaryotic orthologous groups (KOG), Kyoto encyclopedia of genes and genomes (KEGG) and Swiss-Prot.
Identification of lncRNAs, coding sequences (CDSs) and AS variants
LncRNA candidates were identified using the following software: coding potential calculator 2 (CPC2), coding potential assessment tool (CPAT), predictor of long non-coding RNAs and messenger RNAs based on an improved k-mer scheme (PLEK), and the coding–non-coding index (CNCI), respectively. LncRNAs with > 200 nucleotides were selected. TransDecoder version 3.0.0 was used to identify candidate coding sequences (CDSs) in the full-length transcriptome of P. catalpifolia. All non-redundant HQ transcripts were aligned using a previously described method56. Candidate AS events were identified using the selection criteria described by Diao et al.53.
Identification and characterization of SSRs
The microsatellite identification tool (MISA) was used to identify SSRs within the 25,969 HQ transcripts, and the characteristics of the repeated motif types were further analyzed statistically. In this study, the SSR locus were identified according to the criteria below: the repeat number of mononucleotide motifs was ≥ 10 and the repeat numbers of di-, tri-, tetra-, penta- and hexanucleotide motifs were ≥ 6, 5, 5, 5 and 5, respectively.
Data availability
The raw data from SMRT sequencing are accessible at NCBI under bioproject (PRJNA565572).
References
Wu, L. C. et al. Effects of trunk-extension pruning at different intensities on the growth and trunk form of Paulownia fortunei. For. Ecol. Manag. 327, 128–135 (2014).
Manavalan, L. P., Guttikonda, S. K., Phan Tran, L.-S. & Nguyen, H. T. Physiological and molecular approaches to improve drought resistance in Soybean. Plant Cell Physiol. 50(7), 1260–1276 (2009).
Schaart, J. G., van de Wiel, C. C. M., Lotz, L. A. P. & Smulders, M. J. M. Opportunities for products of new plant breeding techniques. Trends Plant Sci. 21(5), 438–449 (2016).
Song, S. L., Sato, T., Ishii, K., Saito, A. & Ohba, K. In vitro mass propagation by meristem culture of two mature trees of Paulownia catalpifolia. J. Jpn. For. Soc. 72(6), 495–498 (1990).
Wang, Y. A. et al. Undescribed C-geranylflavonoids isolated from the fruit peel of Paulownia catalpifolia T. Gong ex D.Y. Hong with their protection on human umbilical vein endothelial cells injury induced by hydrogen peroxide. Phytochemistry 158, 126–134 (2019).
Xiao, C. M. et al. Three new C-geranylated flavonoids from Paulownia catalpifolia T. Gong ex D.Y. Hong seeds with their inhibitory effects on xanthine oxidase. Phytochem. Lett. 36, 162–165 (2020).
Buschiazzo, E. & Gemmell, N. J. The rise, fall and renaissance of microsatellites in eukaryotic genomes. BioEssays 28, 1040–1050 (2006).
Wheeler, G. L., Dorman, H. E., Buchanan, A., Challagundla, L. & Wallace, L. E. A review of the prevalence, utility, and caveats of using chloroplast simple sequence repeats for studies of plant biology. Appl. Plant Sci. 2(12), 1400059 (2014).
Gupta, R. K. & Varshney, P. K. The development and use of microsatellite markers for genetic analysis and plant breeding with emphasis on bread wheat. Euphytica 113, 163–185 (2000).
Thomas, M. R. & Scott, N. S. Microsatellite repeats in grapevine reveal DNA polymorphisms when analysed as sequence-tagged sites (STSs). Theor. Appl. Genet. 86(8), 985–990 (1993).
Pan, L. et al. EST-SSR marker characterization based on RNA-sequencing of Lolium multiflorum and cross transferability to related species. Mol. Breed. 38, 80 (2018).
Bazzo, B. R., de Carvalho, L. M., Carazzolle, M. F., Pereira, G. A. G. & Colombo, C. A. Development of novel EST-SSR markers in the macaúba palm (Acrocomia aculeata) using transcriptome sequencing and cross-species transferability in Arecaceae species. BMC Plant Biol. 18, 276 (2018).
Eujayl, I., Sorrells, M. E., Baum, M., Wolters, P. & Powell, W. Isolation of EST-derived microsatellite markers for genotyping the A and B genomes of wheat. Theor. Appl. Genet. 104, 399–407 (2002).
Jeong, S. W. et al. Development of EST-SSR markers through de novo RNA sequencing and application for biomass productivity in kenaf (Hibiscus cannabinus L.). Genes Genom. 39, 1139–1156 (2017).
Tang, D. Q., Sun, Y., Li, X., Yan, Z. & De Shi, Y. M. novo sequencing of the Freesia hybrida petal transcriptome to discover putative anthocyanin biosynthetic genes and develop EST-SSR markers. Acta Physiol. Plant 40, 168 (2018).
Acuña, C. V. et al. Discovery, validation, and in silico functional characterization of EST-SSR markers in Eucalyptus globulus. Tree Genet. Genom. 8, 289–301 (2012).
Du, F. K. et al. Exploiting the transcriptome of Euphrates Poplar, Populus euphratica (Salicaceae) to develop and characterize new EST-SSR markers and construct an EST-SSR database. PLoS ONE 8(4), e61337 (2013).
Feng, S. P., Li, W. G., Huang, H. S., Wang, J. Y. & Wu, Y. T. Development, characterization and cross-species/genera transferability of EST-SSR markers for rubber tree (Hevea brasiliensis). Mol. Breed. 23, 85–97 (2009).
Guo, Q. et al. Development and evaluation of a novel set of EST-SSR markers based on transcriptome sequences of Black Locust (Robinia pseudoacacia L.). Genes 8, 177 (2017).
Yan, L. P. et al. De novo transcriptome analysis of Fraxinus velutina using Illumina platform and development of EST-SSR markers. Biol. Plant. 61(2), 210–218 (2017).
Du, J., Zhang, Z., Zhang, H. G. & Tang, J. H. EST-SSR marker development and transcriptome sequencing analysis of different tissues of Korean pine (Pinus koraiensisSieb. et Zucc.). Biotechnol. Biotechnol. Equip. 31(4), 679–689 (2017).
Feng, K., Lu, X. Y., Luo, J. & Tang, F. SMRT sequencing of the full-length transcriptome of Odontotermes formosanus (Shiraki) under Serratia marcescens treatment. Sci. Rep. 10, 15909 (2020).
Wu, Q. C. et al. Full-length transcriptome sequencing analysis and development of EST-SSR markers for the endangered species Populus wulianensis. Sci. Rep. 10, 16249 (2020).
Hoang, N. V. et al. A survey of the complex transcriptome from the highly polyploid sugarcane genome using full-length isoform sequencing and de novo assembly from short read sequencing. BMC Genomics 18, 395 (2017).
Kuang, X. J., Sun, S. J., Wei, J. H., Li, Y. & Sun, C. Iso-Seq analysis of the Taxus cuspidatatranscriptome reveals the complexity of Taxol biosynthesis. BMC Plant Biol. 19, 210 (2019).
Chao, Y. H. et al. Analysis of transcripts and splice isoforms in red clover (Trifolium pratense L.) by single-molecule long-read sequencing. BMC Plant Biol. 18, 300 (2018).
Jia, D. et al. SMRT sequencing of full-length transcriptome of flea beetle Agasicles hygrophila (Selman and Vogt). Sci. Rep. 8, 2197 (2018).
Zeng, D. G. et al. Single-molecule long-read sequencing facilitates shrimp transcriptome research. Sci. Rep. 8, 16920 (2018).
Li, W. et al. novo transcriptomic analysis and development of EST-SSRs for Styrax japonicus. Forests 9, 748 (2018).
Tian, X. Y. et al. Transcriptome sequencing and EST-SSR marker development in Salix babylonica and S. suchowensis. Tree Genet. Genom. 15, 9 (2019).
Xia, H. et al. Distinguishing upland and lowland rice ecotypes by selective SSRs and their applications in molecular-assisted selection of rice drought resistance. Euphytica 206, 11–20 (2015).
Wang, P. L. et al. Characterization and development of EST-SSR markers from a cold-stressed transcriptome of centipedegrass by Illumina paired-end sequencing. Plant Mol. Biol. Rep. 35, 215–223 (2016).
Mercer, T. R., Dinger, M. E. & Mattick, J. S. Long non-coding RNAs: Insights into functions. PRogRess 10, 155–159 (2009).
Sammeth, M., Foissac, S. & Guigó, R. A general definition and nomenclature for alternative splicing events. PLoSComput. Biol. 4(8), e1000147 (2008).
Abdel-Ghany, S. E. et al. A survey of the sorghum transcriptome using single-molecule long reads. Nat. Commun. 7, 11706 (2016).
Wang, B. et al. Unveiling the complexity of the maize transcriptome by single-molecule long-read sequencing. Nat. Commun. 7, 11708 (2016).
Chen, S. Y., Deng, F. L., Jia, X. B., Li, C. & Lai, S. J. A transcriptome atlas of rabbit revealed by PacBio single-molecule long-read sequencing. Sci. Rep. 7, 7648 (2017).
Minoche, A. E. et al. Exploiting single-molecule transcript sequencing for eukaryotic gene prediction. Genome Biol. 16, 184 (2015).
Jia, X. P. et al. Single-molecule long-read sequencing of the full-length transcriptome of Rhododendron lapponicum L. Sci. Rep. 10, 6755 (2020).
Sharon, D., Tilgner, H., Grubert, F. & Snyder, M. A single-molecule long-read survey of the human transcriptome. Nat. Biotechnol. 31(11), 1009–1014 (2013).
Zhang, L. et al. Fatty acid profile and unigene-derived simple sequence repeat markers in Tung Tree (Vernicia fordii). PLoS ONE 9(8), e105298 (2014).
Wang, X., Li, S. T., Li, J., Li, C. F. & De Zhang, Y. S. novo transcriptome sequencing in Pueraria lobata to identify putative genes involved in isoflavones biosynthesis. Plant Cell Rep. 34(5), 733–743 (2014).
Feng, Y. Z. et al. Characterization of glycolytic pathway genes using RNA-Seq in developing kernels of Eucommia ulmoides. J. Agric. Food Chem. 64(18), 3712–3731 (2016).
Dong, Y. P., Fan, G. Q., Zhao, Z. L. & Deng, M. J. Transcriptome expression profiling in response to drought stress in Paulownia australis. Int. J. Mol. Sci. 15(3), 4583–4607 (2014).
Fan, G. Q. et al. Transcriptome, microRNA, and degradome analyses of the gene expression of Paulownia with phytoplamsa. BMC Genomics 16, 896 (2015).
Chao, Y. H. et al. Analysis of transcripts and splice isoforms in Medicago sativa L. by single-molecule long-read sequencing. Plant Mol. Biol. 99(3), 219–235 (2019).
Dutta, S. et al. Development of genic-SSR markers by deep transcriptome sequencing in pigeonpea [Cajanus cajan (L.) Millspaugh]. BMC Plant Biol. 11, 17 (2011).
Zhai, L. L. et al. Novel and useful genic-SSR markers from de novo transcriptome sequencing of radish (Raphanus sativus L.). Mol. Breed. 33, 611–624 (2014).
Li, D. J., Deng, Z., Qin, B., Liu, X. H. & De Men, Z. H. novo assembly and characterization of bark transcriptome using Illumina sequencing and development of EST-SSR markers in rubber tree (Hevea brasiliensisMuell. Arg.). BMC Genomics 13, 192 (2012).
Song, X. M., Ge, T. T., Li, Y. & Hou, X. L. Genome-wide identification of SSR and SNP markers from the non-heading Chinese cabbage for comparative genomic analyses. BMC Genomics 16, 328 (2015).
Lagercrantz, U., Ellegren, H. & Andersson, L. The abundance of various polymorphic microsatellite motifs differs between plants and vertebrates. Nucleic Acids Res. 21(5), 1111–1115 (1993).
Wang, H. X., Walla, J. A., Zhong, S. B., Danqiong Huang, D. Q. & Dai, W. H. Development and cross-species/genera transferability of microsatellite markers discovered using 454 genome sequencing in chokecherry (Prunus virginiana L.). Plant Cell Rep. 31, 2047–2055 (2012).
Diao, S., Ding, X. Y., Luan, Q. F. & Jiang, J. M. A complete transcriptional landscape analysis of Pinus elliottii Engelm. using third-generation sequencing and comparative analysis in the Pinus phylogeny. Forests 10, 942 (2019).
Yang, L. F. et al. Full-length transcriptome sequences of ephemeral plant Arabidopsis pumila provides insight into gene expression dynamics during continuous salt stress. BMC Genomics 19, 717 (2018).
Xu, Z. C. et al. Full-length transcriptome sequences and splice variants obtained by a combination of sequencing platforms applied to different root tissues of Salvia miltiorrhiza and tanshinone biosynthesis. Plant J. 82, 951–961 (2015).
Liu, X. X., Mei, W. B., Soltis, P. S., Soltis, D. E. & Barbazuk, W. B. Detecting alternatively spliced transcript isoforms from single-molecule long-read sequences without a reference genome. Mol. Ecol. Resour. 17, 1243–1256 (2017).
Acknowledgements
This research work was supported by the Fundamental Research Funds for the Central Non-profit Research Institution of Chinese Academy of Forestry (CAFYBB2017ZA001-6).
Author information
Authors and Affiliations
Contributions
Y.Z.F., Y.Z. and J.Q. performed the study conception and experiments. J.J.Z., B.P.W. and C.W.Y. collected samples, prepared materials and analyzed the data. Y.Z.F. and H.J.Z. collected and analyzed the data. Y.Z.F. and Y.Z. written the original draft of the manuscript. J.Q. reviewed and edited of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Feng, Y., Zhao, Y., Zhang, J. et al. Full-length SMRT transcriptome sequencing and microsatellite characterization in Paulownia catalpifolia. Sci Rep 11, 8734 (2021). https://doi.org/10.1038/s41598-021-87538-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-87538-8
This article is cited by
-
Full-length transcriptome combined with RNA sequence analysis of Fraxinus chinensis
Genes & Genomics (2023)
-
Development and validation of SSR markers related to flower color based on full-length transcriptome sequencing in Chrysanthemum
Scientific Reports (2022)
-
Microsatellite analysis and polymorphic marker development based on the full-length transcriptome of Camellia chekiangoleosa
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.