Abstract
NTRK fusion-positive tumors are known to be highly sensitive to TRK inhibitors, such as larotrectinib and entrectinib. Therefore, identification of patients who can potentially benefit from these inhibitors is important; however, the frequency of NTRK fusions in Japanese patients with colorectal cancer (CRC) is unknown. We performed pan-TRK staining using TMA-based immunohistochemistry (IHC) on samples from 971 consecutive Japanese CRC cases from a single institution. Positive cases were further analyzed using NanoString and subsequent targeted RNA sequencing. We found three positive cases using TRK-IHC. Furthermore, the Nanostring assay supported the presence of NTRK fusion in these cases. Subsequent targeted RNA-sequencing and RT-PCR revealed two cases with TPM3-NTRK1 and one with TPR-NTRK1. The TNM stages of these cases were stage I, stage IIA, and stage IIIB, and two showed microsatellite instability-high status. Next-generation sequencing analysis using Cancer hotspot panel revealed TP53 and SMAD4 mutations in separate cases. IHC of β-catenin did not show nuclear accumulation. We found three cases (0.31%) of CRC with NTRK1 fusion among 971 consecutive Japanese CRC cases. No potential driver alterations other than NTRK fusion were identified in these three patients.
Similar content being viewed by others
Introduction
The neurotrophin kinase (NTRK) genes NTRK1, NTRK2 and NTRK3 encode the tropomyosin receptor tyrosine kinases TRKA, TRKB and TRKC, respectively. These receptor tyrosine kinases function during normal neuronal tissue development and maintenance. Genome-wide and RNA sequence analyses have identified gene rearrangements involving each of the NTRK genes in malignancies, including colorectal carcinomas (CRCs). To date, various kinds of NTRK fusions have been reported in CRCs, including LMNA-NTRK11,2,3,4, PLEKHA-NTRK15, ETV6-NTRK33, SCYL3-NTRK16, TPM3-NTRK14,7, TPR-NTRK1, and EML4-NTRK34. These fusion genes are thought to promote tumor growth by driving expression of constitutively active fusion proteins through the activation of downstream signal transduction pathways, such as phosphoinositide 3-kinase/protein kinase B (PI3K-AKT) and mitogen-activated protein kinase (MAPK) signaling cascades8,9,10. NTRK fusions have been reported to be infrequent in common cancers11, and pathognomonic in specific rare cancers, such as infantile fibrosarcoma12,13 and mammary analog secretory carcinoma14. NTRK fusion-positive tumors are reported to be highly sensitive to TRK inhibitors such as larotrectinib and entrectinib; therefore, identification of patients who can potentially benefit from these inhibitors is important. In CRC, the frequency of NTRK fusion is reported to be as high as 0.3%4, although its frequency in Japanese patients with CRC is unknown.
In this study, we performed pan-TRK staining using tissue microarray (TMA)-based immunohistochemistry (IHC) in a large cohort of consecutive Japanese CRC samples. Positive cases were further analyzed using NanoString and target RNA and DNA sequencing. This is the first report to investigate the frequency of NTRK fusion in a large cohort of Japanese CRC patients.
Materials and methods
Case selection
Sequential colorectal adenocarcinoma cases were selected from the 2008–2018 pathological records at the Pathology Department of Juntendo University Hospital, Tokyo, Japan. A total of 971 colorectal adenocarcinoma cases from 956 patients (including 13 patients with synchronous double CRCs and two patients with non-synchronous double cancer) were collected. Clinicopathological information, including age, sex, tumor location, tumor depth, stromal volume, lymphatic invasion, vascular invasion, nodal metastasis and survival data, was obtained for these patients. Tissue microarray (TMA) blocks, each consisting of 2 mm cores, were made for these cases.
Immunohistochemistry (IHC)
Immunohistochemical staining was performed using the streptavidin–biotin method11, with antibodies to the following: pan-TRK (clone: EPR17341; Abcam, Cambridge, MA, USA) as previously described3. IHC for p53 (Clone: DO-7; DAKO, Glostrup, Denmark), Ki-67 (Clone: MIB-1; DAKO, Glostrup, Denmark), and β-catenin (Clone: Rabbit Poly; DAKO, Glostrup, Denmark) were also performed. The diffuse staining pattern was judged as positive for pan-TRK, regardless of the staining intensity on TMA-based IHC. Three positive cases for pan-TRK IHC on TMA sections were also stained on the whole FFPE sections. IHC for p53 was performed on TMA sections, while IHC for β-catenin and Ki-67 was performed and evaluated using whole sections for only 3 positive cases. The p53 IHC findings were classified into overexpression, wild-type, and complete absence15,16. The Ki-67 labeling index was evaluated in representative areas that showed the highest immunoreactivity by counting the number of positive cells among 1000 tumor cells.
Nanostring assay
Nanostring (NanoString Technologies, Inc., Seattle, Washington, USA) assays were performed as previously described17,18, to confirm that positive staining by pan-TRK immunohistochemistry led to unbalanced expression of NTRK gene exons, as expected of gene fusions. The custom-designed probe set contained 90 tyrosine kinase genes. For the Nanostring, 400 ng of RNA was hybridized to the probes (a reporter probe and a capture probe) at 65 °C for 18–24 h, using a thermal cycler. Samples were then inserted into the nCounter Prep Station for the removal of excessive probes, purification, and immobilization onto the internal surface of a sample cartridge for 3 h. Finally, the sample cartridge was transferred to the nCounter Digital Analyzer, where color codes were counted and tabulated for each target molecule. The expression number of the base sequence of the probe set was analyzed using nSolver software (Version 4). Using the standard deviation, a graph plotted with 2SD and 3SD lines was prepared.
Next generation sequencing or Targeted RNA sequencing
Total RNA was extracted from formalin fixed paraffin embedded (FFPE) tissue using a Maxwell RSC RNA FFPE Kit (Promega, Madison, WI, USA). Targeted RNA sequencing was performed at RIKEN GENESIS (Tokyo, Japan). To check sample quantity and quality, the concentration of total RNA was determined with the Qubit RNA BR Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA), and quality was examined using an Agilent 4200 TapeStation (Agilent Technologies, Santa Clara, CA, USA). Library preparation was performed using Archer FusionPlex Lung Kit (ArcherDX, Boulder, CO, USA) and Archer Molecular Barcode (MBC) Adapters for next-generation sequencing on an Illumina platform, according to the manufacturer's protocol. In short, cDNA was first synthesized from total RNA using random priming, and RNA quality was then determined by real-time quantitative PCR. After second-strand cDNA synthesis, end repair and A-tailing steps were performed, and the MBC adapters were ligated. Two rounds of PCR with universal and gene-specific primers were then performed. The libraries were quantified using the KAPA Library Quantification Kit for Illumina (KAPA Biosystems, Wilmington, MA, USA). Next-generation sequencing was performed on an Illumina MiSeq platform, and configured to generate 151 bp paired-end reads. Data analysis, including adapter trimming, deduplication, alignment to the reference sequence, and fusion gene detection were performed in Archer Analysis 6.0.4.
RT-PCR
Using extracted total RNA, cDNA was synthesized with SuperScript III First-Strand Synthesis System (Invitrogen/Thermo Fisher Scientific, Waltham, MA, USA) for RT-PCR19. RT-PCR was performed according to the manufacturer’s protocol as follows: initial denaturation at 94 °C for 2 min and 40 cycles at 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s, followed by 72 °C for 2 min. The PCR product was electrophoresed on a 2% agarose gel. PCR products with the appropriate and expected sizes were excised from the agarose. Purified PCR products were sequenced with dideoxynucleotides (BigDye Terminator v3.1; Applied Biosystems, Foster City, CA, USA) and specific primers, subsequently purified using a BigDye X Terminator Purification Kit (Applied Biosystems, Foster City, CA, USA), and analyzed with a capillary sequencing machine (3730xl Genetic Analyzer, Applied Biosystems). The primer sequences were designed based on the NGS results as follows: TPM3 exon8, 5′-GATCGGTAGCCAAGCTGGAA-3 and NTRK1 exon9, 5′-GCACAAGGAGCAGCGTAGAA-3′ for the detection of TPM3-NTRK1 fusion; TPR exon24, 5′-TCTTCAGAGAGCAAGCACAGC-3′ and NTRK1 exon10, 5′-GCACAAGGAGCAGCGTAGA-3′ for the detection of TPR-NTRK1 fusion.
DNA extraction and NGS by cancer hotspot panel
Genomic DNA was extracted from tumoral and corresponding non-tumoral tissues of NTRK fusion-positive cases using the Maxwell RSC DNA FFPE Kit—PKK Custom (Promega). We assessed DNA integrity and NGS using the Ion Ampliseq Cancer Hotspot Panel v2 (Thermo Fisher Scientific). The details of this analysis have been described previously20.
MSI analysis
MSI analysis was performed as previously described21. Briefly, the five microsatellite loci (BAT-25, BAT-26, D2S123, D5S346, and D17S250) recommended in the 1997 NCI-sponsored MSI workshop were amplified in each PCR reaction. PCR products were analyzed by capillary electrophoresis (CE) using an ABI 310 Genetic Analyzer (Applied Biosystems). For interpretation purposes, microsatellite instability at ≥ 2 loci was defined as MSI-high, instability at a single locus was defined as MSI-low, and no instability at any of the loci tested was defined as MSS.
Statistical analysis
Statistical analyses were performed using GraphPad Prism software version 7.0a (GraphPad, San Diego, CA, USA). Smirnov-Grubbs’ test was used to identify outlier.
Ethics approval
This study was reviewed and approved by Juntendo University School of Medicine Institutional Review Board (#2017177). All experiments were performed in accordance with relevant guidelines and regulations of the institution and the Declaration of Helsinki. The informed consents were obtained from all subjects.
Results
Detection of TRK expression by pan-TRK IHC
The clinicopathological characteristics of the 971 CRC samples are summarized in Table 121. Briefly, lymphatic invasion and vascular invasion were observed in 430 (44.3%) and 437 (45.0%) cases, respectively. The observed pathological stages were as follows: stage I, 122 patients; stage II 337; stage III 307; stage IV 189; and unknown, 3. The three unknowns were due to local resection without lymph node dissection.
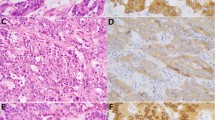
TMA-based pan-TRK IHC identified three positively stained cases. The histologies of cases #1–3 were tub2 > por1 (Fig. 1a), muc > tub2 (Fig. 1b), and tub2 > muc (Fig. 1c), respectively. Two out of three cases showed strong and diffuse membranous and cytoplasmic staining (Cases #1 and #2, Fig. 1d,e), whereas the remaining case showed weak but diffuse cytoplasmic staining (Case #3, Fig. 1f). These staining patterns were also seen in IHC analysis of whole sections.
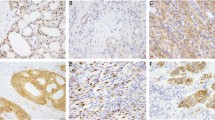
(a–o) Histological feature of Case #1 showing adenocarcinoma with a solid pattern (a). Histology of Case #2 showing mucinous carcinoma with a focal solid area (b). Histology of Case #3 showing moderately differentiated adenocarcinoma with a focal mucinous area (c). Pan-trk IHC images showing diffuse cytoplasmic and membranous staining for Case #1 (d) and Case #2 (e), while weak but diffuse cytoplasmic staining can be seen for Case #3 (f). Insets show higher magnifications. All three cases showed β-catenin membranous staining without nuclear staining (Case #1: (g); Case #2: (h); Case #3: (i). p53 of Case #1 (j) and Case #2 (k) showing wild type pattern, Case #3 showing completely absent pattern (l). Insets show higher magnifications. MIB-1 LI was high in all cases, with levels of approximately 80% in Case #1 (m), 50% in Case #2 (n) and 75% in Case #3 (o).
Nanostring assay for the pan-TRK-IHC positive samples
Before sending the pan-TRK IHC-positive samples for RNA-target sequencing, a Nanostring assay consisting of 90 tyrosine kinase (TK) genes comprising NTRK1-3 was used to evaluate the imbalance in NTRK1-3 expression. One of the three cases (Case #1) was excluded from the statistical analysis after data normalization, probably due to poor sample quality. After the expression level of the NTRK1 exon 14/exon1-2 was checked manually for this case, we observed more than tenfold higher expression of the NTRK1 exon14 compared to that of NTRK1 exon 1–2. The remaining two cases (Cases #2 and #3) showed statistically significant imbalanced expression of NTRK1, with expression more than triple the standard deviation (SD) of our accumulated dataset comprised of various types of malignancies, including our previously reported LMNA-NTRK1 fusion tumor22 (Fig. 2). These accumulated dataset samples other than LMNA-NTRK1 fusion tumor22 analyzed by Nanostring were also examined by pan-trk IHC for the detection of NTRK fusion tumor. Furthermore, expression level for each TK gene was also considered in addition to the imbalanced expression of TK genes to exclude the possibility of fusion formation and the subsequent overexpression.
Nanostring analysis showing an imbalance of NTRK1 gene expression between probes targeted against the 5′-side and 3′-side sequences. Smirnov–Grubbs’ test was used to identify outlier. Blue and red lines represent 2SD and 3SD, respectively. #2 and # 3 correspond to Case #2 and #3, respectively, while #36 is a positive control tumor sample previously shown to have LMNA-NTRK1 fusion22. The samples other than Case #2 and #3 are not colorectal cancers. Data was analyzed with our accumulated dataset comprised of various types of malignancies. Case #21 seems to show statistically significant imbalanced expression (blotted between 2 and 3SD) of NTRK1. This case is an inflammatory myofibroblastic tumor previously shown to have an ETV6-NTRK3 fusion in our previous study20 in which we proposed that imbalanced expression of more than 3SD would be reliable for the presence of a fusion gene.
Targeted RNA sequencing and RT-PCR for NTRK fusion detection
RNA sequencing revealed the presence of TPM3 (exon8)-NTRK1 (exon9) in cases #1 and #2, and TPR (exon24)-NTRK1 (exon10) fusion transcript in case #3. Subsequent RT-PCR confirmed these fusion transcripts in each case (Fig. 3a,b).
Clinicopathological characteristics of CRC with NTRK1 fusion
Clinicopathological information on the three CRC cases harboring NTRK1 fusions are summarized in Table 2. Briefly, the mean age of the three patients was 79 years, which was higher than that of the other CRC patients. The histologies of cases #1–3 included moderately differentiated adenocarcinoma (tub2) > poorly differentiated adenocarcinoma solid type (por1), mucinous carcinoma (muc) > tub2 and tub2 > muc, and the pathological stages were stage I, stage IIIB and stage IIA, respectively. Case #2, who presented with Stage IIIB CRC, experienced liver and lung metastasis one year after surgery and subsequently died of brain vascular disease. Case #3 received chemotherapy after surgery and exhibited no evidence of disease. For Case #1, the patient was lost to follow-up, and the prognostic information was not available at six months after surgery.
Immunohistochemical characteristics of CRC with NTRK1 fusion
All three cases with NTRK fusion showed β-catenin membranous staining without nuclear staining (Fig. 1g,h,i). p53 IHC showed weak to moderate nuclear staining in scattered tumor cells in Case #1 (Fig. 1j) and #2 (Fig. 1k), suggesting a wild-type staining pattern, whereas complete absence was seen in Case #3 (Fig. 1l) 15,16. MIB-1 LI was high in all cases, with levels of approximately 80% in Case #1 (Fig. 1m), 50% in Case #2 (Fig. 1n), and 75% in Case #3 (Fig. 1o).
Mutation analysis by NGS
No mutations were detected in case #1. Mutations in SMAD4 (p.Gly386Asp, c.1157G > A) and TP53 (p.Glu294Ter, c.880G > T) were detected in cases #2 and #3, respectively. This SMAD4 (p.G386D) mutation is described as “Pathogenic/Likely Pathogenic”.
MSI analysis
MSI analysis was performed for the three cases with NTRK fusion. Two (Cases #1 and #2) of the three cases were classified as MSI-H (Fig. 4), whereas the remaining cases were classified as MSS.
Past and family histories
Past and family histories of neoplasms were also examined. Only Case #3 experienced thymoma, while the other two patients had no prior history of neoplasm. Regarding family history, three brothers experienced gastric cancer, leukemia, and a malignant tumor (details unknown), respectively, in Case #1. Case #2 had no particular family history. In Case #3, the patient’ s mother experienced pancreatic cancer.
Discussion
Recent genome-wide and RNA sequence analyses have identified new fusion genes, including NTRK fusions, in CRC. NTRK fusions in CRC are extremely rare, with a reported frequency of up to 0.3%11; the frequency of NTRK fusion in Japanese patients with CRC has not been described so far. In this study, we found three CRC cases with NTRK fusions from examined 971 cases, based on 956 patients. Histologically, two of the three cases exhibited tubular adenocarcinoma, while the remaining case exhibited mucinous carcinoma with a tubular adenocarcinoma component. A recent study demonstrated that in CRC, ALK, ROS1, and NTRK rearrangements occur more frequently in elderly patients23, and this was also the case in our study, with the mean age of these three patients being 79 years. Furthermore, a recent study reported that most of the patients with NTRK fusion colon cancers were women (13/16)4; however, only one out of our three CRC patients with NTRK1 fusions were women. NTRK fusion-positive CRCs have been reported to be located more frequently in the right side of the colon23, whereas another study demonstrated that CRC with NTRK fusion is widely distributed from the cecum to the sigmoid colon4. The three tumors in this study were located in the ascending, transverse, and descending colon.
We employed TMA-based pan-TRK immunohistochemistry as a prescreen for possible CRC harboring NTRK fusions. TMA-based immunohistochemistry is a powerful tool when dealing with thousands of clinical samples, although it has the associated risk of possibly missing positive cases due to limited information from the small cores. To evaluate the heterogeneity of pan-TRK immunohistochemistry in CRC tissue, we also performed pan-TRK IHC using whole sections in positive cases. We confirmed the same staining pattern throughout the sections. In previous reports4, pan-TRK IHC showed a uniform staining pattern within the same CRC sections. Based on these findings, we propose that screening for NTRK fusions using IHC on TMA is reliable.
Recent studies have reported that pan-TRK immunohistochemical staining in tumors with NTRK3 fusion could be weaker than in those with either NTRK1 or NTRK2 fusion-positive tumors, raising the possibility of false negative findings in NTRK3 fusion-positive tumors24. We also experienced a case of a soft tissue tumor with LMNA-NTRK1 fusion showing strong cytoplasmic pan-TRK staining22. In this study, two TPM3-NTRK1 tumors showed strong membranous and cytoplasmic staining, whereas the TPR-NTRK1 tumor showed weak but diffuse cytoplasmic staining in pan-TRK IHC. These findings are consistent with those of previous reports4,23. The expression pattern of pan-TRK IHC may suggest a fusion partner.
The molecular pathological characteristics of the three CRC cases with NTRK1 fusion were also assessed. The p53 IHC results for Cases #1 and #2 were wild type, while Case #3 was completely negative. Using NGS analysis, a TP53 nonsense mutation was detected in one case (Case #3), and this case showed a complete absence pattern for p53 IHC. The remaining two cases showed a TP53 wild-type pattern by p53 IHC15,16, although Case #1 also showed weak cytoplasmic staining. SMAD4 mutations were also detected in one case (case #2). Regarding this point, few SMAD4 mutations have been reported, although mutations of other genes with a role in the transforming growth factor β (TGFβ) signaling pathway have been reported in NTRK fusion-positive CRCs4. No APC/CTNNB1 mutations were detected, which is in accordance with the finding that none of the cases showed nuclear staining of β-catenin, a hallmark of WNT signal activation. Moreover, none of our NTRK-fusion-positive CRCs contained other mutations typical of CRC, such as in KRAS and BRAF. This mutually exclusive pattern between NTRK fusion and KRAS/BRAF mutation has previously been proposed4,23. Based on these molecular characteristics of CRC with NTRK fusions, the tumorigenesis of CRCs harboring activating NTRK1 fusions could be considered different from those of conventional CRC. On the other hand, our results suggest that the TGFβ signaling pathway may be involved in a subset of these tumors. In addition, reports of MSI status in NTRK fusion-positive CRCs are limited23. A previous report on metastatic adenocarcinoma of colorectal origin demonstrated that 10 out of 13 cases with NTRK fusions were MSI-H23. A recent study demonstrated that the prevalence of NTRK gene fusion was around 7% in the MSI-H CRC cohort25. Another study also demonstrated that NTRK fusions were less frequent in MSS tumors than in MSI-H tumors (< 1% vs. 8%) in metastatic colon adenocarcinoma26. In our study, two out of three NTRK fusion-positive CRC patients were MSI-H, which is consistent with prior findings, although that these prior studies dealt only with metastatic CRC23, while our series contained consequent CRC from pStage I to pStage IV.
The prognosis of patients with CRC harboring NTRK fusion is not clear; however, NTRK3 fusion-positive tumors, such as the mammary analog secretory carcinoma of the salivary gland, seem not to be aggressive14. Of the three CRC cases we identified with NTRK1 fusion, one (Case #1, stage I) had been followed up for a short period, one (Case #3, stage IIA) had no evidence of recurrence or metastasis for 100 months, and the remaining patient (Case #2), with Stage IIIB CRC, died one year after surgery. This patient died of brain vascular disease, and it cannot be ruled out that he experienced malignancy-associated Trousseau syndrome. A recent study has shown that all CRC patients with ALK-, ROS1-, or NTRK fusions had shorter overall survival rates than fusion-negative patients23. Prognosis of CRC patients with NTRK fusion seemed to be worse or no different from that of patients with conventional CRC, although an increased sample size of CRC patients with NTRK fusion would be necessary to draw any significant conclusions. A recent study demonstrated that the frequency of NTRK fusion in CRC was 0.31%11. However, this number could be biased by sample selection, as patients with advanced CRC may have been selectively included in the study. Nonetheless, this frequency is similar to that of our study. As our study was a retrospective consecutive study, we could determine the frequency of CRC with NTRK fusion in addition to the natural history of CRC with NTRK fusion without specific target therapy, unlike with conventional CRC.
Finally, we found only three cases with pan-TRK staining from 971 CRC cases, including a weak staining pattern for which NTRK fusions were confirmed by the subsequent procedures, and the remaining cases did not show any staining. Solomon et al.11 showed a sensitivity of 87.5% (7/8) and a specificity of 100% (24/24) for colon cancer, and the cause of the lower sensitivity is the false negative for NTRK3 fusion tumor11. As a limitation of this study, our samples did not include NTRK fusion data; therefore, we could not evaluate the true sensitivity of this IHC-based NTRK fusion screening system. Although our NTRK fusion positive rate was similar to that of a previous study11, some NTRK3 fusions tumor may have been missed due to the lower sensitivity of this test. On the other hand, another study demonstrated a CRC with EML4-NTRK3 fusion showing weak but distinct cytoplasmic staining of pan-TRK IHC4. Molecular test-based screening is difficult in terms of labor and cost; thus, pan-TRK IHC could be a reliable and useful screening tool for the detection of CRC with NTRK fusion.
References
Sartore-Bianchi, A. et al. Sensitivity to entrectinib associated with a novel LMNA-NTRK1 gene fusion in metastatic colorectal cancer. J. Natl. Cancer Inst. https://doi.org/10.1093/jnci/djv306 (2016).
Drilon, A. et al. Safety and antitumor activity of the multitargeted Pan-TRK, ROS1, and ALK inhibitor entrectinib: Combined results from two phase I trials (ALKA-372-001 and STARTRK-1). Cancer Discov. 7, 400–409. https://doi.org/10.1158/2159-8290.CD-16-1237 (2017).
Hechtman, J. F. et al. Pan-Trk immunohistochemistry is an efficient and reliable screen for the detection of NTRK fusions. Am. J. Surg. Pathol. 41, 1547–1551. https://doi.org/10.1097/pas.0000000000000911 (2017).
Lasota, J. et al. Colonic adenocarcinomas harboring NTRK fusion genes: A clinicopathologic and molecular genetic study of 16 cases and review of the literature. Am. J. Surg. Pathol. 44, 162–173. https://doi.org/10.1097/PAS.0000000000001377 (2020).
Kummar, S. & Lassen, U. N. TRK inhibition: A new tumor-agnostic treatment strategy. Target Oncol. 13, 545–556. https://doi.org/10.1007/s11523-018-0590-1 (2018).
Milione, M. et al. Identification and characterization of a novel SCYL3-NTRK1 rearrangement in a colorectal cancer patient. Oncotarget 8, 55353–55360. https://doi.org/10.18632/oncotarget.19512 (2017).
Lee, S. J. et al. NTRK1 rearrangement in colorectal cancer patients: Evidence for actionable target using patient-derived tumor cell line. Oncotarget 6, 39028–39035. https://doi.org/10.18632/oncotarget.5494 (2015).
Arévalo, J. C. & Wu, S. H. Neurotrophin signaling: Many exciting surprises!. Cell Mol. Life Sci. 63, 1523–1537. https://doi.org/10.1007/s00018-006-6010-1 (2006).
Nakagawara, A. Trk receptor tyrosine kinases: A bridge between cancer and neural development. Cancer Lett. 169, 107–114. https://doi.org/10.1016/s0304-3835(01)00530-4 (2001).
Huang, E. J. & Reichardt, L. F. Trk receptors: Roles in neuronal signal transduction. Annu. Rev. Biochem. 72, 609–642. https://doi.org/10.1146/annurev.biochem.72.121801.161629 (2003).
Solomon, J. P. et al. NTRK fusion detection across multiple assays and 33,997 cases: Diagnostic implications and pitfalls. Mod. Pathol. 33, 38–46. https://doi.org/10.1038/s41379-019-0324-7 (2020).
Knezevich, S. R., McFadden, D. E., Tao, W., Lim, J. F. & Sorensen, P. H. A novel ETV6-NTRK3 gene fusion in congenital fibrosarcoma. Nat. Genet. 18, 184–187. https://doi.org/10.1038/ng0298-184 (1998).
Bourgeois, J. M., Knezevich, S. R., Mathers, J. A. & Sorensen, P. H. Molecular detection of the ETV6-NTRK3 gene fusion differentiates congenital fibrosarcoma from other childhood spindle cell tumors. Am. J. Surg. Pathol. 24, 937–946. https://doi.org/10.1097/00000478-200007000-00005 (2000).
Boon, E. et al. Clinicopathological characteristics and outcome of 31 patients with ETV6-NTRK3 fusion gene confirmed (mammary analogue) secretory carcinoma of salivary glands. Oral. Oncol. 82, 29–33. https://doi.org/10.1016/j.oraloncology.2018.04.022 (2018).
Köbel, M. et al. Interpretation of P53 immunohistochemistry in endometrial carcinomas: Toward increased reproducibility. Int. J. Gynecol. Pathol. 38(Suppl 1), S123-s131. https://doi.org/10.1097/pgp.0000000000000488 (2019).
Prall, F. & Hühns, M. Quantitative evaluation of TP53 immunohistochemistry to predict gene mutations: Lessons learnt from a series of colorectal carcinomas. Hum. Pathol. 84, 246–253. https://doi.org/10.1016/j.humpath.2018.10.012 (2019).
Suehara, Y. et al. Identification of KIF5B-RET and GOPC-ROS1 fusions in lung adenocarcinomas through a comprehensive mRNA-based screen for tyrosine kinase fusions. Clin. Cancer Res. 18, 6599–6608. https://doi.org/10.1158/1078-0432.Ccr-12-0838 (2012).
Kurihara, T. et al. Nanostring-based screening for tyrosine kinase fusions in inflammatory myofibroblastic tumors. Sci. Rep. 10, 18724. https://doi.org/10.1038/s41598-020-75596-3 (2020).
Akaike, K. et al. Distinct clinicopathological features of NAB2-STAT6 fusion gene variants in solitary fibrous tumor with emphasis on the acquisition of highly malignant potential. Hum. Pathol. 46, 347–356. https://doi.org/10.1016/j.humpath.2014.11.018 (2015).
Akazawa, Y. et al. Next-generation sequencing analysis for gastric adenocarcinoma with enteroblastic differentiation: Emphasis on the relationship with hepatoid adenocarcinoma. Hum. Pathol. 78, 79–88. https://doi.org/10.1016/j.humpath.2018.04.022 (2018).
Yamashiro, Y. et al. Molecular and clinicopathological features of colorectal adenocarcinoma with enteroblastic differentiation. Histopathology https://doi.org/10.1111/his.14158 (2020).
Kohsaka, S. et al. Pediatric soft tissue tumor of the upper arm with LMNA-NTRK1 fusion. Hum. Pathol. 72, 167–173. https://doi.org/10.1016/j.humpath.2017.08.017 (2018).
Pietrantonio, F. et al. ALK, ROS1, and NTRK rearrangements in metastatic colorectal cancer. J. Natl. Cancer Inst. https://doi.org/10.1093/jnci/djx089 (2017).
Gatalica, Z., Xiu, J., Swensen, J. & Vranic, S. Molecular characterization of cancers with NTRK gene fusions. Mod. Pathol. 32, 147–153. https://doi.org/10.1038/s41379-018-0118-3 (2019).
Guo, Y. et al. Genomic alterations of NTRK, POLE, ERBB2, and microsatellite instability status in Chinese patients with colorectal cancer. Oncologist https://doi.org/10.1634/theoncologist.2020-0356 (2020).
Yaeger, R. et al. Clinical sequencing defines the genomic landscape of metastatic colorectal cancer. Cancer Cell 33, 125-136.e123. https://doi.org/10.1016/j.ccell.2017.12.004 (2018).
Funding
This study was supported in part by a Grant-in-Aid for General Scientific Research from the Ministry of Education, Science, Sports, and Culture (#17K08730 and #20K07415 to TS), Tokyo, Japan. This study was funded by Bayer HealthCare Pharmaceuticals, Inc.
Author information
Authors and Affiliations
Contributions
T.S., T.H., Y.S., and S.K. designed the experiments. T.S., T.H., and T.Y. diagnosed all clinical samples. Y.Y. made TMA blocks and performed the immunohistochemistry and NGS assays. Y.Y. and T.S. evaluated IHC sections. T.K. performed the gene expression analysis. Y.Y. and T.S. wrote the main part of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yamashiro, Y., Kurihara, T., Hayashi, T. et al. NTRK fusion in Japanese colorectal adenocarcinomas. Sci Rep 11, 5635 (2021). https://doi.org/10.1038/s41598-021-85075-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-85075-y
This article is cited by
-
Possible correlation of apical localization of MUC1 glycoprotein with luminal A-like status of breast cancer
Scientific Reports (2023)
-
Incidence and clinical significance of 491 known fusion genes in a large cohort of Japanese patients with colorectal cancer
International Journal of Clinical Oncology (2023)
-
Detection of NTRK fusions by RNA-based nCounter is a feasible diagnostic methodology in a real-world scenario for non-small cell lung cancer assessment
Scientific Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.