Abstract
The aim of this study was to establish a method for geographical origins identification of Panax notoginseng (P. notoginseng) based on abundant chromatographic spectral information. Characteristic fingerprints of P. notoginseng extracts samples were generated by Multi-wavelength Fusion Profiling (MWFP) method based on the HPLC fingerprints established at three wavelengths of 203 nm, 270 nm and 325 nm. The samples grouping results calculated with the averagely linear quantified fingerprint method (ALQFM) and the unsupervised statistical methods based on fusion fingerprints matches with the geographical origins. The Multi-wavelength Fusion Profiling (MWFP) method has been successfully applied to identification of geographical origins of P. notoginseng and shows the advantages compared with single—channel fingerprints. In addition, eight physiologically active components, including four saponins, two flavones and two amino acids, were identified from the most relevant ingredients of P. notoginseng geographical origins by fusion fingerprint-efficacy relationship analysis. Besides the recognized active saponins, other categories of active ingredients such as flavonoids and amino acids should be paid attention to in the producing areas identification or the quality judgment of P. notoginseng.
Similar content being viewed by others
Introduction
Panax notoginseng named “San-qi” and “Tian-qi” is the dried roots and rhizomes of Panax notogitiseng (Burk.) F. H. Chen listed in the Chinese pharmacopoeia. It is a virtual Chinese medicine (TCM) with an annual output value exceeded 70 billion RMB yuan and more than 3600 related drug approval numbers in China1. Its main efficacy is promoting blood circulation, removing blood stasis, detumescence and analgesia2,3,4.
The genuine producing area of P. notoginseng was in Yunnan province, China5. Due to the great demand cannot be met, the production of P. notoginseng expanded to the adjacent region. Suining City, Sichuan Province was the first to introduce successfully, and the only production area outside Yunnan with stable output for three consecutive years up to 2019. The difference of producing area involves the distinction in complicate natural and artificial factors, which will lead to the diversity of the integral composition and eventually the efficacy of Traditional Chinese Medicine6,7. In terms of geographical conditions, the P. notoginseng origin in Yunnan is significantly different from that in Sichuan. The former is plateau mountain area and the latter is hills in the hinterland of the basin. The natural environment and the growing techniques are both various in the two provinces producing area. Therefore, it is necessary to distinguish P. notoginseng produced in the two provinces by origin discriminant technique. Basing on the current legal quality indicators of P. notoginseng regulated in Pharmacopoeia of China, the individual indicators of Ginsenoside Rb1, Ginsenoside Rg1 and Notoginsenoside R1 content, the identification of geographical origins is difficult to achieve. There were some studies of the identification of geographical origins of TCM8,9 including P. notoginseng10 based on stable isotope ratios. Except the strategy related to the inorganic stable isotope the origins identified strategy based on complex organic composition information have been worked out. P. notoginseng geographical discrimination models have been established based on spectral data and various data mining algorithms. In Wang’s study11, the model is established by partial least squares discriminant analysis (PLS-DA) of the optimized UV spectra data of P. notoginseng samples. The Fourier transform-infrared spectrum12 or near-infrared spectrum data of P. notoginseng are used to distinguish the producing area13. In order to exploit the synergetic and complementary information, researchers established models of origin discrimination based on high-level fusion of Fourier transform mid-infrared spectroscopy and near infrared spectroscopy combined with the algorithm of random forest14,15. Different from the methods based on spectral data matrix mentioned above, the discrimination model based on near-infrared spectra image and deep learning strategy is also applied to P. notoginseng16. These discrimination methods based on spectral analysis have an advantage in applying the spectral information in a certain wavelength range of samples comprehensively. However, the information of spectral without separation is complex and could only be used for discriminant analysis after calculation and transformation. At the same time, it is difficult to directly associate the information of components with the spectral and established discriminant model.
At present, researchers discussed Spectral and Chromatographic Overall Analysis technique (SCOA), aiming to extract, integrate and transform the information from complex multi-channel into analyzable forms17. Multi-wavelength Fusion Profiling (MWFP) is a representative of the SCOA. It is obtained by the projection along the wavelength axis of the HPLC chromatograms collected from the same sample analyzed with multiple wavelengths (or DAD). It is chromatogram maximizes the available Multi-wavelength chromatographic information. MWFP could provide more information for origin identification than single wavelength HPLC fingerprint, and the more accurate conclusion may be obtained with MWFP analysis. Compared to the spectral without separation, chromatographic separation reduces the complexity of the spectrum, and the relationship between chromatographic peaks and chemical compositions is more direct. In recent studies, MWFP method was applied for the quality assessment of the single herbs18 and proprietary Chinese medicine with more complex components consisting of a variety of medicines19. These studies based on the MWFP method provide novel and comprehensive strategy for TCM quality assessment.
In this present study, a Multi-wavelength Fusion Profiling (MWFP) method is developed to distinguish the P. notoginseng samples from two producing area with various geographical conditions.
Materials and methods
Chemical and materials
Acetonitrile and methanol (HPLC-grade) were obtained from Sigma-Aldrich (Steinheim, Germen). Formic acid (HPLC-grade) was supplied from the local dealer. Ultra-pure water was made by the pure water machine (Millipore, France, 18.2 MΩcm@25 °C). Reference standards of Ginsenoside Rb1 (G-Rb1), Ginsenoside Rg1 (G-Rg1), Ginsenoside Rd (G-Rd), Notoginsenoside R1 (NG-R1), Notoginsenoside R2 (NG-R2) were acquired from Chengdu Glip Biotechnology Co., Ltd., Chengdu, Sichuan. The structures of the five saponins were presented in Fig. 1.
Sample sources
The P. notoginseng samples S1–S15 were collected in Suining City, Sichuan Province; S16–S19 and S23–S26 were collected in Wenshan Zhuang and Miao Autonomous Prefecture, Yunnan Province, S20–S22 were collected in Zhaotong, Yunnan Province, and S27–S32 were collected in Hani-Yi Autonomous Prefecture of Honghe, Yunnan Province, which are the main producing areas of P. notoginseng.
Sample preparation and HPLC analysis
The P. notoginseng samples were smashed by the pulverizer. About 0.500 g smashed sample was accurately weighed into a 50 mL centrifugal tube. 20 mL methanol/water (60:40, v/v) was added in and the centrifugal tube was leave in an ultrasonic water bath to extraction for 30 min. Afterward the tube was put into a 60 °C water bath for 2 h and centrifuged at 3000 rpm for 5 min. The supernatant was filtered through a 0.22 μm filter membrane and collected as the sample solution. The standards solution was prepared by accurately weighed amount of 1.0 mg and dissolved with methanol in the 10 mL volumetric flask. All solutions were stored at 4 °C until analysis.
Chromatographic separation was operate with Shimadzu LC-20AD equipped with a Poroshell 120 EC-C18 column (100 × 4.6 mm, 2.7 μm). The mobile phase consists of acetonitrile (A) and water (B). Separation was achieved using the following linear gradient program: 0–10 min, 95%B–80%B; 10–15 min, 80%B–60%B; 15–25 min, 60%B; 25–50 min, 60%B–0%B; 50–60 min, 0%B; 60–60.1 min, 0%–95%B, 70 min, stop. The column temperature was maintained at 40 °C. A 10 μL aliquot of each sample was injected into the HPLC–UV system. The flow rate was set at 0.3 mL/min. The detection wavelengths were set at 203 nm, 270 nm and 325 nm. The five saponins concentration of the samples calculated with the chromatography of 203 nm.
Similarity analysis with ALQFM
The fusion fingerprint of each P. notoginseng sample and their similarity were calculated with the software “Digitized Evaluation System for super-information Characteristics of TCM Chromatographic Fingerprints 4.0” (developed by Guoxiang Sun et al., Software certificated NO.0407573, China). The Average Linear Quantitative Fingerprint method (ALQFM) was adopted with the software to analyze the similarity20,21. There were three parameters involved. Average linear qualitative similarity (Sm), average linear quantitative similarity (Pm) and the fingerprint variation coefficient (α). The simplified parameter “Grade” was combined with Sm, Pm and α to evaluate the similarity of samples (Table 1). Generally, samples with the grade value ≤ 5 were recommended as the similar ones to the chosen reference sample.
Statistical analysis
Hierarchical clustering analysis (HCA) was performed according to the contents of the five saponins concentration of the 32 batch P. notoginseng samples to show the relatedness of the samples based on the composition indexes content. Heat map clustering and the principal component analysis (PCA) were performed according to the peak areas of 33 co-possessing peaks of the fusion fingerprints with the samples to show the relatedness of samples based on the fusion fingerprints. The co-possessing peaks’ areas of the fusion fingerprints with the P. notoginseng samples grouped according to their origin (Yunnan province or Sichuan province) were evaluated by Orthogonal Projection to Latent Structure Discriminant Analysis (OPLS-DA) to verify grouping and to find the peaks that can represent the difference between the two origins.
HCA was performed with the SPSS statistical system (version 13.0) using the between group linkage method with squared Euclidean distances. Heat map clustering were performed in R Programming Language and the similarity was measured with the between-groups linkage method and squared Euclidean. PCA and OPLS-DA were performed using SIMCA 14.1 software.
Results and discussion
Methodology evaluation of HPLC analysis
The calibration curves of five saponins standard solutions, as well as the linear ranges and the limit of detection (LOD) were showed in Supporting Information Table S1. All the standard products showed excellent linearity (R2 ≥ 0.999) over the tested concentration ranges. The quantitative accuracy was assessed with the standard addition method. The average recoveries for the five investigated compounds ranged from 91.84 to 105.38%. The fingerprint stability was evaluated with RSD values of each co-possessing fingerprint peak area were, respectively, less than 3.22%, 3.71% and 2.16% for the stability, precision and repeatability tests. Considering these results, the method was accurate and valid enough.
Composition indexes content analysis
According to previous studies, saponins are the important functional components of P. notoginseng, and are often used as its indicator components. The sum of G-Rb1, G-Rg1 and NG-R1 content has been used as a marker for the quality control of P. notoginseng in Chinese Pharmacopoeia. In this study five saponins aboundant in P. notoginseng reported in our previous study22 are chosen as composition indexes. The content of five saponins contents in 32 P. notoginseng samples were showed in Table 2.
The HCA plot based on the five saponins contents is shown in Fig. S1. The failure of origin discrimination begins with the minimum between-groups distance. The result verified that it is impossible to distinguish the origin of P. notoginseng in Yunnan Province (the genuine producing area in plateau mountain area) and Sichuan Province (the new introduced producing area in hills in the hinterland of the basin) by the indicators content.
HPLC fingerprint similarity analysis with ALQFM
The UV spectra of the five saponins were investigated. The UV spectra of sample solution is different with that of the five saponins in 203–350 nm (shown in Fig. 2). In order to fully reflect the characteristics of the P. notoginseng, three absorption bands (203 nm, 270 nm and 325 nm) were chosen to establish the fusion fingerprint, which capable of synthesizing enhancing signal response and rich fingerprint information.
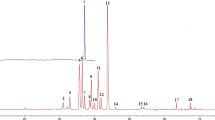
The fingerprints of 32 sample solution with the three absorption bands (203 nm, 270 nm and 325 nm) were accomplished, respectively at first (shown in Supporting Information Fig. S2). Then the fusion fingerprint of each sample (shown in Fig. 3) and the similarity evaluation results (presented in Table 3) were calculated.
In ALQFM, qualitative analysis is performed first, then quantitative analysis is used. As Table 3 showed that all the samples had α < 0.30 and Sm > 0.90, indicating that the samples were similar in the distribution and number of chemical compositions. Based on Table 1, all the samples (α < 0.30, Sm > 0.90) should be in grade 1–5. Because of the difference content in the samples, some of the samples Pm < 70% or Pm > 130% which made their grades between 6 and 8.
If consider only qualitative similarity Sm, origin distinguish is not showed in any wavelength fingerprints nor in the fusion fingerprints. As further elaborated in follows, the addition of Pm makes the “Grade” have the function of origin discrimination, and the fusion fingerprints reflects an advantage in this respect.
Correlation between Grade and origins
“Grade” is a simplified parameter for evaluating qualitative and quantitative similarity of samples of ALQFM method. The grades distribution of samples from different producing area were reflected with the scatter plots (Fig. S3). As shown in Fig. S3A–C the samples of the two producing areas cannot distinguish with grades distribution at any single wavelength, while as shown in Fig. S3D most of the samples (12 of 15) from Sichuan Province had a grade between 1 and 4, and most of the samples (15 of 17) from Yunnan Province had a grade between 5 and 8 calculated with fusion fingerprints. It can be seen that the fusion fingerprint is better than the single channel fingerprint in the respect of the producing areas distinguish capacity of Sichuan and Yunnan with Grade. This can be interpreted as the fusion fingerprints can reflect the sample characteristics more completely than the single-channel fingerprints because of the comprehensive utilization of the chromatographic information of the three bands of ultraviolet spectrum. The error frequency of using Grade to distinguish the origins is about 5 of 32.
Relationship between fusion fingerprints and origins
The HCA heat-map (Fig. 4) shows the relatedness of 32 P. notoginseng samples based on the fusion fingerprints. Except S8, the samples are clustered into two groups, one group is S1–S15 and another group is S16–S32. The result is consistent with the origin distinguish (S1–S15 were collected from Sichuan province and S16–S32 were from Yunnan province). The PCA score plots (Fig. 5A) showed the classify result consistent with that of HCA heat-map.
That is to say, while the conventional unsupervised similarity evaluation analysis of the fusion fingerprints used to distinguish the origins between the two provinces the error frequency rate would reduce to 1 of 32.
To analyze the characteristic compounds most reflected to the difference of the P. notoginseng samples from the two provinces, Orthogonal Projection to Latent Structure Discriminant Analysis (OPLS-DA) were used to analyze the peaks which contributed to classification of the fusion fingerprints (Fig. 5B). Variable influence on projection statistics (VIP) values were calculated to reflect the contribution of each variable to the established model. The 15 peaks with VIP value > 1 in Fig. 5C were considered to be relevant for the classification significantly.
Qualitative analysis of peaks relevant for the origin classification
High performance liquid chromatography combined with high resolution mass spectrometry (Orbitrap LC–MS, Thermo Scientific) was involved to analysis the sample. Data were processed with the software Exactive 2.8 with database searching (MZ Vault, Mass List, MZ Cloud). The results were shown in Table 4.
In Table 4 ginsenoside Rg1, ginsenoside Rb1, ginsenoside Rd are the selected compounds to investigate the concentration in this study, and the significant difference of their content in the P. notoginseng samples from two origins (P < 0.05) had been verified. The flavonoid and the amino acids identified as the characteristics compounds relevant with origin classification reflected the importance of non-saponin compounds in the quality control of the P. notoginseng.
The results in Table 4 are consistent with those in Table 2. In Table 2 there were significant differences in the G-Rb1, G-Rg1, G-Rd and NG-R2 content of the P. notoginseng samples from two producing areas and three of the four compounds with higher content were in Table 4. This result could be a verification of this geographical origin identification method.
Conclusion
Compared with the single wavelength fingerprint, the fusion fingerprint has a significant advantage in geographical origin identification. This method comprehensive utilized of the chromatographic information of the three bands of ultraviolet spectrum, and has an advantage in the discrimination of geographical origin than single-channel fingerprints method. In this study, five saponins, two flavonoids and two amino acids showed significant correlation with the geographical origins. This suggested that in addition to the recognized active saponins, categories of active ingredients such as flavonoids and amino acids should also be paid attention to in the work of producing areas identification or the quality judgment of P. notoginseng.
References
Pang, H. H. et al. Effect of compatible herbs on the pharmacokinetics of effective components of Panax notoginseng in Fufang Xueshuantong Capsule. J. Zhejiang Univ. Sci. B 18, 343–352 (2017).
Zhang, Z. et al. Steamed Panax notoginseng attenuates anemia in mice with blood deficiency syndrome via regulating hematopoietic factors and JAK-STAT pathway. Front. Pharmacol. 10, 1578 (2019).
Xu, C. et al. Analytical methods and biological activities of Panax notoginseng saponins: Recent trends. J. Ethnopharmacol. 236, 443–465 (2019).
Yin, S. J. et al. Antithrombotic effect and action mechanism of Salvia miltiorrhiza and Panax notoginseng herbal pair on the zebrafish. Chin. Med. 15, 35 (2020).
Cui, X. M. et al. Analysis on the gelogic background and physicochemical properties of soil for the cultivation of Panax notoginseng in Yunnan province, China. J. Chin. Materia Med. 30, 332–335 (2005).
Bai, H. et al. Metabolomics study of different parts of licorice from different geographical origins and their anti-inflammatory activities. J. Sep. Sci. 43, 1593–1602 (2020).
Miao, L. et al. Discrimination of the geographical origin of the lateral roots of Aconitum carmichaelii using the fingerprint, multicomponent quantification, and chemometric methods. Molecules, 24, 4124 (2019).
Li, G. et al. Identification of geographical origins of Schisandra fruits in China based on stable carbon isotope ratio analysis. Eur. Food Res. Technol. 232, 797–802 (2011).
Tian, Z. et al. Identification of geographical origins of raw American ginseng and tablets based on stable isotope ratios. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 1009, 73–79 (2016).
Liang, S. W., et al. Origin traceability of main root of spring Panax notoginseng based on stable isotope fingerprint. China J. Chin. Materia Medica 46, 560–566 (2020).
Wang, Y. Z. et al. Study on the identification of Panax notoginseng from different areas by ultraviolet fingerprint and stoichiometry. Spectrosc. Spectral Anal. 36, 1789–1793 (2016).
Fei, L. et al. Study on the genuineness and producing area of Panax Notoginseng based on infrared spectroscopy combined with discriminant analysis. Spectrosc. Spectral Anal. 35, 108–112 (2015).
Chen, H., Lin, Z. & Tan, C. Fast discrimination of the geographical origins of notoginseng by near-infrared spectroscopy and chemometrics. J. Pharm. Biomed. Anal. 161, 239–245 (2018).
Li, Y. et al. FT-MIR and NIR spectral data fusion: A synergetic strategy for the geographical traceability of Panax notoginseng. Anal. Bioanal. Chem. 410, 91–103 (2018).
Zhou, Y. et al. Origin identification of Panax notoginseng by multi-sensor information fusion strategy of infrared spectra combined with random forest. Spectrochimica Acta Part A Mol. Biomol. Spectrosc. 226, 117619 (2020).
Dong, J. et al. Deep learning for geographical discrimination of Panax notoginseng with directly near-infrared spectra image. Chemometr. Intell. Lab. Syst. 197, 103913 (2020).
Zhang, J., Chen, S. & Sun, G. Spectral and chromatographic overall analysis: An insight into chemical equivalence assessment of traditional Chinese medicine. J. Chromatogr. A 1610, 460–556 (2020).
Yang, F. et al. Quality assessment of licorice (Glycyrrhiza glabra L.) from different sources by multiple fingerprint profiles combined with quantitative analysis, antioxidant activity and chemometric methods. Food Chem. 324, 126–154 (2020).
Hanxiao Chi, G. S. Integrated evaluation of HPLC fingerprints for the quality control of Ginseng Yang Rong Wan by multi-wavelength fusion fingerprint method. Asian J. Tradit. Med. 14, 171–182 (2019).
Zhang, Y. et al. A strategy for qualitative and quantitative profiling of glycyrrhiza extract and discovery of potential markers by fingerprint-activity relationship modeling. Sci. Rep. 9, 1309 (2019).
Zhang, Y. et al. Evaluation of the quality consistency of powdered poppy capsule extractive by an averagely linear-quantified fingerprint method in combination with antioxidant activities and two compounds analyses. J. Sep. Sci. 40, 4511–4520 (2017).
Yue, P. et al. Multi-dimensional evaluation of the quality of Panax Notoginseng in Suining, Sichuan. J. Chin. Med. Mater. 43, 2729–2733 (2020).
Acknowledgements
This study was supported by the Young Science and Technology Innovation Team of Sichuan Province (2019CXTD0055), the Natural Sciences Foundation of China Science (81872959, 81373920, 30801522) and the China scholarship foundation (201708510027). We sincerely thank Professor Sun Guoxiang's research group of Shenyang Pharmaceutical University for their technical assistance in this study.
Author information
Authors and Affiliations
Contributions
J.B. designed the study. J.B., P.Y. and Q.D. performed the experiments and analyzed the raw data. F.W. and C.H. supervised the study. Y.L. and J.G. provided the technical support and adviced for the study. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bai, J., Yue, P., Dong, Q. et al. Identification of geographical origins of Panax notoginseng based on HPLC multi-wavelength fusion profiling combined with average linear quantitative fingerprint method. Sci Rep 11, 5126 (2021). https://doi.org/10.1038/s41598-021-84589-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-84589-9
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.