Abstract
Precocious puberty (PP) is an important endocrine disorder affecting children globally. Several genes, SNPs and comorbidities are reported to be associated with PP; however, this data is scattered across scientific literature and has not been systematically collated and analysed. In this study, we present PrecocityDB as the first manually curated online database on genes and their ontology terms, SNPs, and pathways associated with PP. A tool for visualizing SNP coordinates and allelic variation on each chromosome, for genes associated with PP is also incorporated in PrecocityDB. Pathway enrichment analysis of PP-associated genes revealed that endocrine and cancer-related pathways are highly enriched. Disease enrichment analysis indicated that individuals with PP seem to be highly likely to suffer from reproductive and metabolic disorders such as PCOS, hypogonadism, and insulin resistance. PrecocityDB is a useful resource for identification of comorbid conditions and disease risks due to shared genes in PP. PrecocityDB is freely accessible at http://www.precocity.bicnirrh.res.in. The database source code and content can be downloaded through GitHub (https://github.com/bic-nirrh/precocity).
Similar content being viewed by others
Introduction
Disease-centric databases significantly contribute to advancement of health research as they offer curated and well-annotated data amenable for further analysis. While many of the diseases such as cancers, autism, Alzheimers, PCOS and endometriosis have dedicated databases, it would be worthwhile to include more diseases in this list. In the present study, we have reviewed scientific literature on precocious puberty (PP) and developed a manually curated database with information on genes associated with PP, their ontology terms, SNPs and pathways.
Puberty is characterized by growth and maturation of primary sexual organs leading to development of secondary sexual characteristics. Puberty attained before age of 8 years in girls and 9 years in boys is considered as precocious1. PP is statistically defined as early pubertal development at an age that is 2.5 standard deviations lower than the mean age of pubertal onset2.
The onset of puberty is initiated by the maturation of hypothalamic–pituitary–gonadal (HPG) axis that ultimately leads to sexual dimorphism and fertility3,4. Activation of the HPG axis, nearing the pubertal age, stimulates the anterior pituitary to secrete gonadotropins such as FSH and LH, in response to which gonads develop. This process is termed as gonadarche. Gonadarche is accompanied by increased secretion of gonadal sex steroid hormones such as estrogen and testosterone. Prior to and independent of gonadarche, adrenarche occurs when there is a surge in the secretion of adrenal androgens such as DHEA and DHEA-S5 leading to growth and appearance of sexual hair6,7,8.
PP can be sub-classified as (i) gonadotropin dependent or central precocious puberty (CPP); (ii) gonadotropin independent or peripheral precocious puberty (PPP); and (iii) incomplete precocious puberty (IPP). CPP is caused due to premature activation of HPG axis whereas PPP is associated with increased gonadal secretion of steroidal sex hormones9. The overall prevalence of PP is in the range of 1: 5000 to 1: 10,000 children7,10. CPP is more common in girls with a female to male ratio that ranges from 3:1 to 23:1. Some rare cases of CPP triggered due to organic causes are more common among boys11,12. IPP comprises of variants of pubertal development like premature adrenarche, premature thelarche, and premature menarche that may lead to PP7.
PP is diagnosed based on physical and biochemical changes that are associated with puberty. Assessment is done using detailed family history, bone X-ray, brain MRI, and hormonal profiling7.
The incidence of PP is higher in individuals with central nervous system (CNS) disorders or CNS lesions10. Early onset of puberty profoundly impacts the psychosocial well-being of individuals13,14. Children with early puberty experience higher levels of behavioural and psychological disturbances as compared to children with normal puberty. Individuals with PP suffer from short stature due to premature fusion of the growth plates15. PP is often present with other morbidities such as McCune-Albright syndrome, congenital adrenal hyperplasia, neurofibromatosis type 1, Sturge–Weber syndrome, adrenal and ovarian tumors12. Premature puberty has been found to be associated with cervical, ovarian and thyroid cancers16,17,18. Few studies have reported that early menarche increases the risk of breast cancer in girls19,20.
The cause of PP can be ascribed to genetic, metabolic, or environmental factors. 70–80% of the variance in pubertal timing can be attributed to genetic factors21. Genetic studies suggest that PP has an autosomal dominant mode of inheritance22. Mutations in genes that are involved in sexual development such as MKRN3, KISS1, GPR54, CYP19A1, and LHCGR contribute to PP. Apart from genetic factors, intrauterine growth retardation and low birth weight are linked with early menarche23. It is unclear if childhood obesity is cause or effect of PP23. Exposure to endocrine-disrupting chemicals such as 1,1,1-trichloro-2,2-bis(4-chlorophenyl)ethane (DDT), monobutyl phthalate (MBP), n-nonyl phenol (n-NP), t-octylphenol (t-OP), and isoflavones like equol, genistein, and daidzein can influence hormonal dysregulation leading to PP23,24.
Several research groups have published valuable information on the causal factors and comorbid conditions associated with PP; this data is worthwhile to collate and analyse systematically to gain further insights on PP.
Results
Database content
PrecocityDB has an interactive, user-friendly web interface with options to search, browse, and visualize data. The database has curated information of 44 genes and associated 26,874 ontology terms, 235 pathways, and 199 SNPs associated with PP.
This database can be easily explored using navigation tabs on the top panel. A brief description of these tabs is given below.
-
1.
Home: Short description of PP and information available in PrecocityDB is provided.
-
2.
Search:
-
a.
Quick search: Data available in PrecocityDB for a keyword can be retrieved.
-
b.
Advanced search: Specific queries, through keywords, can be built to retrieve precise information.
-
a.
-
3.
Browse: Datasets on genes, SNPs, pathways and ontology terms can be screened.
-
4.
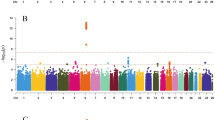
SNP visualizer: Allelic variants and SNP coordinates on each chromosome can be visualized. Chromosomes 1 and 6 are associated with the maximum number of SNPs that are associated with PP (Fig. 1).
-
5.
Enrichment analysis: Graphical representations of outcomes of pathway and disease enrichment analyses are provided.
-
6.
Help: Detailed explanation of all the features in PrecocityDB and its usage, with examples, is provided.
Data analysis
Pathway enrichment analysis of genes in PrecocityDB identified 32 statistically significant pathways (Fig. 2A1, Supplementary File S1, Supplementary Fig. S1A). Pathways related to endocrine system, signal transduction, and cancers were amongst the highly enriched (Fig. 2A1). Clustering analysis of these enriched pathways resulted in nine pathway clusters, of which three were interconnected and six were independent pathway clusters (Fig. 2A2). The most significant pathway clusters comprised of ovarian steroidogenesis and FoxO signalling pathway. The most significant independent pathway clusters were neuroactive ligand-receptor interactions, regulation of lipolysis in adipocytes and long-term depression. Pathway enrichment analysis of transcription factors of these genes led to identification of 45 statistically enriched pathways (Supplementary File S1, Supplementary Fig. S1B). Pathways related to viral infectious diseases, cancers, and endocrine systems were highly enriched (Fig. 2B1). Clustering analysis of the enriched pathways captured nine pathway clusters. Pathways related to cancers formed the largest clusters followed by parathyroid synthesis and thyroid hormone signalling pathways (Fig. 2B2). Disease enrichment analysis revealed that reproductive, endocrine, metabolic and cancer-related disorders were enriched in PP (Fig. 3, Supplementary File S1, Supplementary Fig. S2).
Pathway enrichment analysis using PrecocityDB gene-set (A1, A2) and transcription factors of PrecocityDB gene-set (B1, B2). In the bubble plot (generated using ggplot2 R package25), the x-axis represents gene overlap score (provided by Enrichr) and y-axis indicates enriched KEGG pathway terms. Bubble size is proportional to adjusted P value of enriched pathways. Bubble color represents parent pathway term. In the pathway networks (generated using ClueGO v2.5.7), the node size is proportional to adjusted P value. Edge represents at least one gene shared between two connecting nodes. Nodes within a cluster are colored based on the most significant pathway term (labelled in same color).
Disease enrichment analysis using enriched diseases obtained using PrecocityDB gene-set and GS2D. In the bubble plot (generated using ggplot2 R package), the x-axis represents gene overlap score (as given by GS2D) and y-axis indicates enriched disease terms. Bubble size is proportional to P value of enriched diseases. Bubble color represents parent disease term. In the disease-disease association network (generated using D3.js javascript library26), edges represent at least one gene shared between two connecting nodes. Node color represents parent disease term and node size is proportional to P value of disease enrichment analysis.
Discussion
PP is a manifestation of hormonal dysregulation; specifically, growth hormone, gonadotropins and steroidal sex hormones27. These hormones are majorly responsible for endocrine and reproductive disorders such as PCOS, endometriosis, hypogonadism and infertility. These hormones are also known to influence cancer progression28,29. Hence, it is highly likely that individuals with PP have higher risk of developing these disorders in their life span.
In order to systematically assess the risk of co-occurrence of diseases, it is important to have a database of manually curated genes associated with PP. Since databases on PP were not available in the public domain, we developed PrecocityDB for providing researchers a high quality database on information related to PP that is curated from scientific publications.
Through the curation process, we identified 44 genes and 199 SNPs associated with PP. The highest number of SNPs associated with PP was reported for MKRN3 (Makorin ring finger protein 3) followed by LHCGR (luteinizing hormone (LH)/choriogonadotropin receptor gene), KISS1 (Kisspeptin), KISS1R (Kisspeptin receptor), and LIN28B (Lin-28 Homolog B). MKRN3 gene is the most widely studied gene for its association with precocious puberty30. It has ubiqutin protein ligase activity. Mutation in MKRN3 inhibits proteolysis of proteins involved in activation of GnRH neurons31. Mutations in MKRN3, KISS1 and KISS1R contribute to precocious puberty by activating gonadotropic axis32. Inactivating LHCGR mutations in females lead to amenorrhea and infertility, whereas activating LHCGR mutations in males have been associated with familial male-limited precocious puberty (FMPP)33. LIN28B expression influences the timing of major developmental events and is thus associated with pubertal onset. Loss of function mutations in LIN28B gene are known to contribute to precocious development34,35.
To evaluate the comorbidity risk, disease and pathway enrichment analyses of gene-set in PP were performed. Some of the top pathways identified were ovarian steroidogenesis, GnRH signalling, estrogen signalling, and thyroid hormone signalling pathways (Fig. 2); these pathways are known to be critical for PP, PCOS and cancers36,37,38,39,40. In order to further validate these observations, disease enrichment analysis was performed based on significant co-occurrences of PP gene-set and diseases using GS2D tool57. It is noteworthy that reproductive, endocrine, metabolic and cancer-related disorders were enriched (Fig. 3).
The above findings are in good agreement with clinical observations of patients with PP. Franceschi et al. investigated the prevalence of PCOS in a cohort of young women with previous idiopathic central precocious puberty (ICPP) and observed that patients with ICCP were prone to develop PCOS in their adulthood40. Bodicoat et al., through their large cohort study, had investigated the association of pubertal age and risk of developing breast cancer. An increased risk of breast cancer was associated with incidences of earlier thelarche, menarche, early regular periods, a longer time between thelarche and menarche, or a shorter time between menarche and the onset of regular periods41. Bonilla et al., using a Mendelian randomization approach, observed that attainment of sexual maturity at a younger age than normal poses a significant threat of developing prostate cancer in adolescent boys42.
Enrichment analysis performed using the genes present in PrecocityDB helped to identify the well-studied pathways of PP (Fig. 2). Additionally, enrichment analysis could also identify genes and pathways that are not yet reported to be associated with PP based on human studies. For example, although there is evidence suggesting the role of FOXO3 in early pubertal development in rodents43,44,45, FOXO gene family is not included in PrecocityDB as its association with PP based on human studies is not yet reported. Interestingly, FoxO signalling pathway was found to be one of the enriched pathways based on enrichment analysis of PrecocityDB genes because of the overlapping genes from related pathways such as insulin signalling pathway, MAPK signalling and PI3K-Akt signalling pathway.
PrecocityDB will be updated with time and as more genes get validated for its association with PP; this will lead to more accurate results for enrichment analysis. However, even with the present data the enrichment analysis does give out interesting insights and testable hypothesis for the scientific community.
Conclusion
PP adversely affects the quality of life of adolescents and could have long-term health consequences. In order to comprehend the comorbidity risk in PP due to shared genes and biochemical pathways, it is essential to have a high quality database of genes and SNPs that are associated with PP. PrecocityDB is the first online database that catalogues genetic and polymorphism data associated with PP along with relevant reference literature. Pathway enrichment analysis using the PrecocityDB gene-set indicated that individuals with PP are at higher risk of developing reproductive and metabolic disorders such as PCOS, hypogonadism, insulin resistance, metabolic syndrome, glucose intolerance and hyperinsulinism. Pathways related to prostate cancer, breast, endometrial and hormone-dependent neoplasms were also found to be enriched in the PP gene-set. The hypotheses generated based on gene enrichment analysis are consistent with existing clinical observations. The clinical data on longitudinal prognosis for individuals with PP and diseases that are more likely to emerge in adulthood is scarce. This research gap needs to be addressed for evidence-based management of PP.
Methodology
Creation of gene dataset
PubMed was searched on 3rd July 2020 using MeSH (Medical Subject Headings) terms associated with PP such as, "Precocious Puberties", “Premature Pubarche” , "Puberties, Precocious", "Pubertas Praecox", "Praecox, Pubertas", "Precocious Puberty", "Precocious Puberty, Central", "Central Precocious Puberties", "Central Precocious Puberty", "Precocious Puberties, Central", "Puberties, Central Precocious", "Puberty, Central Precocious", "Sexual Precocity", "Precocities, Sexual", "Precocity, Sexual", "Sexual Precocities", "Idiopathic Sexual Precocity", "Idiopathic Sexual Precocities", "Precocities, Idiopathic Sexual", "Precocity, Idiopathic Sexual", "Sexual Precocities, Idiopathic", "Sexual Precocity, Idiopathic", "Familial Precocious Puberty", "Familial Precocious Puberties", "Precocious Puberties, Familial", "Precocious Puberty, Familial", "Puberties, Familial Precocious", "Puberty, Familial Precocious", "Precocious Puberty, Male-Limited", "Male-Limited Precocious Puberties", "Male-Limited Precocious Puberty", "Precocious Puberties, Male-Limited", "Puberties, Male-Limited Precocious", "Puberty, Male-Limited Precocious", "Precocious Puberty, Male Limited", "Testotoxicosis" and "gene". Using this query, 487 literature records were retrieved from PubMed. These publications were reviewed to confirm gene association with PP (Fig. 4).
The studies were included in PrecocityDB if (a) gene association with PP was confirmed based on experiments such as Real Time-Polymerase Chain Reaction(RT-PCR), Restriction fragment length polymorphism (RFLP), Multiplex ligation-dependent probe amplification (MLPA), Single-Strand Conformation Polymorphism(SSCP) and (b) puberty was attained at age less than 8 years for girls and less than 9 years for boys. Studies were excluded if (a) samples were not from human source; and (b) disease terms were not directly associated with PP; such as “pseudo-precocious puberty”, “premature/precocious adrenarche”, “premature/precocious thelarche”, “congenital adrenal hyperplasia”, “McCune Albright Syndrome”, “21-hydroxylase deficiency”, “androgen excess syndrome”, “aromatase excess syndrome”, “hyperandrogenism”, and “hirsutism” (Fig. 4).
Relevant data, such as nature of study population, ethnicity, hereditary information, and mutations/SNPs were appended to gene records based on evidence available in literature. The gene records were annotated for additional information such as unique identifiers for gene and protein, protein structures, family and ontology term details, metabolic pathway information by mapping them to databases such as NCBI46, dbSNP47, Ensembl48, UniProt49, GO50, KEGG51, PDB52 and, Reactome53.
Pathway enrichment analysis
Pathway enrichment analysis was performed on the manually curated gene-set and its transcription factors using Enrichr54, a gene list enrichment analysis tool which is independent of expression data and based on Fisher’s exact test. The transcription factors of the gene-set were identified using TRRUST V255. KEGG database was used as pathway resource. Pathways with adjusted P value < 0.05 were selected. Enriched pathways were mapped to parent pathway groups using KEGG. The pathways were clustered based on KEGG pathway terms using ClueGO v2.5.7 with a minimum of two genes per term56.
Disease enrichment analysis
Gene-disease associations were derived for PP gene-set based on statistically significant co-occurrences of genes and diseases in PubMed by using the GS2D tool57. Diseases with P value and FDR < 0.05 were selected. Enriched diseases were mapped to parent disease terms using a rule-based method adapted from PCOSKBR258.
Database interface development
PrecocityDB was developed using PHP 7.2.24, MariaDB Server 10.1.44, JavaScript, ideogram.js v1.22.0, and XHTML 1.0 and hosted on Apache webserver.
SNP visualizer
This tool can be used for visualization of chromosomal coordinates of SNPs associated with PP. Ideogram.js JavaScript library59 was used for creation of SNP visualizer. Chromosomes, in haploid state, are shown as vertical bars. Black and grey bands represent heterochromatin region; lighter bands represent euchromatin region; and blue represents variable region. Centromere is marked in pink. Users can select a shape from list of shapes (circles, triangles or rectangles) to display SNPs associated with PP. The gene name, SNP ID and chromosomal location of each SNP can be perused by hovering over the SNP pointer.
References
Merke, D. P. & Cutler, J. Evaluation and management of precocious puberty. Arch. Dis. Child. 75, 269–271 (1996).
Aguirre, R. S. & Eugster, E. A. Central precocious puberty: From genetics to treatment. Best Pract. Res. Clin. Endocrinol. Metab. 32, 343–354 (2018).
Jeong, H. R. et al. Inhibin B as a screening tool for early detection and treatment monitoring of central precocious puberty. Gynecol. Endocrinol. https://doi.org/10.1080/09513590.2020.1718642 (2020).
Skakkebæk, N. E. Endocrine disrupters and testicular dysgenesis syndrome. in Hormone Research vol. 57 43 (S. Karger AG, 2002).
Breehl, L. & Caban, O. Physiology (StatPearls (StatPearls Publishing, Puberty, 2019).
Osman, H. A. et al. Precocious puberty: An experience from a major teaching hospital in Central Saudi Arabia. Sudan. J. Paediatr. 17, 19–24 (2017).
Pallavee, P. & Samal, R. Precocious puberty: A clinical review. Int. J. Reprod. Contracept. Obstet. Gynecol. 7, 771 (2018).
Partsch, C. J., Heger, S. & Sippell, W. G. Management and outcome of central precocious puberty. Clin. Endocrinol. 56, 129–148 (2002).
Abreu, A. P. & Kaiser, U. B. Pubertal development and regulation. Lancet Diabetes Endocrinol. 4, 254–264 (2016).
Partsch, C. J. & Sippell, W. G. Pathogenesis and epidemiology of precocious puberty. Effects of exogenous oestrogens. Hum. Reprod. Update 7, 292–302 (2001).
Lee, J., Kim, J., Yang, A., Cho, S. Y. & Jin, D. K. Etiological trends in male central precocious puberty. Ann. Pediatr. Endocrinol. Metab. 23, 75–80 (2018).
Chittwar, S. & Ammini, A. C. Precocious puberty in girls. Indian J. Endocrinol. Metab. 16, S188–S191 (2012).
Cavanagh, S. E., Riegle-Crumb, C. & Crosnoe, R. Puberty and the education of girls. Soc. Psychol. Q. 70, 186–198 (2007).
Wang, H., Lin, S. L., Leung, G. M. & Schooling, C. M. Age at onset of puberty and adolescent depression: ‘children of 1997’ birth cohort. Pediatrics 137(6), e20153231. https://doi.org/10.1542/peds.2015-3231 (2016).
Carel, J. C., Lahlou, N., Roger, M. & Chaussain, J. L. Precocious puberty and statural growth. Hum. Reprod. Update 10, 135–147 (2004).
Soriano-Guillén, L. & Argente, J. Central precocious puberty, functional and tumor-related. Best Pract. Res. Clin. Endocrinol. Metabol. 33, 101262 (2019).
Yoo, J.-H. Effects of early menarche on physical and psychosocial health problems in adolescent girls and adult women. Korean J. Pediatr. 59, 355 (2016).
Gong, T. T., Wang, Y. L. & Ma, X. X. Age at menarche and endometrial cancer risk: A dose-response meta-analysis of prospective studies. Sci. Rep. 5, 1–10 (2015).
Hsieh, C.-C., Trichopoulos, D., Katsouyanni, K. & Yuasa, S. Age at menarche, age at menopause, height and obesity as risk factors for breast cancer: Associations and interactions in an international case-control study. Int. J. Cancer 46, 796–800 (1990).
Hamajima, N. et al. Menarche, menopause, and breast cancer risk: Individual participant meta-analysis, including 118 964 women with breast cancer from 117 epidemiological studies. Lancet Oncol. 13, 1141–1151 (2012).
Parent, A. S. et al. The timing of normal puberty and the age limits of sexual precocity: Variations around the world, secular trends, and changes after migration. Endocr. Rev. 24, 668–693 (2003).
De Vries, L., Kauschansky, A., Shohat, M. & Phillip, M. Familial central precocious puberty suggests autosomal dominant inheritance. J. Clin. Endocrinol. Metab. 89, 1794–1800 (2004).
Parent, A. S. et al. Early onset of puberty: Tracking genetic and environmental factors. In Hormone Research Vol. 64 41–47 (Horm Res, 2005).
Yum, T., Lee, S. & Kim, Y. Association between precocious puberty and some endocrine disruptors in human plasma. J. Environ. Sci. Heal. Part A Toxic Hazard. Subst. Environ. Eng. 48, 912–917 (2013).
Wickham, H. ggplot2. ggplot2 (Springer New York, 2009). https://doi.org/10.1007/978-0-387-98141-3.
McLean, M., Davis, A. J. & Reindollar, R. H. Abnormalities of Female Pubertal Development. Endotext (MDText.com Inc, South Dartmouth, 2000).
Bhateja, B., Synd, E. M., Bohra, A. & Bhateja, S. Citation: Bohra A, Bhateja S. Carcinogenesis and sex hormones: A review. Endocrinol. Metab. Synd.s 4, 156 (2015).
Henderson, B. E. & Feigelson, H. S. Hormonal carcinogenesis. Carcinogenesis 21, 427–433 (2000).
Canton, A. P. M., Seraphim, C. E., Brito, V. N. & Latronico, A. C. Pioneering studies on monogenic central precocious puberty. Arch. Endocrinol. Metabol. 63, 438–444 (2019).
Valadares, L. P. et al. MKRN3 mutations in central precocious puberty: A systematic review and meta-analysis. J. Endocr. Soc. 3, 979–995 (2019).
Pagani, S. et al. MKRN3 and KISS1R mutations in precocious and early puberty. Ital. J. Pediatr. 46(1), 39. https://doi.org/10.1186/s13052-020-0808-6 (2020).
Narayan, P. Genetic models for the study of luteinizing hormone receptor function. Front. Endocrinol. (Lausanne) 6, 152 (2015).
Tommiska, J. et al. LIN28B in constitutional delay of growth and puberty. J. Clin. Endocrinol. Metab. 95, 3063–3066 (2010).
Tommiska, J. et al. LIN28B, LIN28A, KISS1, and KISS1R in idiopathic central precocious puberty. BMC Res. Notes 4, 363 (2011).
Chaudhari, N., Dawalbhakta, M. & Nampoothiri, L. GnRH dysregulation in polycystic ovarian syndrome (PCOS) is a manifestation of an altered neurotransmitter profile. Reprod. Biol. Endocrinol. 16, 37 (2018).
Hu, M. et al. Uterine glycolytic enzyme expression is affected by knockout of different estrogen receptor subtypes. Biomed. Reports 11, 135–144 (2019).
Magoffin, D. A. Ovarian steroidogenic abnormalities in the polycystic ovary syndrome. In Androgen Excess Disorders in Women 203–211 (Humana Press, 2007). https://doi.org/10.1007/978-1-59745-179-6_18.
Shaaban, Z., Khoradmehr, A., Jafarzadeh Shirazi, M. R. & Tamadon, A. Pathophysiological mechanisms of gonadotropins- and steroid hormones-related genes in etiology of polycystic ovary syndrome. Iran. J. Basic Med. Sci. 22, 3–16 (2019).
Holmes, D. Reproductive endocrinology: Puberty timing and cancer risk. Nat. Rev. Endocrinol. 13, 377 (2017).
Bodicoat, D. H. et al. Timing of pubertal stages and breast cancer risk: The Breakthrough Generations Study. Breast Cancer Res. 16, R18 (2014).
Bonilla, C. et al. Pubertal development and prostate cancer risk: Mendelian randomization study in a population-based cohort. BMC Med. 14, 66 (2016).
Hosaka, T. et al. Disruption of forkhead transcription factor (FOXO) family members in mice reveals their functional diversification. Proc. Natl. Acad. Sci. U. S. A. 101, 2975–2980 (2004).
Li, J., Zhang, W., Zhu, S. & Shi, F. Nitric Oxide Synthase Is Involved in Follicular Development via the PI3K/AKT/FoxO3a Pathway in Neonatal and Immature Rats. Animals 10, 248 (2020).
Wu, H.-C. et al. Effects of electroacupuncture on expression of PI3K/Akt/Foxo3a in granulosa cells from women with shen (kidney) deficiency syndrome undergoing in vitro fertilization-embryo transfer. Chin. J. Integr. Med. 25, 252–258 (2019).
Agarwala, R. et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. https://doi.org/10.1093/nar/gkx1095 (2018).
Sherry, S. T. et al. DbSNP: The NCBI database of genetic variation. Nucleic Acids Res. https://doi.org/10.1093/nar/29.1.308 (2001).
Yates, A. D. et al. Ensembl 2020. Nucleic Acids Res. https://doi.org/10.1093/nar/gkz966 (2020).
The UniProt Consortium. UniProt: A worldwide hub of protein knowledge The UniProt Consortium. Nucleic Acids Res. https://doi.org/10.1093/nar/gky1049 (2019).
Ashburner, M. et al. Gene ontology: Tool for the unification of biology. Nat. Genet. https://doi.org/10.1038/75556 (2000).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. https://doi.org/10.1093/nar/28.1.27 (2000).
Berman, H. M. et al. The Protein Data Bank. Nucleic Acids Res. https://doi.org/10.1093/nar/28.1.235 (2000).
Fabregat, A. et al. The reactome pathway knowledgebase. Nucleic Acids Res. https://doi.org/10.1093/nar/gkx1132 (2018).
Chen, E. Y. et al. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics https://doi.org/10.1186/1471-2105-14-128 (2013).
Han, H. et al. TRRUST v2: An expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. https://doi.org/10.1093/nar/gkx1013 (2018).
Bindea, G. et al. ClueGO: A cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics https://doi.org/10.1093/bioinformatics/btp101 (2009).
Fontaine, J. F. & Andrade-Navarro, M. A. Gene set to diseases (GS2D): Disease enrichment analysis on human gene sets with literature data. Genomics Comput. Biol. https://doi.org/10.18547/gcb.2016.vol2.iss1.e33 (2016).
Sharma, M. et al. PCOSKBR2: A database of genes, diseases, pathways, and networks associated with polycystic ovary syndrome. Sci. Rep. 10, 14738 (2020).
Ideogram.js: A JavaScript library for chromosome visualization. https://www.biostars.org/p/305698/.
Acknowledgements
We are grateful to Ms. Ulka Gawde for providing technical assistance and Mr.Tanmay Shirsat for assisting in the literature review.
Funding
This work [RA/969/09-2020] was supported by grants received from Indian Council of Medical Research Department of Biotechnology [BT/PR26350/BID/7/810/2017] and Department of Health Research [R. 12020/01/2019-HR].
Author information
Authors and Affiliations
Contributions
S.I-T. planned, designed, and supervised the study. S.B., M.S., K.D. curated the data. M.S. annotated the data. R.S.B., and K.P. developed the interface. Data analysis and interpretation were performed by I.K., M.S and S.I-T. M.S., I.K., and S.I-T., wrote the manuscript. All authors approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sharma, M., Kundu, I., Barai, R.S. et al. Enrichment analyses of diseases and pathways associated with precocious puberty using PrecocityDB. Sci Rep 11, 4203 (2021). https://doi.org/10.1038/s41598-021-83446-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-83446-z
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.