Abstract
The repeatability and reproducibility of radiomic features extracted from CT scans need to be investigated to evaluate the temporal stability of imaging features with respect to a controlled scenario (test–retest), as well as their dependence on acquisition parameters such as slice thickness, or tube current. Only robust and stable features should be used in prognostication/prediction models to improve generalizability across multiple institutions. In this study, we investigated the repeatability and reproducibility of radiomic features with respect to three different scanners, variable slice thickness, tube current, and use of intravenous (IV) contrast medium, combining phantom studies and human subjects with non-small cell lung cancer. In all, half of the radiomic features showed good repeatability (ICC > 0.9) independent of scanner model. Within acquisition protocols, changes in slice thickness was associated with poorer reproducibility compared to the use of IV contrast. Broad feature classes exhibit different behaviors, with only few features appearing to be the most stable. 108 features presented both good repeatability and reproducibility in all the experiments, most of them being wavelet and Laplacian of Gaussian features.
Similar content being viewed by others
Background
Medical images are routinely used for cancer staging, treatment planning and evaluation. Radiological findings are mainly evaluated in a qualitative or semi-qualitative fashion guided predominantly by visual inspection1. However, human interpretation of images is open to subjectivity and potentially misses some of the quantitative and objective information that could otherwise be retrieved from patients’ scans through computer-assisted methods2.
The field of “radiomics” aims to address the above-mentioned issues by objectively quantifying visual information in the images as a vast set of numerical metrics known as “features”. Radiomics hypothesizes that a certain subset of features, analyzed with the aid of machine learning algorithms due to high dimensionality, may have some predictive/prognostic value. Such subsets of features denote a “signature”, i.e. a digital image phenotype of the target disease, which opens the way towards personalized treatment in oncology3.
One of the most challenging problems for translating radiomic studies into clinical decision support systems is to evaluate the robustness of radiomic-based models and hence their potential generalizability across multiple datasets from different institutions4. Different institutions commonly acquire scans with different settings (e.g. scanner manufacturers, slice thickness, signal-to-noise ratio) according to largely self-defined imaging protocols, which add unwanted variation in the resulting radiomic features that are not related to the disease phenotype. A feature that is useful on one dataset may therefore lose its value on another dataset, since the feature may be sensitive to different methods of acquisition5.
When discussing robustness of radiomic studies two concepts need to be considered: “repeatability” and “reproducibility”. Repeatability refers to features that remain the same when imaged multiple times in the same subject, be that a human or a suitable phantom, using the same image acquisition methods. Reproducibility refers to features that remain the same when extracted using different equipment, different software, different image acquisition settings, or different operators (e.g. other clinics), be that in the same subject or in different subjects6. Repeatability and reproducibility concerns have been raised as major source of uncertainties in radiomic models7.
Most of the studies that investigated the reproducibility of radiomic features with respect to different image acquisition settings, demonstrate a strong dependence of radiomic features on such settings. Texture features appear to be more vulnerable to reproducibility/repeatability issues. There is a strong connection between reproducibility/repeatability and prognostic values8. In a study about time series classification, the investigators concluded that poorly reproducible/repeatable features were usually accompanied by poor discriminative performances9.
Recent publications have also investigated the presence of correlations between radiomic features and tumor volume9,10. The latter has been shown to be one of the most generalizable features. Therefore, there is the need to investigate if the most reproducible features were also strongly correlated with tumor volume.
Several studies have investigated the repeatability/ reproducibility of radiomic features on phantom as well as well as clinical cohort6,7,8,9,10,11,12,13,14,15,16,17. Few publications have also investigated the disease specific dependency of radiomic feature repeatability/ reproducibility and presented the results. These studies have either performed repeatability or reproducibility study alone; or performed repeatability and reproducibility study only on phantoms13,14 or clinical15,16 cohorts, which (1) limits the possibility to isolate a subset of features that are both repeatable and reproducible, and (2) does not allow comparing differences in the results because of using only phantom or human data. There remains a need to evaluate reproducibility and repeatability of radiomic features, not only on phantoms datasets, but also on human cohorts in the same study. The risk is that phantom studies do not have sufficiently high complexity and heterogeneity within the synthetic “tumors” to be a fair test of feature robustness. In our study, stable feature refers to both repeatable and reproducible features at the same time. With our study, we provide an extension to currently available literature by performing a comprehensive evaluation of the reproducibility and repeatability of 1080 radiomic features considering not only different groups of features, but also features extracted using digital filtering both with phantoms and human data. In this study, we also investigated how the correlations between radiomic features and tumor volume impact the reproducibility and repeatability results.
Results
Phantom—repeatability
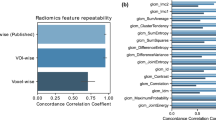
The percentage of radiomic features presenting good repeatability (ICC ≥ 0.9) were 58% (624/1080) for scanner1 (Philips Gemini TF16), 43% (464/1080) for scanner2 (Philips Gemini TF64), 61% (661/1080) for scanner3 (GE Discovery NM 570) and 45% (488/1080) for the three scanners overall. Results are shown in Fig. 1 for each feature category.
Repeatability analysis using repeated phantom scans for all the different radiomic feature classes. The median ICC values for all the 6 protocols is reported, separately for scanner1, scanner2, scanner3 and the union of the three. Repeatability analysis on RIDER (clinical cohort) was also performed. Three different levels of repeatability are defined: good (ICC ≥ 0.9), medium (0.75 < ICC < 0.9), and poor (ICC ≤ 0.75) (FO = First Order Feature; TF = Textural Feature; LOG = Total LoG Feature; WF = Total Wavelet Feature; LOG-FO = LoG First Order Feature; LOG-TF = LoG Textural Feature; WF-FO = Wavelet First Order Feature; WF-TF = Wavelet Textural Feature).
RIDER (clinical cohort)—repeatability
The percentage of radiomic features presenting good, moderate, and poor repeatability were 82% (888/1080), 15% (164/1080), and 3% (28/1080) respectively for the RIDER clinical cohort. The results per feature categories are shown in Fig. 1.
Phantom—reproducibility—intra and inter scanner variability
For the intra-scanner study, 30% (322/1080), 31% (332/1080) and 39% (426/1080) features presented good, moderate, and poor reproducibility (Fig. 2A) for all the scanners. For the inter-CT scanner study, 14% (154/1080), 19% (204/1080) and 67% (722/1080) features presented good, moderate, and poor reproducibility respectively (Fig. 2B) for all the 6 protocols. Reproducibility of the features individually for the six protocols are shown in the Supplementary material S1.
Results of the reproducibility experiments: (A) intra-scanner experiment using the phantom. By taking the median of all ICCs computed on the three scanners; (B) inter-scanner experiment using the phantom across all three scanners; (C) stability of radiomic features with respect to three different clinical protocols in the clinical study; (D) impact of IV (intravenous) contrast medium presence (WBCECT2)/ absence (NCCTT2) and difference in current (WBCECT2:Auto mA = 100–200; NCCTT2: fixed mA = 300 ) in the clinical study, and E) impact of slice thickness (2 vs 5 mm) in the human study. Three different levels of reproducibility are defined: good (ICC ≥ 0.9), medium (0.75 < ICC < 0.9), and poor (ICC ≤ 0.75).
Clinical cohort—reproducibility
Among the features tested, 19% (199/1080) good, 29% (315/1080), moderate and 52% (556/1080) had poor, reproducibility when comparing the 3 different imaging protocols on the Gemini TF16 scanner (Fig. 2C).
When comparing IV contrast (WBCECT2) versus non-contrast (NCCTT2) protocols, 45% (483/1080) of the features had poor, 41% (442/1080) moderate, and 14% (155/1080) good reproducibility (Fig. 2D). When comparing slice thickness, using the BLDCT5 protocol (slice thickness = 5 mm) versus the WBCECT2 protocol (slice thickness = 2 mm), 37% (398/1080) of the features had poor, 17% (179/1080) moderate, and 47% (503/1080) good reproducibility (Fig. 2E).
Volume correlations
In the clinical cohort, 7% (73/1080), 5% (57/1080) and 88% (950/1080) of the radiomic features had good (ρ ≥ 0.9), moderate (0.75 < ρ < 0.9) and poor (ρ ≤ 0.75) correlation with the GTV.
Overall summary
Median ICC was calculated for all the reproducibility studies performed using the phantom and clinical cohorts. A total of 22.5% (243/1080) features had good reproducibility (ICC > 0.9) in clinical cohort. When the median of ICC was calculated for repeatability study performed with phantom and clinical cohorts (RIDER); 46.1% (498/1080) of features had good repeatability (ICC > 0.9). For repeatability study on phantom and clinical cohort together 55% (599/1080) features had good stability (ICC > 0.9) (Fig. 3A). For reproducibility study on phantom and clinical cohort together 15% (164/1080) features had good stability (ICC > 0.9) (Fig. 3B). For repeatability and reproducibility study together on clinical cohort 18% (189/1080) features had good stability (Fig. 3C). For all the experiments, 13% (138/1080) of the features presented both high (median ICC > 0.9) repeatability and high reproducibility (Fig. 3D). Tumor volume was again confirmed to be the most repeatable and reproducible feature with a median ICC of 0.99. When considering volume collinearity, 21% of these stable features presented strong Spearman correlations (ρ > 0.9). If we removed the features with strong correlations with GTV, then the final number of repeatable and reproducible features was 108: 59 WF (Wavelet) (8% of total WF), 46 LOG (Laplacian of Gaussian) (17% of total LOG), and 3 TA (Texture Analysis) (3% of total TA) features (Table 1). Overall, TA had the largest median ICC (0.933 ± 0.024) followed by LOG (0.923 ± 0.017) and WF (0.917 ± 0.014) features (p < 0.05). The topmost robust feature per feature types were: GLRLM-Non-Uniformity (LOG-2 mm kernel); LLH-GLCM-JointEnergy (WF) and Gray Level Dependence Matrix (GLDM) Non-Uniformity (TA).
Common features in various studies showing good stability (ICC > 0.9): (A) Venn diagram shows the overlap of repeatability (RIDER) study and Phantom repeatability study. (B) Venn diagram shows the overlap of Phantom reproducibility study and reproducibility study in clinical cohort. (C) Venn diagram shows the overlap of repeatability (RIDER) study and reproducibility study in clinical cohort. (D) Overall summary of all the experiments. The Venn diagram shows the overlap of the repeatability experiment (phantom + clinical [RIDER] data) with the reproducibility experiments (phantom + clinical data).
It is interesting to notice how the top 50 repeatable features presented strong inter Spearman correlations, with Wavelet and Laplacian of Gaussian features being strongly clustered together (heatmap on Fig. 4). Overall, the number of features with good repeatability was found to be significantly larger than the number of reproducible features. Reproducibility experiments using phantom data (IntraCT experiment) led to more features being found reproducible compared to experiments performed using the clinical cohort (30% vs 19% of features with ICC ≥ 0.9, p < 0.05). Around 57% (138/243) of the robust features overlapped with features from repeatability and reproducibility study. The remaining 67 features being 36% Wavelet and 74% Laplacian of Gaussian were reproducible, but not repeatable.
Discussion
In this study, we investigated: (A) radiomic feature repeatability in a test–retest scenario using a NEMA IQ phantom; (B) radiomic feature reproducibility with respect to different tube currents, slice thickness as well as dependencies to different scanner models using an image quality phantom, and (C) radiomic feature reproducibility in a clinical cohort comparing three different acquisition protocols as well as the impact of slice thickness and the presence of IV contrast medium. We isolated a list of repeatable and reproducible features for all the experiments. Furthermore, we computed the correlations between radiomic features and tumor volume with the aim of investigating if the most repeatable and reproducible features also presented strong correlations. In fact, tumor volume was found to be the most robust feature and we wanted to assess if this could be a reason for a feature to present high reproducibility and repeatability. As shown in the results, only a relatively small percentage of radiomic features (around 13% of the total) presented both good repeatability and reproducibility across all the experiments. However, differences were found between repeatability and reproducibility. The number of features with good repeatability was larger than the number of reproducible features in the phantom experiment. Unfortunately, because we did not have any test–retest clinical data it was not possible to draw the same conclusion. Nevertheless, to obtain a fair comparison, we used the publicly available dataset RIDER to investigate the repeatability of radiomic features in NSCLC patients. The Venn Diagram in Fig. 3D shows that most of the repeatable and reproducible features in human data overlap with features from the phantom studies. This clearly shows that features computed on phantom are a superset of features computed on real human data. Our experiments also showed that there are some features extracted from human data that are robust but do not overlap with phantom results. Two main reasons could be associated with this: (A) statistical fluctuations because of the large number of computed features; (B) differences in the dynamic range of the features between phantom and human data. Point (B) is strictly related to the fact that the image quality phantom with spherical homogenous inserts are still not advanced enough to replicate tumor complexity seen in patients’ data. Therefore, our study should be improved by including several types of imaging phantoms or considering new types of plugs that can better mimic tumor heterogeneity. In the last years, attention has been devoted to produce more realistic inserts by using 3D printing techniques18,19. The above-mentioned hypothesis seem to be confirmed by the fact that the features that did not overlap were only wavelet and Laplacian of Gaussian features, which might indicate that some real tumors’ texture patterns are still difficult to be reproduced with imaging phantoms.
We found large variation of radiomic feature in repeatability study even within a short time gap of 30 min “coffee-break”. Overall, less than 50% of features had a good repeatability (ICC > 0.9) using phantom scans, in agreement with previously published literature19,20,21. When considering time-series analysis of radiomic features (e.g. for monitoring treatment response), temporal stability of radiomic features becomes imperative to be investigated. As mentioned in the introduction section, poor repeatability seems to be associated with poor prognostic/predictive power, while the reverse might not be equally true9. Therefore, our results can be taken by other radiomic studies to reduce the dimensionality of computed features by excluding poorly repeatable features.
When considering radiomic reproducibility, the presence or absence of IV contrast medium had a stronger impact than differences in slice thickness in the human study: 14% (155/1080) versus 47% (503/1080) (p < 0.05) of features with good reproducibility.
From the overall summary section in the results, it emerges that the different feature categories are sensitive with different degrees to reproducibility and repeatability. Our results are in line with the previous literature. The usage of image filtering could enhance the quality of the images even when acquired with different protocols and thus improve reproducibility. It is important to point out that this study did not investigate the robustness of shape metrics, since the contours were co-registered from PET to CT images and the same contour was used for all sets of CT series. However, shape metrics have been shown to be strongly affected by inter-observer variability in tumor delineations and this aspect was not investigated in this study.
We investigated how correlations between tumor volume and radiomic features could impact the repeatability and reproducibility. In line with other studies, not only tumor volume was the most repeatable and reproducible feature (median ICC = 0.99), but most of the top reproducible features showed strong Spearman correlations (ρ > 0.9) with tumor volume. This opens the debate whether their robustness could be an effect of an underlying “volume effect”. However, more investigation is needed to isolate and further explain this effect. Therefore, in Table 1 we proposed the final list of most repeatable and reproducible features with lower correlations with tumor volume.
Finally, the list provided in Table 1 represents a starting point to isolate repeatable and robust features, but this is not enough to conclude about their prognostic predictive performance. Furthermore, as shown in Fig. 4, most of these features present strong intercorrelations and might produce redundant information if all are injected into a classifier for radiomic-based models. The results presented in this study needs to be validated in additional multi-institutional studies and considering additional parameters that can affect features’ reproducibility and repeatability. First, in our analysis we only considered two different scanner manufacturers. We did not investigate the role of other acquisition parameters such as reconstruction kernels or tube voltage. These results are intended to be shared within the radiomic community for confirmation.
Methods
This study was approved by the hospital Institutional Ethics Committee (Institutional Ethics Committee-I, Tata Memorial Centre [IEC, TMC], Mumbai, India) as a retrospective study, with waivers of informed consent from involved patients as per IEC policy of our hospital by the same Ethics Committee. All methods were carried out in accordance with relevant guidelines and regulations. This study comprises PET/CT images from a polymer phantom as well as from a clinical cohort. Our study has focused only on CT radiomic features stability. PET images were used to delineate the tumor (using SUV threshold of 40%) and this delineation was transferred to the corresponding CT images included in this study.
Phantom
The National Electrical Manufacturers Association (NEMA) Image Quality (IQ)PET/CT phantom (Data Spectrum Inc., NJ, USA) was used for this study22. The external dimensions of the phantom are 241 mm × 305 mm × 241 mm with interior length of 180 mm and volume of 9.7L. It has six fillable spheres and one central cylinder. The largest insert with a diameter of 37 mm was used for radiomic feature analysis study. The phantom was filled with distilled water containing 18F-FDG. The concentration of 18F-FDG was adjusted until a target to background signal ratio of 4:1 was created between the active sphere and water background.
Clinical cohort
Patients with non-small cell lung cancer (NSCLC) (n = 104) who underwent pre-treatment PET/CT scans in our department were included in this study. There were 85 males and 19 females. The median age was 66 (36–90) and 53 (35–72) years respectively for males and females. The median tumor volume was 92 (14–486) cm3 for men and 86 (22–432) cm3 for women. Population demographics and clinical information are provided in Supplementary table S2.
RIDER: The Reference Image Database to Evaluate Therapy Response (RIDER) data base was used in this study to perform repeatability study. All the 32 patients DICOM data (i.e. Images and RTSTRUCTs) of the RIDER data set were included in this study23.
Scanners
Three different scanners were used in the study. Two scanners were from the same manufacturer (Philips Medical, Eindhoven, The Netherlands) but different models, and the last scanner was from another manufacturer (General Electric Medical System, Milwaukee, USA). For simplicity of reading we will refer to the scanners as follows: scanner 1 is the Philips Gemini TF16 PET/CT, scanner 2 is the Gemini TF64 PET/CT, and scanner 3 is the General Electric Discovery NM 670 pro SPECT/CT.
Scanning protocols
NEMA IQ phantom
The NEMA IQ phantom was scanned twice, 30 min apart (‘coffee break’) without repositioning, one the same scanner and within the same conditions. This procedure was performed for all the three scanners and considering six different acquisition protocols. They had the same tube voltage (120 kV for all three scanners), pitch (0.46 for scanner 1 and 2 and 2.5 for Scanner 3) and reconstruction kernel based on filtered back projection for scanner 1, 2 and adaptive statistical iterative reconstruction (ASiR) (40% ASiR setting and a noise index of 13.75) for scanner 3, but different tube currents (ranging from 100 to 300 mA) slice thicknesses (ranging from 2 to 5 mm for scanner 1& 2 and 2.5 to 5 for scanner 3). These protocols are listed in Table 2.
Clinical cohort
Patients were scanned using three different clinical protocols on the Philips Gemini TF64 PET/CT (previously referred to as scanner 2). The three protocols had the same tube voltage (120 kV), pitch (0.46) and reconstruction kernel, but different slice thicknesses, tube current and presence or absence of an intravenous contrast medium, namely, one whole body contrast CT with 2 mm slice thickness (referred as WBCECT2), one whole body contrast CT with 5 mm slice thickness (referred as BLDCT5), and one non contrast thoracic CT with 2 mm slice thickness (referred as NCCTT2). Modulated tube current (between 100 and 200 mA) as per dose care automated system was used for BLDCT5 and WBCECT2. The protocols are listed in Table 3.
RIDER
The RIDER data set comprises of 32 NSCLC patient’s test–retest CT imaging performed with a time lag of 15 min and two sets of delineations (RTSTRUCT) (i.e. tumor delineated by manual and automatic methods). Imaging parameters of RIDER database is summarized in Table 4. Radiomic extraction and statistical analysis was performed as per the study protocol.
Study design
In this study we investigated both reproducibility and repeatability of radiomic features. The repeatability of radiomic features was evaluated using the test retest scans acquired with the IQ phantom on three different scanners and for all the 6 protocols listed in Table 2 and on the publicly available clinical cohort RIDER data set. The reproducibility of radiomic features with respect to different acquisition protocols but within the same scanner (intra-scanner variability) was evaluated comparing radiomic feature values using the test scans acquired with the IQ phantom across the 6 different protocols. This analysis was repeated for all the three scanners. The reproducibility of radiomic features with respect to different scanner models was evaluated comparing radiomic feature values extracted from the test scans acquired with the IQ phantom for each protocols on the three different scanners (inter-scanner variability). The reproducibility of radiomic features with respect to presence/absence of intravenous contrast medium and slice thickness in clinical data was investigated comparing radiomic features using the images acquired with the NSCLC patients (clinical study). Figure 5 summarizes the overall study design.
In this study we investigated both reproducibility and repeatability of radiomic features. The repeatability of radiomic features was evaluated using the test retest scans acquired with the IQ phantom on three different scanners and with 6 protocols and online available RIDER data set. The reproducibility of radiomic features with respect to different acquisition protocols but within the same scanner (intra-scanner variability) was evaluated comparing radiomic feature values using the test–retest scans acquired with the IQ phantom across the 6 different protocols. A clinical cohort of NSCLC patients was used to investigate the reproducibility of radiomic features with respect to 3 different clinical acquisition protocols, with a focus on the impact of slice thickness and IV contrast medium.
ROIs (Region of Interest) definition
PET and CT series of all the studies were loaded on a GE Advantage image processing workstation (GE Healthcare, Waukesha, WI, USA) from our hospital PACS. Standardized Uptake Value (SUV)-based auto-segmentation using a threshold of 40% from the maximum value was used to delineate the primary lung tumor and active phantom insert on PET images for scanners 1 and 2. Manual delineation of the phantom insert was performed by an experienced physicist for phantom images acquired with scanner 3, since PET series were not available for this scanner. These delineations were performed using the AdvantageSimMD software installed on the Advantage image processing workstation and stored as RTSTRUCT. This RTSTRUCT creates a ROI instance corresponding to each PET and CT series in the study24. As all the PET and CT series belongs to same study it automatically accounts for differences in resolution between PET and CT images when the RTSTRUCT is saved. The stored RTSTRUCT has the location of the ROI instance for corresponding image sets (series) about matrix size and slice thickness of that series. Images and ROIs, in form of DICOM and RTSTRUCT files, respectively, were transferred to a research workstation where radiomic features were extracted.
Image pre-processing
Images and ROIs are saved in Digital Imaging and Communications in Medicine (DICOM) format. However, the Pyradiomics software uses images and ROIs in Nearly Raw Raster Data (NRRD) format for radiomic feature extraction. We used an in-house developed python Script to perform batch processing to convert images and ROIs from a DICOM CT and RTSTRUCT into an NRRD format using 3DSlicer v4.10.225. An in-house python script based on the image processing toolkit simpleITK v1.2.0 was used to convert contours to binary masks26.
All images were re-sampled to isotropic voxel of 2 × 2 × 2 cubic millimeters prior to 3D radiomic feature extraction using the default b-spline interpolation function in simpleITK. A fixed-bin width of 25 was used for grey level binning of the images. Radiomic features were extracted from the original CT images as well as from images with the following filters: (A) wavelet transformed images using the standard wavelets transforms implemented in Pyradiomics v2.2.0; (B) Laplacian of Gaussian with sigma values 1, 2 and 3 mm. All the features were extracted in 3D, with texture features were aggregated with the method 3D Average.
Radiomic feature definitions
The following radiomic features were extracted as per the definition provided in Pyradiomics documentation (https://pyradiomics.readthedocs.io/en/latest/): First Order Statistics (FO-17 features); Gray Level Co-Occurrence Matrix (GLCM-22 features); Gray Level Run Length Matrix (GLRLM-16 features); Gray Level Size Zone Matrix (GLSZM-16 features); Neighboring Gray Tone Difference Matrix (NGTDM-5 features); Gray Level Dependence Matrix (GLDM-14 features); plus corresponding features with Laplacian of Gaussian filters (LOG-270 features) and with wavelet (WF-720 features). A total of 1080 radiomic features were extracted.
Statistical analysis
The ICC (Intraclass Correlation Coefficient) based on a two-way mixed effect, consistency, single rater/measurement was used to measure the repeatability/reproducibility of features for our experiments, as per Eq. (1)27. Three different level of repeatability/reproducibility were defined: good (ICC ≥ 0.90); moderate (0.75 < ICC < 0.90); poor (ICC < = 0.75)28.
Definition of the ICC used as reproducibility metric. Where, MSE = mean square for error, MSR = mean square for rows, k = number of raters/measurements.
For the repeatability experiment, the ICC was computed between test and re-test scans for all the 6 protocols, separately and together for all the scanners. For the repeatability study with the RIDER dataset, the ICC was computed between test and re-test scans. For the intra-scanner reproducibility experiment, ICC values were computed separately for the three scanners and the median ICC value is reported in the results. For the inter-scanner reproducibility experiment, the ICC values were computed comparing radiomic features separately for the six protocols between the three scanners. The median ICC values for the 6 protocols is reported. For the clinical study, the ICC values comparing radiomic features between the three protocols are reported, as well as only comparing protocols BLDCT5 versus NCCTT2 (same slice thickness but with and without intravenous contrast medium) and NCCTT2 versus WBCECT2 (both with intravenous contrast medium, but different slice thicknesses).
Commonality study
In the clinical cohort common good stable features were found between repeatability study of RIDER data set and reproducibility study of our clinical cohort. Median ICC of repeatability study (Phantom and clinical cohort [RIDER]) was as well as reproducibility study (Phantom and clinical cohort) was calculated. Median ICC of repeatability and Reproducibility study was compared to find common good (ICC > 0.9) stable features.
Volume collinearity analysis
Using the clinical cohort, we assessed the correlation between the GTV and radiomic features using the Spearman correlation coefficient (ρ) to account for possible nonlinear dependencies. The median Spearman correlation coefficient between the 3 different protocols is used in the analysis.
Statistical analysis was performed using R (version 3.2.3) using the package psych. p values were corrected for multiple comparisons using the false-discovery rate corrections method and statistical significance after correction was set at p < 0.05.
References
Beaton, L., Bandula, S., Gaze, M. N. & Sharma, R. A. How rapid advances in imaging are defining the future of precision radiation oncology. Br. J. Cancer 120, 779–790 (2019).
Gillies, R. J., Kinahan, P. E. & Hricak, H. Radiomics: images are more than pictures, they are data. Radiology 278, 563–577 (2016).
Gardin, I. et al. Radiomics: principles and radiotherapy applications. Crit. Rev. Oncol. Hematol. 138, 44–50 (2019).
Kumar, V. et al. Radiomics: the process and the challenges. Magn. Reson. Imaging 30, 1234–1248 (2012).
Yip, S. S. F. & Aerts, H. J. W. L. Applications and limitations of radiomics. Phys. Med. Biol. 61, R150–R166 (2016).
O’Connor, J. P. B. et al. Imaging biomarker roadmap for cancer studies. Nat. Rev. Clin. Oncol. 14, 169–186 (2017).
Traverso, A., Wee, L., Dekker, A. & Gillies, R. Repeatability and reproducibility of radiomic features: a systematic review. Int. J. Radiat. Oncol. Biol. Phys. 102, 1143–1158 (2018).
Gudmundsson, S., Runarsson, T. P. & Sigurdsson, S. Test–retest reliability and feature selection in physiological time series classification. Comput. Methods Programs Biomed. 105, 50–60 (2012).
Zhovannik, I. et al. Volume bias in textural radiomics. Int. J. Radiat. 105(1), S118–S119 (2019).
van Griethuysen, J. J. M. et al. Computational radiomics system to decode the radiographic phenotype. Can. Res. 77, e104–e107 (2017).
Larue, R. T. H. M. et al. Quantitative radiomics studies for tissue characterization: a review of technology and methodological procedures. Br. J. Radiol. 90, 20160665 (2017).
Kim, H. G. et al. Quantitative analysis of the effect of iterative reconstruction using a phantom: determining the appropriate blending percentage. Yonsei Med. J. 56, 253–261 (2015).
Shafiq-Ul-Hassan, M. et al. Intrinsic dependencies of CT radiomic features on voxel size and number of gray levels. Med. Phys. 44, 1050–1062 (2017).
van Timmeren, J. E. et al. Test-retest data for radiomics feature stability analysis: generalizable or study-specific?. Tomography 2, 361–365 (2016).
Balagurunathan, Y. et al. Reproducibility and prognosis of quantitative features extracted from CT images. Transl. Oncol. 7, 72–87 (2014).
Parmar, C. et al. Robust radiomics feature quantification using semiautomatic volumetric segmentation. PLoS ONE 9, e102107 (2014).
Welcha, M. L. et al. Vulnerabilities of radiomic signature development: the need forsafeguards. Radiother. Oncol. 130, 2–9 (2019).
Mitsouras, D. et al. Medical 3D printing for the radiologist. Radiographics 35(7), 1965–1988 (2015).
Samei, E., Hoye, J., Zheng, Y., Solomon, J. B. & Marin, D. Design and fabrication of heterogeneous lung nodule phantoms for assessing the accuracy and variability of measured texture radiomics features in CT. J. Med. Imaging (Bellingham) 6(2), 021606. https://doi.org/10.1117/1.JMI.6.2.021606 (2019).
Traverso, A. et al. Stability of radiomic features of apparent diffusion coefficient (ADC) maps for locally advanced rectal cancer in response to image pre-processing. Phys. Med. 61, 44–51 (2019).
Sanduleanu, S. et al. Tracking tumor biology with radiomics: a systematic review utilizing a radiomics quality score. Radiother. Oncol. 127, 349–360 (2018).
Jha, A. K. et al. Performance characteristic evaluation of a bismuth germanate-based high-sensitivity 5-ring discovery image quality positron emission tomography/computed tomography system as per National Electrical Manufacturers Association NU 2–2012. World J. Nucl. Med. 18, 351–360 (2019).
Armato, S. G. 3rd. et al. The Reference Image Database to evaluate response to therapy in lung cancer (RIDER) project: a resource for the development of change-analysis software. Clin. Pharmacol. Ther. 84(4), 448–456 (2008).
CT/PET FUSION DICOM CONFORMANCE STATEMENT for DICOM V3.0, Technical Publications Direction 2290660-100 Revision A, GE Medical Systems. https://www.gehealthcare.com/-/jssmedia/5337d686cfe442b2a75083038a877029.pdf?la=en-us. Accessed 17 July 2020.
Fedorov, A. et al. 3D slicer as an image computing platform for the quantitative imaging network. Magn. Reson. Imaging 30, 1323–1341 (2012).
Lowekamp, B. C., Chen, D. T., Ibáñez, L. & Blezek, D. The design of simple ITK. Front. Neuroinform. 7, 45 (2013).
Bartko, J. J. The intraclass correlation coefficient as a measure of reliability. Psychol. Rep. 19, 3–11 (1966).
Koo, T. K. & Li, M. Y. A guideline of selecting and reporting intraclass correlation coefficients for reliability research. J. Chiropract. Med. 15, 155–163 (2016).
Acknowledgements
Author A.T was supported by the STW-Strategy research grant (P14-19_1.2), the authors V.R.J. and U.B.S. were supported by the Ministry of Electronics and Information technology, research grant 13(2)-2015-CC-BT. This work was supported Indo-Dutch NWO research grant BIONIC (629.002.205) and MietY-13(2)-2015-CC-BT.
Author information
Authors and Affiliations
Contributions
A.K.J., S.M., V.J., U.B.S. and V.R. are responsible for the study design. A.K.J. performed the experiments, analyses, and manuscript writing. S.M., V.J. assisted in image extraction and ROI generation and transfer of image on research workstation. U.B.S. assisted with the radiomic computations. L.W. and A.T. assisted in statistical analysis, manuscript writing and senior authorship. N.C.P., K.P. provided clinical support and data management. V.R., A.D. are the sponsors and PIs of these study. All co-authors contributed to proof-reading of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jha, A.K., Mithun, S., Jaiswar, V. et al. Repeatability and reproducibility study of radiomic features on a phantom and human cohort. Sci Rep 11, 2055 (2021). https://doi.org/10.1038/s41598-021-81526-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-81526-8
This article is cited by
-
Quantitative MRI-based radiomics analysis identifies blood flow feature associated to overall survival for rectal cancer patients
Scientific Reports (2024)
-
Machine learning-based radiomic analysis and growth visualization for ablation site recurrence diagnosis in follow-up CT
Abdominal Radiology (2024)
-
Feasibility and intra-and interobserver reproducibility of quantitative susceptibility mapping with radiomic features for intracranial dissecting intramural hematomas and atherosclerotic calcifications
Scientific Reports (2023)
-
Criteria for the translation of radiomics into clinically useful tests
Nature Reviews Clinical Oncology (2023)
-
Magnetic resonance radiomics-derived sphericity correlates with seizure in brain arteriovenous malformations
European Radiology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.