Abstract
The in vivo function of cell-derived extracellular vesicles (EVs) is challenging to establish since cell-specific EVs are difficult to isolate and differentiate. We, therefore, created an EV reporter using truncated CD9 to display enhanced green fluorescent protein (EGFP) on the EV surface. CD9truc-EGFP expression in cells did not affect EV size and concentration but enabled co-precipitation of EV markers TSG101 and ALIX from the cell-conditioned medium by anti-GFP immunoprecipitation. We then created a transgenic mouse where CD9truc-EGFP was inserted in the inverse orientation and double-floxed, ensuring irreversible Cre recombinase-dependent EV reporter expression. We crossed the EV reporter mice with mice expressing Cre ubiquitously (CMV-Cre), in cardiomyocytes (αMHC-MerCreMer) and renal tubular epithelial cells (Pax8-Cre), respectively. The CD9truc-EGFP positive mice showed Cre-dependent EGFP expression, and plasma CD9truc-EGFP EVs were immunoprecipitated only from CD9truc-EGFP positive CD9truc-EGFPxCMV-Cre and CD9truc-EGFPxαMHC-Cre mice, but not in CD9truc-EGFPxPax8-Cre and CD9truc-EGFP negative mice. In urine samples, CD9truc-EGFP EVs were detected by immunoprecipitation only in CD9truc-EGFP positive CD9truc-EGFPxCMV-Cre and CD9truc-EGFPxPax8-Cre mice, but not CD9truc-EGFPxαMHC-Cre and CD9truc-EGFP negative mice. In conclusion, our EV reporter mouse model enables Cre-dependent EV labeling, providing a new approach to studying cell-specific EVs in vivo and gaining a unique insight into their physiological and pathophysiological function.
Similar content being viewed by others
Introduction
Extracellular vesicles (EVs) are nanosized membrane-bound vesicles that may function as mediators of cell–cell communication by transfer of cellular proteins, lipids, and nucleic acids1. The presence of EVs in biological fluids, such as urine and plasma, has also gained clinical interest since it facilitates monitoring physiological and pathophysiological processes using minimally invasive techniques. While EVs potentially enable the non-invasive interrogation of cells buried deep within an organism, the cell-specific isolation and tracking of EVs in vivo remain challenging since EVs are secreted from almost every cell types2,3,4,5,6,7,8,9. This hampers our ability to investigate the role of EVs in a physiologically relevant context.
EVs are a heterogeneous group of cell-derived vesicles, and the two main forms are exosomes and microvesicles. Their biogenesis differs: microvesicles directly bud off the plasma membrane, while exosomes are of endosomal origin and accumulate in intraluminal vesicles (ILVs) in multivesicular bodies (MVBs). EVs are, however, enriched in tetraspanins, such as CD63 and CD910,11,12, which are transmembrane proteins widely distributed in the plasma membrane13. The tetraspanins are considered valid markers of EVs14 and have been widely used for the isolation and tracking of EVs.
The tetraspanins have intracellular N- and C-terminals that enable genetic fusion of fluorescent reporter proteins and luciferase15,16,17,18,19. This strategy allows for tissue-specific signals exclusively from EVs and thereby overcome major limitations of previous approaches using radioisotopes, fluorescent dyes, and magnetic conjugated nanoparticles conjugated to lipophilic reagents to label EVs7,8,9. The lipophilic reagents may be released from the EVs, resulting in the distribution of non-EV-associated fluorescent signal12,13. Furthermore, infusing labeled EVs generated in vitro into mice may not be at physiologically relevant concentrations. While N and C terminal fusion of reporter proteins to the tetraspanins enables tracking of cell-specific EVs, these reporter proteins do not allow for affinity isolation of the labeled EVs from biological fluids in that the tetraspanin terminals are located inside EVs12,13.
Tetraspanins, however, can be used to display fluorescent proteins on the EV surface17, and we, therefore, hypothesized that the fusion of EGFP to the C-terminal of a truncated form of the tetraspanin CD9, devoid of the large extracellular domain and the last transmembrane domain, would create fluorescence-labeled EVs with affinity tags. Furthermore, flanking the inverted EV reporter with loxP sites20 would establish a genetic switch that enabled Cre recombinase-dependent EV reporter expression in vivo and allow easy and reliable tracking and isolation of cell-specific EVs. Using this novel transgenic mouse model enables the assimilation of new knowledge regarding EVs' physiological and pathophysiological functions.
Results
Fusion of EGFP to CD9 enable affinity isolation of EVs
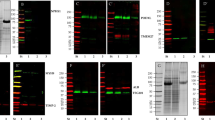
We designed an EV reporter protein by fusion of mouse CD9 truncated after the first 117 amino acids (the third transmembrane domain) to EGFP (Fig. 1A). The predicted molecular weight of the fusion protein is 41.5 kDa. Stably transfected epithelial M1 cells–termed M1-CD9truc-EGFP cells-showed green fluorescent intracellular vesicles co-localized with CD9 (Fig. 1B and C). Transfected fluorescent markers for the endoplasmic reticulum (mCherry-ER), early endosome marker Rab5 (mRFP-Rab5), and the tetraspanin CD81 (mTagBFP2-CD81) showed co-localization with EGFP in CD9truc-EGFP cells (Supplemental Figure S1A). Western blotting revealed EGFP expression restricted to transfected cells (Fig. 1D, the full-length blot is shown in Supplemental Figure S2A). While polyethylene glycol (PEG) precipitated cell-conditioned medium from M1 and M1-CD9truc-EGFP cells revealed enriched EV markers ALIX and flotillin, and not \(\upbeta\)-actin or nuclear Lamin A/C, EGFP was detected only in cell-conditioned medium from M1-CD9truc-EGFP cells (Fig. 1D, the full-length blots are shown in Supplemental Figure S2B-F). We noted that without the addition of protease inhibitors to the cell-conditioned medium, EGFP was present as two bands: one at ~ 37 kDa, slightly below the expected molecular weight of the fusion protein, and one at ~ 27 kDa, indicating partial proteolytic shedding of EGFP (Supplemental Figure S2B). Conditioned medium from M1 and M1-CD9truc-EGFP cells showed similar extracellular particle concentration and size distribution (Fig. 1E). Size-exclusion chromatography (SEC) of the cell-conditioned medium from M1-CD9truc-EGFP cells showed that full-length CD9truc-EGFP band was only present in the fractions 1–3 with a low protein content, while shedded EGFP was present in fractions 10–12 characterized by a higher protein content (Fig. 1F, the full-length blot is shown in Supplemental Figure S2G). This indicates that full-length CD9truc-EGFP is associated with EVs. Consistent with this, EGFP immunoprecipitation from cell-conditioned medium isolated EGFP and EV markers TSG101 and ALIX only from M1-CD9truc-EGFP cells (Fig. 1G, the full-length blot is shown in Supplemental Figure S3A-C). Thus, EVs from CD9truc-EGFP expressing cells can be isolated using GFP immunoprecipitation.
Expression of EGFP and associated EV markers M1-CD9truc-EGFP cells and their conditioned medium. (A) Illustration of our EV reporter gene and protein. The CD9truc-EGFP coding sequence is driven by the CMV promoter and encodes a fusion protein consisting of the first 3 transmembrane domains of CD9 and EGFP, enabling genetic labeling and EV surface display of EGFP. (B) M1 cells stable transfected with CD9truc-EGFP are green fluorescent in contrast to non-tansfected M1 cells. Nuclei (blue) and GFP (green). n = 3, ×200 magnification (C) Immunofluorescence labeling of CD9 in M1-CD9truc-EGFP cells using Anti-hCD9 AB shows colocalization between CD9 (red), EGFP (green) and nuclei (blue). ×200 magnification (n = 3) (D) M1 cells stable transfected with CD9truc-EGFP expressing a band reacting with an anti-GFP antibody in cells and PEG-precipitated conditioned medium. Actin and Lamin A/C were detected in cell lysates only, and EVs markers ALIX and Flotillin were detected in conditioned medium from transfected and non-transfected M1 cells (n = 3). Full-length blots are shown in Supplementary Figure S2A-F. (E) Tunable resistive pulse sensing on the conditioned medium from non-transfected M1 cells and stable transfected CD9truc-EGFP cells indicated that the size distribution of EVs is not affected the expression reporter proteins (n = 3). (F) Size-exclusion chromatography fractions of CD9truc-EGFP cell-conditioned medium showed full-length CD9truc-EGFP in fractions 1–3, while shedded EGFP was present in fractions 10–12 (n = 3). Full-length blots are shown in Supplementary Figure S2G. Below the western blot, the protein concentration in each fraction is shown (G) Anti-GFP immunoprecipitation of cells conditioned medium co-isolates CD9truc-EGFP and EV markers ALIX and TSG101 only in M1 cells stable transfected with CD9truc-EGFP (n = 3). Full-length blots are shown in Supplementary Figure S3A-C.
Cre recombinase-dependent CD9truc-EGFP expression
To obtain cell-specific CD9truc-EGFP expression, we inverted the coding sequence of EV reporter protein CD9truc-EGFP and flanked it by double loxP sites, allowing for CD9truc-EGFP expression driven by a CAG promoter only in Cre recombinase expressing cells (Fig. 2A). HEK293T cells were transiently transfected with CD9truc-EGFP, which only was expressed and yielded green fluorescent cells when co-transfected with Cre recombinase (Fig. 2B). CD9truc-EGFP expression was observed in Cre recombinase co-transfected cells and their corresponding condition medium (Fig. 2C, the full-length blot is shown in Supplemental Figure S3D and E). Next, we used our CD9truc-EGFP plasmid to generate transgenic mice through pro-nuclear injection of the linearized construct. We selected a founder mouse that showed specific and substantial expression of EGFP in Cre-expressing cardiomyocytes when crossed with the tamoxifen-treated αMHC-MerCreMer mouse (Supplemental Figure S1B). After backcrossing onto a C57Bl/6 background, we isolated genomic DNA from the liver of a transgene CD9truc-EGFP mouse. Nanopore sequencing revealed three reads (approximately 30 kb) containing mouse genomic sequence and CD9truc-EGFP insert sequence. The alignment of the sequenced reads was compatible with an integration of the insert in the mouse genome at approximately chromosomal location 4:99.620.973 (GRCm38.p6). The insert was present in two copies immediately adjacent to each other in opposite directions. The insert does not disrupt any known mouse genes at the location where it is embedded. The area of insertion is, however, an annotated constrained conserved region between multiple eutherian mammals. We designed primers for genotyping (Table 1) that produced PCR products of 449 bp in wild-type mice and 324 bp in homozygotes (Fig. 2D, the full-length gel is shown in Supplemental Figure S3F), enabling us to identify transgenic mice and distinguish heterozygotes from homozygotes.
Cre recombinase-dependent expression of CD9truc-EGFP. (A) Our EV reporter CD9truc-EGFP was inverted and flanked by double lox sites. Upon Cre recombinase expression, the CD9truc-EGFP is inverted and yields CD9truc-EGFP expression driven by the CAG promoter. (B) HEK293T cells transiently transfected with double-floxed and inverted CD9truc-EGFP only express EGFP when co-transfected with Cre recombinase (n = 3). ×200 magnification (C) HEK293T cells co-transfected with double-floxed and inverted CD9truc-EGFP expressed CD9truc-EGFP in cells and conditioned medium (n = 3. Full-length blots are shown in Supplementary Figures S3D and E. (D) The double-floxed and inverted CD9truc-EGFP construct are inserted into chromosome 4 of the EVRep mouse, and mice homo- (Tg/Tg) and heterozygous (Tg/WT) can be identified by PCR. The full-length gel is shown in Supplementary Figure S3F.
Tissue-specific EGFP signal in different Cre-positive transgenic mice
We next used established transgenic Cre recombinase mice to enable tissue-specific activation of the EV reporter protein CD9truc-EGFP. Initially, we crossed the transgene CD9truc-EGFP mice with CMV-Cre mice21 expressing Cre recombinase under the control of the ubiquitously active human cytomegalovirus (CMV) promoter. CD9truc-EGFP positive CD9truc-EGFPxCMV-Cre mice showed EGFP expression in kidney, liver, lung spleen, and heart by anti-GFP immunohistochemistry (IHC) (Fig. 3A). EGFP was undetectable in any of these tissues in CD9truc-EGFP negative littermates (Fig. 3A). To show cell-specific expression of our CD9truc-EGFP construct, we crossed our transgene CD9truc-EGFP mice with mice harboring tamoxifen-inducible cardiomyocyte-specific Cre activity (αMHC-MerCreMer)22 and Pax8-Cre mice expressing Cre recombinase kidney tubular epithelium23. In CD9truc-EGFP positive, tamoxifen-treated CD9truc-EGFPxαMHC-MerCreMer mice, EGFP was explicitly detected in the heart by western blotting and direct fluorescence microscopy (Fig. 4A-B, the full-length blot is shown in Supplemental Figure S4A). EGFP was undetectable in kidneys, liver, lung, and spleen in tamoxifen-treated CD9truc-EGFP positive and negative CD9truc-EGFPxαMHC-Cre mice and the hearts of tamoxifen-treated CD9truc-EGFP negative CD9truc-EGFPxαMHC-Cre littermates (Fig. 4A). Likewise, we found EGFP expression specifically in the kidney of CD9truc-EGFP positive CD9truc-EGFPxPax8-Cre mice using western blotting (Fig. 5A, the full-length blot is shown in Supplemental Figure S4B). By fluorescence microscopy, however, green fluorescence was not directly detectable in the kidney epithelium of kidney cryo-sections, and only low EGFP signals were associated with glomeruli (Fig. 5B). The preparation of cryo-sections involved exposure of the tissue to large osmotic gradients, and we tested whether epithelial EGFP expression was present in paraffin-embedded formalin-fixed kidneys. Using an anti-EGFP antibody, we detected kidney-specific EGFP expression in CD9truc-EGFP positive but not negative, CD9truc-EGFPxPax8-Cre mice (Fig. 5C). Thus, these data demonstrate that our transgene CD9truc-EGFP mouse, which we term EVRep, allows for cell-specific expression of CD9truc-EGFP.
Expression of EGFP in tissue from CD9truc-EGFP positive CD9truc-EGFPxCMV-Cre mice. (A) In contrast to the CD9truc-EGFP negative CD9truc-EGFPxCMV-Cre mice, EGFP expression is detected by anti-GFP (brown) immunohistochemical staining of paraffin-embedded kidneys, liver, lung, spleens, and hearts of CD9truc-EGFP positive CD9truc-EGFPxCMV-Cre mice. Scalebar 20 µm. (n = 2).
Expression of EGFP in cardiomyocytes from tamoxifen-treated CD9truc-EGFP positive CD9truc-EGFPxαMHC-MerCreMer mice. (A) Tissue homogenates display CD9truc-EGFP expression only in the heart from tamoxifen-treated CD9truc-EGFP positive CD9truc-EGFPxαMHC-Cre mice. The full-length blot is shown in Supplementary Figure S4A. (B) Tissue sections only show EGFP expression (green) in cardiomyocytes from tamoxifen-treated CD9truc-EGFP positive CD9truc-EGFPxαMHC-Cre. Nuclei (blue). Scalebar 20 µm. (n = 4).
Expression of EGFP in kidney epithelial cells from CD9truc-EGFP positive CD9truc-EGFPxPax8-Cre mice. (A) Tissue homogenates from CD9truc-EGFP positive CD9truc-EGFPxPax8-Cre mice display CD9truc-EGFP expression in kidneys. Full-length blots are shown in Supplementary Figure S4B. (B) Direct fluorescence imaging of kidneys only shows EGFP expression (green) in glomerular cells of CD9truc-EGFP positive CD9truc-EGFPxPax8-Cre mice. Nuclei (blue). Scalebar 20 µm. (n = 4). (C) Immunohistochemical staining of paraffin-embedded kidneys, liver, lung, spleen, and heart from CD9truc-EGFPxPax8-Cre mice, EGFP expression is detected by anti-GFP labeling (brown) in epithelial cells in the kidney from only CD9truc-EGFP positive CD9truc-EGFPxPax8-Cre but not CD9truc-EGFP negative littermates. Scalebar 20 µm. (n = 2).
Isolation of EGFP-positive extracellular vesicles from plasma and urine
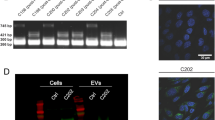
Since EVs have been detected in most body fluids, we next used plasma and urine samples from CD9truc-EGFP positive and negative CD9truc-EGFPxCMV-Cre, tamoxifen-treated CD9truc-EGFPxαMHC-MerCreMer, and CD9truc-EGFPxPax8-Cre mice to determine the cell-specific contribution of EVs to plasma and urine fluid compartments. EVs were precipitated and isolated from plasma using anti-GFP nanobody conjugated beads. CD9truc-EGFP was detected in plasma from CD9truc-EGFP positive CD9truc-EGFPxCMV-Cre and tamoxifen-treated CD9truc-EGFPxαMHC-Cre mice, but not from CD9truc-EGFPxPax8-Cre and CD9truc-EGFP negative littermates (Fig. 6A, the full-length blot is shown in Supplemental Figure S5A). The EV marker ALIX co-precipitated with CD9truc-EGFP only in plasma from tamoxifen-treated CD9truc-EGFP positive CD9truc-EGFPxαMHC-Cre mice (Fig. 6A, the full-length blot is shown in Supplemental Figure S5B). In urine samples, CD9truc-EGFP was observed in CD9truc-EGFP positive CD9truc-EGFPxCMV-Cre, and CD9truc-EGFPxPax8-Cre mice, whereas tamoxifen-treated CD9truc-EGFPxαMHC-Cre urine samples were devoid of detectable CD9truc-EGFP (Fig. 6B, the full-length blot is shown in Supplemental Figure S5C). The EV marker CD81 co-precipitated with CD9truc-EGFP only in urine samples from CD9truc-EGFP positive CD9truc-EGFPxPax8-Cre (Fig. 6B, the full-length blot is shown in Supplemental Figure S5D). Together, these data suggest that the EVRep mouse allows for the isolation of cell-specific EVs from biological fluids.
Isolation of EVs from plasma and urine by anti-GFP precipitation of plasma and urine samples from CD9truc-EGFP transgene mice. (A) CD9truc-EGFP was only isolated from CD9truc-EGFP positive CD9truc-EGFPxCMV-Cre and tamoxifen-treated CD9truc-EGFPxαMHC but not CD9truc-EGFPxPax8-Cre mice or CD9truc-EGFP negative littermates. The EV marker ALIX was co-isolated with CD9truc-EGFP in plasma samples from CD9truc-EGFP positive CD9truc-EGFPxαMHC-Cre only. Full-length blots are shown in Supplementary Figure S5A and B. (B) CD9truc-EGFP was only detected by anti-GFP immunoprecipitation of urine samples from CD9truc-EGFP positive CD9truc-EGFPxCMV-Cre and CD9truc-EGFPxPax8-Cre mice, but not tamoxifen-treated CD9truc-EGFPxαMHC-Cre or CD9truc-EGFP negative littermates. The EV marker CD81 co-precipitated with CD9truc-EGFP only in urine samples from CD9truc-EGFP positive CD9truc-EGFPxPax8-Cre mice. Full-length blots are shown in Supplementary Figures S5C and D.
Discussion
Using a genetically encoded CD9truc-EGFP fusion protein that displays EGFP on EVs combined with a Cre-dependent switch, we have created a transgenic mouse enabling EV detection at the cellular level as well as rapid and selective isolation of cell-specific EVs from plasma and urine. This EVRep mouse model is thus the first genetic EV reporter model that allows easy tracking and isolation of EVs in vivo in mice. Therefore, we believe that this novel tool can be utilized to generate novel insights into EV biology.
Cell-to-cell EV communication has already been demonstrated in several settings. In vitro, the transport of miRNAs in EVs act as a neuron-to-astrocyte communication pathway in the central nervous system24, and stem cell-derived EVs appear to target bone marrow and peripheral sites25. In vivo, intravascular administration of mesenchymal stem cell-derived EVs (MSC-EV) labeled with PKH26 dye closely resembles the positive effect of MSCs on postischemic recovery after acute tubular injury26. Another example includes intravenous injection of endothelial progenitor cell-derived EVs labeled with PKH26 dye isolated from cell-conditioned medium that protects from complement-mediated mesangial injury after experimental anti-Thy1.1 glomerulonephritis27. Furthermore, similar positive effects have been obtained in the myocardium where intravenous administration of EVs isolated from plasma protected against Ischemia–Reperfusion Injury28. Altogether, this suggests that EVs retain regenerative abilities. However, for most EV studies, such injections cause a supra-physiological concentration of EVs from a single cell type, and the naturally occurring in vivo concentrations might not be sufficient to cause similar effects. Furthermore, important paracrine and autocrine effects might be altered or unobserved with this approach and affect the in vivo distribution and the biological function of the EVs. Importantly, injected EVs often accumulate in the liver, spleen, and lungs regardless of the origin26,27,29. We did not observe CD9truc-EGFP accumulation in these organs under normal physiological circumstances. These observations emphasize some of the hurdles with injections of labeled EVs30,31, which may be overcome using the EVRep mouse developed to enable a robust EV fate mapping.
EVs isolated from biological fluid provide access to cell-derived lipids, RNAs, and proteins; however, the representation of EVs from different cell types in, for example, urine and plasma is not well described. Consistent with a previous study32, we found that EVs from cardiomyocytes were readily detectable in plasma samples. Intravascular injection of labeled EVs isolated from serum have been detected in urine33; however, we did not detect cardiomyocyte-derived EVs in urine, indicating that EVs are not freely filtered across the glomerular filtration barrier in the kidney, probably as a result of their size and negative charge34,35,36. On the other hand, we detected kidney epithelial EVs in urine. This is consistent with our previous findings using proteomic database analysis showing that 99.96% of urinary EV-associated proteins are likely to originate from the kidney, the urinary tract epithelium, and the male reproductive tract in humans37. It should be noted that, for example, the CD9truc-EGFPxPax8-Cre mice only showed CD9truc-EGFP in some of the tubular epithelial cells; thus, methods with higher sensitivity than western blotting may be able to detect cardiomyocyte and renal tubular epithelial-derived EVs in plasma and urine. Nonetheless, the distribution of EVs appears to be fluid compartment restricted, and EVs isolated from different fluid compartments, e.g., plasma versus urine, may only represent a subset of cell types in the body.
We found that kidney epithelial-derived EV signal was significantly affected by preparation of frozen kidney cross-sections before fluorescence microscopy, while cardiomyocyte EV tissue abundance was less affected. Western blotting suggested that CD9truc-EGFP was abundantly expressed in CD9truc-EGFP positive CD9truc-EGFPxPax8-Cre mice, but in frozen kidney sections, the EGFP was only slightly visible in the glomerulus of kidneys by directed fluorescence microscopy. The reason for this discrepancy is not known, but renal epithelial cells are, however, highly water permeable38, and we speculate that rapid osmotic changes (e.g., 25% sucrose) imposed during tissue preparation are involved. Recently, it was shown that the handling and preparation of tissue samples significantly affect tissue EV abundance through release to, e.g., washing buffers39. Our observations agree with this and suggest that careful and specific tissue preparation is essential when analyzing tissue EV levels and intercellular communication.
Similar to our approach, three other studies have used genetic labeling of EVs15,18,40 to track the faith of endogenous EVs. While their use of an intravesicular localization of the reporter proteins prevents affinity isolation, our approach shares the limitation that there is a risk that only a subpopulation of EVs is labeled. Notably, the number of identified EV subtypes is continuously growing, and there is no consensus on which protein markers represent the different populations. Thus, we cannot exclude that genetic labeling using CD9 may add a bias in the downstream analysis, as it may only represent EV fractions. Moreover, we used a truncated version of CD9 in which the large extracellular loop was deleted. The large extracellular loop mediates many lateral CD9 interactions, and its deletion might reduce potential adverse effects of its overexpression; however, the CD9truc-EGFP may not show the exact same behavior as wild-type CD9. Nonetheless, the ease at which cell-specific EVs can be isolated using the EVRep mouse will benefit a more comprehensive characterization and identification of EV subtypes and enable other cell-specific EV markers to be identified.
In summary, our novel transgenic EVRep mouse allows easy in vivo tracking and isolation of EVs and can be used to elucidate EV biology and their further use.
Methods
EV track data
We have submitted all relevant data of our experiments to the EV-TRACK knowledgebase (EV-TRACK ID: EV210297)41.
Plasmids
The genes encoding truncated CD9 fused to EGFP, and the doubled-floxed and inverted version were synthesized and cloned into plasmid pcDNA3.1 by BioCat GMBH. The coding sequence is shown in Supplemental Figure S1C. The double-floxed, inverted CD9truc-EGFP from the pcDNA3.1 plasmid was amplified by PCR and ligated into pCAG-Cre (a gift from Connie Cepko; Addgene plasmid # 13775) digested with EcoRI and NotI (New England Biolabs). All constructs were verified by Sanger sequencing at Eurofins Genomics. mRFP-Rab5 was a gift from Ari Helenius (Addgene plasmid # 14437; http://n2t.net/addgene:14437; RRID:Addgene_14437), mCherry-ER (Addgene plasmid # 55041; http://n2t.net/addgene:55041; RRID:Addgene_55041) and mTagBFP2-CD81-10 (Addgene plasmid # 55281; http://n2t.net/addgene:55281; RRID:Addgene_55281) were gifts from Michael Davidson.
Cell cultures
Mouse epithelial M1 (ATCC, Virginia, United States) cells were cultured as previously described42. For analysis of cell-conditioned medium, PC1 serum-free medium (Lonza, No. 344018, Walkersville, MD, USA) was used. The M1 cells with stable CD9truc-EGFP expression were generated by transfection with pcDNA3.1 CD9truc-EGFP using Metafectene Pro (Biontex, München, Germany) and selection with G418 (InvivoGen). HEK293T (ATCC, Virginia, United States) cells were maintained in Dulbecco's Modified Eagle Medium, F-12 Nutrient Mixture (Gibco, Sigma Aldrich, Denmark) containing 10% fetal bovine serum (FBS, Fisher Scientific, DK) and 1% Penicillin–Streptomycin in a 5% CO2 humidified incubator at 37 °C. Harvesting of cells and medium were performed when cells reached 80% confluence in 25 cm2-flasks.
Animal models
We linearized pCAG-DIO-CD9truc-EGFP with HindIII and SalI (New England Biolabs) and gel extracted (New England Biolabs) the 3649-bp fragment containing the transgene. The purified fragment was used to generate transgenic mice with genomic integration of the CAG-promotor driven double-floxed inverted CD9truc-EGFP by pro-nucleus injection43. The αMHC-MerCreMer strain (JAX stock #005650, The Jackson Laboratory, Bar Harbor, ME, USA) expresses inducible Cre in cardiomyocytes upon tamoxifen treatment, and it is maintained by homozygous breeding22. The CMV-Cre (JAX stock #006054), The Jackson Laboratory, Bar Harbor, ME, USA) expresses Cre in all tissues and is maintained by homozygous breeding21. The B6.129P2(Cg)-Pax8tm1.1(cre)Mbu/J (JAX stock #028196), The Jackson Laboratory, Bar Harbor, ME, USA) strain referred to as Pax8-Cre expresses Cre recombinase in known Pax8 expression domains including the developing thyroid gland, inner ear, kidney, and mid-hindbrain region23. The animal experiment was performed in accordance with Danish Law and approved by the Danish Animal Experimentation Council (#2019-15-0201-01644 and 2019-15-0202-00052). The study was performed in accordance with ARRIVE guidelines.
Sample collection
7–9 weeks old mice were transferred individually to metabolic cages (12:12 h light–dark cycle, 28 ± 1 °C) for three days with free access to water and regular rodent diet (LabDiet® 5001, Forth Worth, TX, USA). After 24 h acclimation, urine samples were collected on days 2 and 3. Subsequently, mice were used for perfusion fixation or direct organ harvest, as described below. Urine was stored at − 80 °C and a protease inhibitor (1:1000, P8340, Protease Inhibitor Cocktail, Sigma Aldrich, Denmark) were added when thawed. Mice used for immunohistochemistry and fluorescent microscopy were anesthetized by i.p. injection with 10 mg/kg Xylazine (Rompun, Bayer Healthcare, Shawnee Mission, KS) and 50 mg/kg Ketamine (Ketalar, Pfizer, Sandwich, Kent, UK). Blood samples were taken by cardiac puncture through the apex before mice were flushed with 1 × PBS and fixed with 4% paraformaldehyde by retrograde perfusion via the left ventricle. Subsequently, mice were fixed for an additional 6 h in paraformaldehyde and transferred to 1 × PBS. Mice used for western blotting were likewise anesthetized, blood samples were taken, and relevant organs were removed and snap-frozen in liquid nitrogen.
Immunohistochemistry and fluorescence microscopy
Staining was performed on 2 μm paraffin-embedded tissue samples deparaffined in xylene and rehydrated in decreasing ethanol solutions (99–70%) followed by target retrieval in heated TEG buffer (1 mmol/L Tris, 0.5 mM EGTA, pH 9.0). After cooling, slides were exposed to a 50 mM NH4Cl and 0.3% H2O2 solution for 10 min, washed in 1 × PBS, and incubated in 1 × PBS with 0.3% Triton for 30 min at room temperature. Next, the slides were incubated with primary antibody (Anti-GFP) diluted in 1 × PBS with 0.3% Triton × 100 at 4 °C overnight.
Next, slides were incubated for 30 min at room temperature, washed in 1 × PBS + 0.05% tween five times, and incubated with secondary HRP antibody in PBS + 0.05% tween for 1 h at room temperature. Slides were then washed in PBS, and HRP was visualized by DAB-staining (3,3’-diaminobenzidine). Lastly, slides were stained in hematoxylin mounted with Aquatex (Merck KGaA, Darmstadt, Germany).
For fluorescence microscopy, snap-frozen organs were removed from mice and placed in 1 × PBS with 0.05% acid and 25% sucrose overnight. Next, organs were placed in 1 × PBS with 0.05% acid and 50% sucrose for 2 h, embedded in OCT tissue freezing medium and 5 μm cross-sectioned on a Cryostat (Leica CM3050 S, Leica Biosystems, USA). Cross-sections were placed in 1 × TBS for 10 min, dried, and one drop of Slowfade™ Gold antifade reagent with DAPI (Invitrogen, Thermo Fisher Scientific, Eugene, OR USA) was added before mounting. All tissue and cell samples were obtained using an Olympus BX51 Fluorescence microscope.
Isolation of extracellular vesicles from cells and medium
48 h before, the cell medium was changed to PC1 serum-free medium (Lonza, No. 344018, Walkersville, MD, USA). Cells were lysed in RIPA Lysis buffer for 1 h at 4 °C on an orbital shaker. Next, 25 cm cell scrapers were used to release cells from the flask into the RIPA buffer. The solution was then centrifugated at 13,000g at 4 °C for 10 min, and the supernatant was stored in new tubes at − 80 °C. The medium was transferred to 15 mL tubes and centrifugated for 10 min at 5000 g. The supernatant was then transferred to new tubes and stored at − 80 °C. To isolate EVs, the thawed medium was centrifugated for 15 min at 5000 g at 4 °C. The supernatant was mixed with an equal amount of freshly made ExtraPEG44 (16% PEG-6000, Sigma Aldrich, 1 M NaCl, Milli-Q water) and left in a Multi-Rotator at 4 °C overnight. Next, samples were centrifugated for 15 min at 5000 g at 4 °C, and the supernatant was discarded. The remaining pellet was resuspended in 100 μl × 1 RIPA buffer and stored at − 20 °C until further use.
Immunofluorescence of cells
Cells were seeded on coverslips in multiple 12-wells plates (Biocoat Cell environments, Poly-D-Lysine Cellware) and given 24 h to attach to the coverslips. In the experiments with transfection of fluorescent organelle markers, the cells were transfected the day after seeding on coverslips using Metafectene Pro (Biontex, München, Germany) and grown for 24 h. Next, cells were fixed in 4% paraformaldehyde for 10 min and rinsed 2 times in 1 × PBS with 1 mM MgCl2 and 0.1 mM CaCl2, followed by permeabilization with 0.3% Triton in 1 × PBS for 15 min and another wash in 1 × PBS with 1 mM MgCl2 and 0.1 mM CaCl2. Cells where then incubated in 300 μl primary antibody (Table 2) at 4 °C overnight washed 3 times in 1xPBS and incubated with 300 μl secondary antibody for 1 h at room temperature. Afterward, cells were incubated with 4’,6-diamidino-2-phenylindole (D9542-10MG DAPI, Sigma Aldrich, Denmark) to stain DNA followed by 5 washes in 1 × PBS. Lastly, coverslips containing cells were mounted with fluorescent mounting media (DAKO, Carpinteria, CA, USA) on coverslips. ImageJ, version 2.0.0-RC-43/1.10e) was used to analyze pictures.
Immunoblotting
Samples were mixed with LDS sample buffer (NuPAGE, Invitrogen, Thermo Fischer Scientific, Van Allen Way, Carlsbad, USA) and sample reducing agent (NuPAGE, Invitrogen, Thermo Fischer Scientific, Van Allen Way, Carlsbad, USA) and heated for 10 min before they ran on a gel. For cell and tissue fractions, 10 μg was loaded per well. The protein was then transferred to a membrane activated in 99% Ethanol. Afterward, the membrane was blocked for 30 min in 5% skimmed milk and incubated overnight with primary antibodies at 4 °C (Table 2). Following, membranes were washed 3 times in Tris-Buffered saline with Tween-20 (TBST, 20 mM Tris-base, 137 nM NaCl, 0.05% Tween-20 (Merck), pH 7.6) and incubated with secondary antibody (Table 2) for 1 h at room temperature. Next, membranes were washed 3 times in TBS-T, and proteins were visualized by ECL plus a Molecular Imager (ChemiDoc XRS + , BIO-RAD) with Image Lab software (BIO-RAD).
Immunoprecipitation of EGFP associated EVs
EVs containing EGFP were precipitated from 200 μl plasma and 750 μl urine with extraPEG as described above, except that the pellet was resuspended in 100 μl 1 × PBS to keep EVs intact. EVs immunoprecipitated using ChromoTek GFP-trap Magnetic Beads (Chromotek GmbH, Germany). 25 μl beads were added to a 1.5 ml tube and rinsed with 1000 μl ice-cold 1 × PBS. Next, plasma or urine was added to the equilibrated beads, and tubes were placed in a Multi-Rotator (Grant-bio, PTR-35) at 4 °C for 1 h. Samples were when placed back in the magnet and allowed to attach before the supernatant was discarded. Subsequently, beads were resuspended in 1 mL ice-cold 1 × PBS-T, and the samples were placed back in the magnet. The supernatant was discarded after 3 min when the beads were attached to the magnet, and this step was repeated 4 times.
Tunable resistive pulse sensing
A qNano platform with a Nanopore NP150 (Izon Science, Oxford, UK) and polystyrene calibration beads CPC200 (Izon Science), Oxford, UK) were used for calibration of relative particle size and speed. 35 μl cell media was loaded, and analyses were performed according to manufactory instructions.
Size exclusion chromatography
A qEVoriginal/35 nm SMART column (Izon Science, Oxford, UK) with optimal separation size between 35–350 nm was used to isolate EVs from 0.5 ml from M1-CD9-EGFP medium according to manufacturer's instructions. 0.5 mL fractions were collected, and western blotting was used to verify the presence of EGFP. To verify that proteins mainly were eluted in the last fractions, proteins concentrations were measured in each fraction with DC Protein Assay (Biorad, California, U. S.) according to the manufacturer's guidelines.
Nanopore sequencing
Mouse liver from a CD9truc-EGFP positive EVRep mouse was homogenized, and DNA was extracted using the Nanobind Tissue Big DNA Kit (Circulomics, Baltimore, MD, USA) according to the manufacturer's instructions. The DNA was prepared for Oxford Nanopore sequencing using the Ligation Sequencing Kit (Oxford Nanopore Technologies, Oxford, UK). The prepared library was then sequenced using one flow cell on the Oxford Nanopore PromethION sequencing platform. Base-calling was performed with MinKNOW software, and FASTQ files were aligned to a custom mouse genome (GRCm38.p6) containing the CD9truc-EGFP insert and accessory sequences as an extra chromosome. Alignment was performed using Minimap245. Data were visualized using IGV ver2.8.1 (Broad Institute), Genome Ribbon46, and NCBI BLAST.
References
Yáñez-Mó, M. et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 4, 1–60 (2015).
Akers, J. C. et al. miR-21 in the extracellular vesicles (EVs) of cerebrospinal fluid (CSF): A platform for glioblastoma biomarker development. PLoS ONE 8, e78115 (2013).
Goto, T. et al. An elevated expression of serum exosomal microRNA-191, −21, −451a of pancreatic neoplasm is considered to be efficient diagnostic marker. BMC Cancer 18, 116 (2018).
Pisitkun, T., Shen, R. F. & Knepper, M. A. Identification and proteomic profiling of exosomes in human urine. Proc. Natl. Acad. Sci. U. S. A. 101, 13368–13373 (2004).
Vojtech, L. et al. Exosomes in human semen carry a distinctive repertoire of small non-coding RNAs with potential regulatory functions. Nucleic Acids Res. 42, 7290–7304 (2014).
Zlotogorski-Hurvitz, A. et al. Human saliva-derived exosomes: Comparing methods of isolation. J. Histochem. Cytochem. 63, 181–189 (2015).
Grigor’eva, A. E. et al. Exosomes in tears of healthy individuals: Isolation, identification, and characterization. Biochem. Suppl. Ser. B Biomed. Chem. 10, 165–172 (2016).
Yoon, S. B. & Chang, J. H. Extracellular vesicles in bile: A game changer in the diagnosis of indeterminate biliary stenoses?. HepatoBiliary Surg. Nutr. 6, 408–410 (2017).
Yamamoto, H. Detection of DNA methylation of gastric juice-derived exosomes in gastric cancer. Integr. Mol. Med. 1, 105 (2014).
Van Niel, G., Porto-Carreiro, I., Simoes, S. & Raposo, G. Exosomes: A common pathway for a specialized function. J. Biochem. 140, 13–21 (2006).
Escola, J. M. et al. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J. Biol. Chem. 273, 20121–20127 (1998).
Umeda, R. et al. Structural insights into tetraspanin CD9 function. Nat. Commun. 11, 7 (2020).
Andreu, Z. & Yáñez-Mó, M. Tetraspanins in extracellular vesicle formation and function. Front. Immunol. 5, 442 (2014).
Bobrie, A., Colombo, M., Krumeich, S., Raposp, G. & Clotilde, T. Diverse subpopulations of vesicles secreted by different intracellular mechanisms are present in exosome preparations obtained by differential ultracentrifugation. J. Extracell. vesicles 1, 18397 (2012).
Luo, W. et al. Spatial and temporal tracking of cardiac exosomes in mouse using a nano-luciferase-CD63 fusion protein. Commun. Biol. 3, 1–9 (2020).
Sung, B. H., Ketova, T., Hoshino, D., Zijlstra, A. & Weaver, A. M. Directional cell movement through tissues is controlled by exosome secretion. Nat. Commun. 6, 1–14 (2015).
Sung, B. H. et al. A live cell reporter of exosome secretion and uptake reveals pathfinding behavior of migrating cells. Nat. Commun. 11, 1–15 (2020).
Frederik Verweij, A. J. et al. Live tracking of inter-organ communication by endogenous exosomes in vivo. Dev. Cell 48, 573-589.e4 (2019).
Yoshimura, A. et al. The Sox2 promoter-driven CD63-GFP transgenic rat model allows tracking of neural stem cell-derived extracellular vesicles. Dis. Model. Mech. 11, 028779 (2018).
Schnütgen, F. et al. A directional strategy for monitoring Cre-mediated recombination at the cellular level in the mouse. Nat. Biotechnol. 21, 562–565 (2003).
Schwenk, F., Baron, U. & Rajewsky, K. cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 23, 5080–5081 (1995).
Sohal, D. S. et al. Temporally regulated and tissue-specific gene manipulations in the adult and embryonic heart using a tamoxifen-inducible Cre protein. Circ. Res. 89, 20–25 (2001).
Bouchard, M., Souabni, A., Mandler, M., Neubüser, A. & Busslinger, M. Nephric lineage specification by Pax2 and Pax8. Genes Dev. 16, 2958–2970 (2002).
Morel, L. et al. Neuronal exosomal mirna-dependent translational regulation of astroglial glutamate transporter glt1. J. Biol. Chem. 288, 7105–7116 (2013).
Salvucci, O. et al. MicroRNA126 contributes to granulocyte colony-stimulating factor-induced hematopoietic progenitor cell mobilization by reducing the expression of vascular cell adhesion molecule 1. Haematologica 97, 818–826 (2012).
Bruno, S. et al. Mesenchymal stem cell-derived microvesicles protect against acute tubular injury. J. Am. Soc. Nephrol. 20, 1053–1067 (2009).
Cantaluppi, V. et al. Endothelial progenitor cell-derived extracellular vesicles protect from complement-mediated mesangial injury in experimental anti-Thy1.1 glomerulonephritis. Nephrol. Dial. Transpl. 30, 410–422 (2015).
Vicencio, J. M. et al. Plasma exosomes protect the myocardium from ischemia-reperfusion injury. J. Am. Coll. Cardiol. 65, 1525–1536 (2015).
Choi, H. & Lee, D. S. Illuminating the physiology of extracellular vesicles. Stem Cell Res. Ther. 7, 1 (2016).
Wiklander, O. P. B. et al. Extracellular vesicle in vivo biodistribution is determined by cell source, route of administration and targeting. J. Extracell. Vesicles 4, 1–13 (2015).
Li, P. et al. PKH26 can transfer to host cells in vitro and vivo. Stem Cells Dev. 22, 340–344 (2013).
Sluijter, J. P. G. et al. Extracellular vesicles in diagnostics and therapy of the ischaemic heart: Position Paper from the Working Group on Cellular Biology of the Heart of the European Society of Cardiology. Cardiovasc. Res. 114, 19–34 (2018).
Cheng, Y. et al. A translational study of urine miRNAs in acute myocardial infarction. J. Mol. Cell. Cardiol. 53, 668–676 (2012).
Midekessa, G. et al. Zeta potential of extracellular vesicles: Toward understanding the attributes that determine colloidal stability. ACS Omega 5, 16701–16710 (2020).
Rodewald, R. & Karnovsky, M. J. Porous substructure of the glomerular slit diaphragm in the rat and mouse. J. Cell Biol. 60, 423–433 (1974).
Costanzo, L. S. & Preceded by: Costanzo, L. S. Physiology. (Elsevier, 2015).
Svenningsen, P., Sabaratnam, R. & Jensen, B. L. Urinary extracellular vesicles: Origin, role as intercellular messengers and biomarkers; efficient sorting and potential treatment options. Acta Physiol. 228, e13346 (2020).
Verkman, A. S. Mechanisms and regulation of water permeability in renal epithelia. Am. J. Physiol. Cell Physiol. 257, C837–C850 (1989).
Gupta, M. P. et al. Non-reversible tissue fixation retains extracellular vesicles for in situ imaging. Nat. Methods 16, 1269–1273 (2019).
Zomer, A., Steenbeek, S. C., Maynard, C. & Van Rheenen, J. Studying extracellular vesicle transfer by a Cre-loxP method. Nat. Protoc. 11, 87–101 (2016).
Van Deun, J. et al. EV-TRACK: Transparent reporting and centralizing knowledge in extracellular vesicle research. Nat. Methods 14, 228–232 (2017).
Svenningsen, P., Granhoj, J. & Dimke, H. Proteolytic activation of the epithelial sodium channel: Role of pro-protein convertases and prostasin. FASEB J. 34, 1–1 (2020).
Hansen, L. A. & Füchtbauer, E. M. Genome editing in mice. In Methods in Molecular Biology vol. 1961, 249–254 (Humana Press Inc., 2019).
Rider, M. A., Hurwitz, S. N. & Meckes, D. G. ExtraPEG: A polyethylene glycol-based method for enrichment of extracellular vesicles. Sci. Rep. 6, 1–14 (2016).
Heng, L. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018).
Nattestad, M., Aboukhalil, R., Chin, C.-S. & Schatz, M. C. Ribbon: Intuitive visualization for complex genomic variation. Bioinformatics 37, 413–415 (2021).
Acknowledgements
We thank Lene B. Andersen, Inger Nissen, Charlotte Nielsen, Natasha Stanic, Seham Bouziri, Kristoffer Rosenstrand, and Mohamed A. Ahmed for excellent technical assistance. The work was supported by research grants from A. P. Møller Foundation (#20-L-0112 to MØN), Nyreforeningen (to MØN), Novo Nordisk Foundation (#NNF19OC0058222 to PS), the Carlsberg Foundation (#CF-180404 to PS), and Innovation Fund Denmark (#9122-00058B to PS).
Author information
Authors and Affiliations
Contributions
Study conception and design: M.Ø.N., D.C.A., and P.S. Acquisition of data: M.Ø.N., D.R.H., and M.B.E. Analysis and interpretation of data: M.Ø.N., D.R.H., L.B.S., and P.S. Article draft: M.Ø.N. and P.S. Review for important intellectual content: L.B.S., D.R.H., E.M.F., M.B.E., H.D., D.C.A. All authors have reviewed and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nørgård, M.Ø., Steffensen, L.B., Hansen, D.R. et al. A new transgene mouse model using an extravesicular EGFP tag enables affinity isolation of cell-specific extracellular vesicles. Sci Rep 12, 496 (2022). https://doi.org/10.1038/s41598-021-04512-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-04512-0
This article is cited by
-
Exosomes define a local and systemic communication network in healthy pancreas and pancreatic ductal adenocarcinoma
Nature Communications (2024)
-
Extracellular vesicles in kidney disease
Nature Reviews Nephrology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.