Abstract
Managing postprandial glycemic response, or the increase in blood sugar following a meal, is a crucial component to maintaining healthy blood sugar in patients with diabetes. To test whether oral probiotics can impact postprandial glycemic response, E. coli Nissle 1917 (EcN) was evaluated in an oral glucose tolerance test. Oral gavage of EcN concurrent with a glucose bolus reduced the post-gavage glycemic response in mice. However, there was no difference in glycemic response when comparing EcN to a mutant deficient in glucose metabolism. This suggests that while EcN can alter glycemic response to a glucose bolus, this effect is not mediated by direct uptake of glucose. Of the possible indirect effects EcN could have, gastric emptying rate was highlighted as a likely cause, but EcN had no effect on gastric emptying rate in mice. This leaves many more possible indirect explanations for the interaction between EcN and host glucose metabolism to be explored in future work.
Similar content being viewed by others
Introduction
One of the key aspects of maintaining healthy blood sugar in patients with diabetes is managing the postprandial glycemic response1,2,3. Shortly after a carbohydrate rich meal, free sugars reach the small intestine where they are rapidly absorbed into the intestinal lumen and into the bloodstream4,5. Rising blood sugar levels cause a glucose stimulated insulin secretion (GSIS), stimulating glucose uptake in peripheral tissue6. However, in patients with diabetes, GSIS is impaired, either by reduced function of the insulin producing pancreatic beta islets (as in type I diabetes and MODY genetic diabetes), or by reduced sensitivity of peripheral tissue to insulin signals (as in type II diabetes)7,8. This leads to impaired postprandial glycemic response, causing long periods of elevated blood sugar after meals, and often requiring pharmaceutical intervention to manage9.
The study of the human gut microbiome has uncovered many promising targets for “functional probiotics”' with experimentally verified effects. For example, Akkermansia muciniphila has been targeted as an anti-obesogenic microbe, with supporting data in mice and in clinical trials10,11,12. Bacteroides thetaiotaomicron has received similar attention as a potential probiotic for reducing weight gain in a high fat diet context13,14,15. And recently our group has identified Veillonella atypica as a candidate for an endurance-enhancing microbe in the context of exercise16. Microbes are a potentially powerful tool for impacting health, as they exist naturally in the gut at high abundance, have biological connections to a massive number of diseases and biological processes, and can be isolated, cultured, and formulated for diet supplementation17.
In the context of altering postprandial glycemic response, we hypothesized a clear mechanism by which the microbiome could impact glucose uptake and improve glycemic profiles. Many bacteria have high affinity for glucose, and in the gut microbiome, bacteria could feasibly compete with the host for glucose absorption and degradation. To combat this, the sites of carbohydrate degradation and glucose uptake, the stomach and upper small intestine, are harsh environments with low colonization rates due to high acidity, fast lumen flow rate, antimicrobial compounds, and a loose mucus layer that impairs adhesion18,19. We hypothesized that by boosting the local concentration of glucose-degrading bacteria in the stomach and small intestine, the microbiome could compete with the host for glucose uptake, reducing the amount of glucose that makes it to the bloodstream. To this end, we chose Escherichia coli Nissle 1917 , a probiotic strain that is used commercially as a probiotic and is closely related to model strains of E. coli that are well characterized for their fast growth rates, high affinity for glucose, and relative ease of engineering20.
In this study, we propose that oral supplementation of probiotic bacteria with high affinity for glucose could impact glycemic response to an oral glucose bolus. We show that concurrent administration of probiotic E. coli Nissle 1917 with a glucose bolus causes a reduction in the glycemic response. We then tested whether E. coli glucose consumption is necessary to see this change in glycemic response but found that a glucose-null mutant had no effect when compared to wild type. We then hypothesized that bacterially-induced changes in gastric emptying rate could explain the change in glycemic response, however we observed no change in gastric emptying in E. coli Nissle supplemented mice versus PBS control. This suggests that while E. coli Nissle appears able to modify the glycemic response, the mechanism is complex. Importantly, we demonstrate that the intuitive explanation of simple bacterial absorption of dietary glucose is likely not responsible for this effect.
Results
We first set out to test the hypothesis that the glucose metabolic capacity of live probiotics can impact glucose uptake rate in the gastrointestinal tract (Fig. 1a). 8-week-old C57-BL/6J mice were gavaged with 1010 CFU of E. coli Nissle 1917, concurrently with 2 mg/kg glucose, and blood glucose was monitored over 2 h (Fig. 1b). At each time point measured, Nissle-treated mice showed a trend of lower average blood glucose than vehicle control, though this difference was not significant. However, the area under the blood glucose curve (AUC) was significantly lower in Nissle-treated mice compared to vehicle control (Fig. 1c). We then explored whether other probiotic bacteria could elicit a similar response. However, administration of a cocktail of probiotic Lactobacillus strains showed no impact on average blood glucose at any time point measured nor the average AUC (Supplementary Fig. S1). These data suggest that while probiotic bacteria can impact the glycemic response to a bolus of glucose, this response is species specific, prompting further investigation into what causes E. coli Nissle specifically to have this effect.
E. coli Nissle 1917 improves glycemic response in healthy mice. (a) Experimental design. (b) Oral glucose tolerance test (OGTT) showing blood glucose over time following oral glucose gavage. Points represent mean and bars represent SEM (n = 5). (c) Area under the curve (AUC, in mg/dL ∗ s) of b (n = 5). Points represent individual mice and bars represent mean and SEM. p values calculated in (b) using repeated measures ANOVA with Tukey’s post-hoc test, and in (c) using student’s T-test.
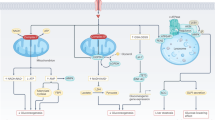
Escherichia coli strains are well characterized for their high rate of glucose uptake. To test whether glucose uptake was necessary for the improvement of glycemic response, we first generated a glucose-null mutant of E. coli Nissle. E. coli metabolizes glucose via central carbon metabolism, primarily through glycolysis and the pentose phosphate pathway (Fig. 2a). Hexose transporters take up glucose and phsophorylate to glucose-6-phosphate, which is incorporated into glycolysis via phosphoglucoisomerase (pgi), or into the pentose phosphate pathway via glucose-6-phosphate dehydrogenase (G6PDH, encoded by gene zwf)21. In E. coli strains lacking both pgi and zwf, glucose-6-phosphate accumulates in the cell, inhibiting hexokinase transporters via feedback inhibition. Using E. coli Nissle 1917 as a base strain, we generated the double knockout EcN ∆pgi∆zwf and confirmed that it was unable to grow in minimal media with glucose as the sole carbon source, while growth on glycerol was unaffected (Supplementary Fig. S2). In an OGTT, mice supplemented with EcN ∆pgi∆zwf showed no significant change in glycemic profile compared to wild type EcN, and the AUC was unchanged as well (Fig. 2b,c). Thus, while EcN appears to affect the glycemic profile in OGTT, this effect is not dependent on EcN possessing functional glucose metabolism, suggesting that this effect is dependent on some other property of the bacteria.
A glucose-null mutant of E. coli Nissle 1917 has no impact on blood glucose AUC compared to wild type. (a) Simplified view of glucose metabolism in E. coli. Hexokinase transports glucose from extracellular space into the cytoplasm and phosphorylates, generating glucose-6-phosphate. Glucose-6P is processed via zwf into the pentose phosphate pathway or via pgi into glycolysis. Knocking out both genes causes buildup of glucose-6P, eliminating further glucose uptake via feedback inhibition. (b) OGTT showing blood glucose over time following oral glucose gavage (n = 14). Points represent mean and bars represent SEM (c) AUC of the OGTT (n = 14). Points represent individual mice and bars represent mean and SEM. p values calculated in (b) using repeated measures ANOVA with Tukey’s post-hoc test, and in c using student’s T-test.
For E. coli Nissle to impact glycemic response without directly interacting with glucose, it would have to have some effect on host physiology, impacting glycemic response indirectly. A prime candidate for this indirect effect is gastric emptying, as microbiota and their small molecule effectors have been shown to impact gastric emptying rate, and gastric emptying rate has a direct effect on glucose absorption rate22. To test whether Nissle impacts gastric emptying rate, we used acetaminophen as a tracer, co-administering a bolus of acetaminophen with either Nissle or PBS as an oral gavage. By monitoring concentration of acetaminophen in the blood, gastric emptying rate can be estimated as the rate of influx of acetaminophen into the bloodstream from the stomach (Fig. 3a). However, we observed no clear effect of Nissle on gastric emptying rate. Inter-mouse variability was high, as was the batch effect between the two trials conducted (Fig. 3b). Aggregated, we observed no difference in average blood concentration of acetaminophen at any time point measured (Fig. 3c). These data suggest that Nissle has no observable impact on gastric emptying rate in mice, and thus gastric emptying cannot explain our initial observation.
Impact of E. coli Nissle on gastric emptying rate. (a) Experimental design. Acetaminophen is co-administered with bacteria or PBS as an oral gavage. Acetaminophen acts as a tracer to monitor rate of gastric emptying and further clearance from the blood. (b,c) Concentration of acetaminophen in the blood over time following oral gavage (n = 9). In (b), points represent individual mice, while in c points represent mean and bars represent SEM. p values calculated in b and c using repeated measures ANOVA with Tukey’s post-hoc test.
Discussion
In this study, we proposed the hypothesis that E. coli Nissle could act as a glucose “sponge” by competing with the host for glucose uptake. Promisingly, we observed that Nissle reduced the glycemic response in mice compared to PBS vehicle control. However, we acknowledged that PBS was not an adequate control to conclude that this effect was caused by direct competition. To test this further, we generated a glucose-null mutant of Nissle (EcN ∆pgi∆zwf), which is unable to grow on or metabolize glucose. However, in a mouse OGTT, EcN ∆pgi∆zwf showed no change in glycemic response compared to wild type, meaning that functional glucose metabolism was not necessary for the glycemic impact we initially observed. Furthermore, this observation meant that in absence of a direct effect, the next best explanation for our initial findings would be an indirect effect: bacterial gavage impacting an aspect of host physiology which alters glycemic response in a separate downstream response. We then tested one likely candidate for this indirect effect via gastric emptying rate, however we found that E. coli Nissle had no effect on gastric emptying.
Eliminating this possibility does little to whittle down the long list of potential bioprocesses a probiotic microbe could be interacting with that alter glycemic response. Microbiome signals have been shown to affect the secretion of several endocrine hormones, including GLP-1 and GIP22 which are both involved in postprandial insulin secretion23. Other affected signals include PYY, which has a more complicated role in glucose homeostasis24, and amylin, which affects blood glucose via delaying gastric emptying25. Microbiome signals have also been shown to interact with the neurological system, via the gut-brain axis26. Neurological connections to glucose sensing and homeostasis represent another possible explanation27. The microbiome has also been shown to interact with metabolism directly via small molecule secretion. Microbiome derived succinate absorbed from the intestinal lumen can induce intestinal gluconeogenesis, which improves glucose homeostasis and reduces glycemic response28. There is even emerging research suggesting that there may be a resident pancreatic microbiome, and oral supplementation of E. coli specifically has been observed entering the pancreatic ducts at high concentrations29. Direct pancreatic stimulation could affect glucose homeostasis via a unique mechanism, making this avenue particularly hazy and unknown. We are left without a clear explanation for our initial finding and a wide expanse of possible avenues towards an explanation which lies outside the scope of this study.
The key finding of this work is the observation that probiotic bacteria given just prior to a glucose bolus can impact the body’s response to the glucose. Our initial assumption was that this effect was caused by direct competition for glucose, as this was the most likely and most exciting possibility. However, our further experimentation showed that competition could not explain the initial observation, demonstrating that the microbiome and microbe-host interactions are much more complicated than they appear.
Methods
Bacterial strains and growth
To generate material for gavage, bacterial strains were grown in LB at 37 °C overnight, then subinoculated into LB at 1:100 dilution and incubated at 37 °C until OD600 0.4–0.8. Cultures were then pelleted at 4000 g, 4 °C, and washed once in 1 × PBS. Pellets were resuspended in PBS to a final concentration of ~ 5 × 1010 CFU/mL and flash frozen in liquid nitrogen. Aliquots were stored at -80 °C. CFU/mL was determined by plating a dilution series of the aliquots onto LB agar plates and counting colonies after 24 h incubation at 37 °C.
To generate the glucose-null mutant, we utilized the lambda red recombineering system encoded on plasmid pREDTKI30. Deletion cassettes were designed for ∆pgi and ∆zwf by PCR amplifying FRT-CatR-FRT (chloramphenicol resistance gene) from pKD3 with 60 bp homology to the upstream and downstream regions of each gene. EcN cultures were made electrocompetent by growing cultures to OD600 0.4–0.6 and washing with 10% glycerol twice and resuspending in 1/100th the original volume. Electrocompetent cultures were transformed with 10 ng plasmid DNA or 100 ng linear DNA at 3 kV 2 mm gap with 1 h recovery in SOC media before plating on LB agar plates. EcN pREDTKI cells were grown in the presence of 0.1% arabinose and electrotransformed with the ∆pgi deletion cassette, then plated on LB Chlor. Activation of the FRT recombinase at 42 °C removed the CatR gene, then the process was repeated for the ∆zwf deletion. Deletions were confirmed by sequencing across the gene and glucose-null growth phenotype confirmed by streaking overnight LB cultures on M9 minimal media with glucose or glycerol as the sole carbon source.
Animal procedures
Protocols for mouse experiments were reviewed and approved by the Joslin Diabetes Center Animal Care and Use Committee (Protocol #2016-05). The study was carried out in accordance with the ARRIVE guidelines. All experiments were performed according the relevant guidelines and recommendations. All mouse experiments were performed using 8–12 week old male C57/B6J mice ordered from Jackson Labs. Male mice were chosen to eliminate sex variation, as well as to keep in line with most comparable studies. To reduce confounding effects, mice were distributed between groups from different litters and cages. No individuals were removed from downstream data analysis post hoc.
Oral glucose tolerance test
For OGTT, standard best practice guidelines were followed31. Mice were individually caged and fasted for 5 h prior to gavage. Roughly 1 h prior to gavage, bacterial aliquots were thawed on ice. Roughly 15 min before gavage, baseline blood glucose was measured. Immediately before gavage, 670µL of 50% w/v glucose was added to 1 mL bacteria maintained at ~ 1010 CFU/mL and gavaged at 10µL per gram body weight. This gives final dosages of 2 g/kg BW glucose and 109 CFU/g BW bacteria. Gavages were timed exactly 1 min apart, then blood glucose was measured via tail vein bleed and a handheld glucose meter at 15, 30, 45, 60, 90, and 120 min post-gavage. Area under the curve was calculated using the trapezoid rule with 0 mg/dL as baseline. For probiotic blend gavage, a commercial probiotic blend was obtained, consisting of 109 CFU per capsule of Bifidobacterium bifidum, Bifidobacterium breve, Bifidobacterium longum, Lb. acidophilus, Lb. casei, Lb. paracasei, Lb. rhamnosus, and Streptococcus thermophilus. (CVS) This probiotic was suspended in PBS to a concentration of 1010 CFU/mL, as measured by CFU plating. Probiotic blend was administered identically to the frozen bacterial aliquots. For all mouse experiments, the administering researcher was blinded to the aliquot contents.
Gastric emptying
Gastric emptying rate was measured as previously described32,33. Briefly, mice were fasted overnight for 20 h. Gavage mixture was prepared by adding equal volumes 2% w/v acetaminophen and either PBS or bacteria maintained at 1010 CFU/mL, thawed on ice as above. Baseline blood samples were collected via tail vein bleed, and mice were gavaged 250µL gavage mixture, giving final dosage of 2.5 mg acetaminophen and ~ 109 CFU bacteria per animal. Blood samples were collected at 15, 30, and 60 min into heparinized tubes and stored on ice. Blood samples were centrifuged at 4 °C for 20 min at 2000×g. Plasma was collected and stored in separate tubes at − 20 °C. Acetaminophen was quantified in the plasma using an enzymatic colorimetric assay specific to acetaminophen (Cambridge Life Sciences).
Statistics
Data were analyzed using repeated measures ANOVA with Tukey’s post-hoc test in Prism 11.0. Significance threshold set at p = 0.05.
Data were analyzed using student’s T test in Excel. Specifically, two-sample unequal variance two-tailed T-test was used. Significance threshold set at p = 0.05.
References
American Diabetes Association 2001 Postprandial blood glucose Clin. Diabetes 19 127 130
A Ceriello 2005 Postprandial hyperglycemia and diabetes complications: Is it time to treat? Diabetes 54 1 7
B Gallwitz 2009 Implications of postprandial glucose and weight control in people with type 2 diabetes: Understanding and implementing the International Diabetes Federation guidelines Diabetes Care 32 Suppl 2 S322 S325
RP Ferraris S Yasharpour KC Lloyd R Mirzayan JM Diamond 1990 Luminal glucose concentrations in the gut under normal conditions Am. J. Physiol. 259 G822 G837
NW Read 1980 Transit of a meal through the stomach, small intestine, and colon in normal subjects and its role in the pathogenesis of diarrhea Gastroenterology 79 1276 1282
M Komatsu M Takei H Ishii Y Sato 2013 Glucose-stimulated insulin secretion: A newer perspective J. Diabetes Investig. 4 511 516
G Thanabalasingham KR Owen 2011 Diagnosis and management of maturity onset diabetes of the young (MODY) BMJ 343 d6044
NG Forouhi NJ Wareham 2010 Epidemiology of diabetes Medicine 38 602 606
American Diabetes Association 2003 Standards of medical care for patients with diabetes mellitus Diabetes Care 26 Suppl 1 S33 50
A Hänninen 2018 Akkermansia muciniphila induces gut microbiota remodelling and controls islet autoimmunity in NOD mice Gut 67 1445 1453
A Everard 2013 Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity Proc. Natl. Acad. Sci. U. S. A. 110 9066 9071
C Depommier 2019 Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: A proof-of-concept exploratory study Nat. Med. https://doi.org/10.1038/s41591-019-0495-2
L Wrzosek 2013 Bacteroides thetaiotaomicron and Faecalibacterium prausnitzii influence the production of mucus glycans and the development of goblet cells in the colonic epithelium of a gnotobiotic model rodent BMC Biol. 11 61
A Modasia 2020 Regulation of enteroendocrine cell networks by the major human gut symbiont bacteroides thetaiotaomicron Front. Microbiol. 11 575595
R Hansen 2020 A double-blind, placebo-controlled trial to assess safety and tolerability of (Thetanix) bacteroides thetaiotaomicron in adolescent Crohn’s disease Clin. Transl. Gastroenterol. 12 e00287
J Scheiman 2019 Meta-omics analysis of elite athletes identifies a performance-enhancing microbe that functions via lactate metabolism Nat. Med. 25 1104 1109
JA Gilbert 2018 Current understanding of the human microbiome Nat. Med. 24 392
GP Donaldson SM Lee SK Mazmanian 2016 Gut biogeography of the bacterial microbiota Nat. Rev. Microbiol. 14 20 32
AJ Kastl Jr NA Terry GD Wu LG Albenberg 2020 The structure and function of the human small intestinal microbiota: Current understanding and future directions Cell Mol. Gastroenterol. Hepatol. 9 33 45
Samie, A. Escherichia coli: Recent Advances on Physiology, Pathogenesis and Biotechnological Applications. (BoD—Books on Demand, 2017).
IM Brockman KLJ Prather 2015 Dynamic knockdown of E. coli central metabolism for redirecting fluxes of primary metabolites Metab. Eng. 28 104 113
HV Lin 2012 Butyrate and propionate protect against diet-induced obesity and regulate gut hormones via free fatty acid receptor 3-independent mechanisms PLoS ONE 7 e35240
W Kim JM Egan 2008 The role of incretins in glucose homeostasis and diabetes treatment Pharmacol. Rev. 60 470 512
D Boey A Sainsbury H Herzog 2007 The role of peptide YY in regulating glucose homeostasis Peptides 28 390 395
WA Scherbaum 1998 The role of amylin in the physiology of glycemic control Exp. Clin. Endocrinol. Diabetes 106 97 102
AC Heijboer 2006 Gut-brain axis: Regulation of glucose metabolism J. Neuroendocrinol. 18 883 894
BE Levin AA Dunn-Meynell VH Routh 1999 Brain glucose sensing and body energy homeostasis: Role in obesity and diabetes Am. J. Physiol. 276 R1223 R1231
F Vadder De 2016 Microbiota-produced succinate improves glucose homeostasis via intestinal gluconeogenesis Cell Metab. 24 151 157
S Pushalkar 2018 The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression Cancer Discov. 8 403 416
J Yang 2014 High-efficiency scarless genetic modification in escherichia coli by using lambda red recombination and I-SceI cleavage Appl. Environ. Microbiol. 80 3826 3834
JE Ayala 2010 Standard operating procedures for describing and performing metabolic tests of glucose homeostasis in mice Dis. Model. Mech. 3 525 534
BJ Lamont 2012 Pancreatic GLP-1 receptor activation is sufficient for incretin control of glucose metabolism in mice J. Clin. Invest. 122 388 402
A Maida JA Lovshin LL Baggio DJ Drucker 2008 The glucagon-like peptide-1 receptor agonist oxyntomodulin enhances beta-cell function but does not inhibit gastric emptying in mice Endocrinology 149 5670 5678
Acknowledgements
We acknowledge Mary Elizabeth Patti and Kristala J. Prather for assistance with experimental design, and Braden T. Tierney and Marsha Wibowo for valuable discussion.
Funding
This work was funded by a Smith Family Foundation Award for Excellence in Biomedical Research (A.D.K.); an American Diabetes Association (ADA) Pathway to Stop Diabetes Initiator Award (A.D.K.), National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases (NIH/NIDDK) Diabetes Research Center (DRC) Grant P30DK036836-30 (T.A.C, L.D.P, A.D.K; P.I. King, George L.); and NIH/NIDDK Grant T32 DK007260 (T.A.C.; PI: Blackwell, Thomas K.).
Author information
Authors and Affiliations
Contributions
T.A.C. and A.D.K. conceived the project. T.A.C. generated bacterial samples and mutants. T.A.C. and L.D.P. performed glucose tolerance tests. T.A.C. wrote the manuscript and generated all images and model figures using PowerPoint. A.D.K. supervised the project. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chavkin, T.A., Pham, LD. & Kostic, A. E. coli Nissle 1917 modulates host glucose metabolism without directly acting on glucose. Sci Rep 11, 23230 (2021). https://doi.org/10.1038/s41598-021-02431-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-02431-8
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.