Abstract
Wheat, one of the major cereal crops worldwide, get adversely affected by rising global temperature. We have identified the diploid B genome progenitor of wheat, Aegilops speltoides (SS), as a potential donor for heat stress tolerance. Therefore, the present work was planned to study the total transcriptome profile of heat stress-tolerant Ae. speltoides accession pau3809 (AS3809) and compare with that of tetraploid and hexaploid wheat cultivars PDW274 and PBW725, respectively. The comparative transcriptome was utilized to identify and validate heat stress transcription factors (HSFs), the key genes involved in imparting heat stress tolerance. Transcriptome analysis led to the identification of a total of 74 K, 68 K, and 76 K genes in AS3809, PDW274, and PBW725, respectively. There was a high uniformity of GO profiles under the biological, molecular, and cellular functions across the three wheat transcriptomes, suggesting the conservation of gene function. Twelve HSFs having the highest FPKM value were identified in the AS3809 transcriptome data, while six of these HSFs namely HSFA3, HSFA5, HSFA9, HSFB2a, HSFB2b, and HSFC1b, were validated with qRT PCR. These six HSFs were identified as an important component of thermotolerance in AS3809 as evident from their comparative higher expression under heat stress.
Similar content being viewed by others
Introduction
Wheat is the world's second most important source of nutritional energy and nutrients, after rice. Despite being the world's second-largest wheat producer, yields have been decreasing for several years, notably in India’s central regions1. Climate change on a global scale, particularly the current anomalous temperature regimes, poses a serious threat to the agricultural environment. Given future climatic uncertainty and resource limits, providing food and nutritional security for an expanding population will be a huge challenge2.
With an increase in ambient temperature since the turn of the century, global climate models project a rise in mean ambient temperatures ranging from 1.8 to 5.8 °C by the end of the century3. Heat stress has the most adverse effect on productivity as wheat flowers at cool temperatures between 0 and 5 °C. Although heat stress affects wheat growth to varying degrees at different phenological stages, its effects are more prominent on reproductive development (also called terminal heat stress) than on vegetative growth as an increase in temperature during grain filling leads to a reduction in grain filling duration, which directly impacts grain number and yield4. Previous studies5 have estimated grain filling duration and grain yield to decrease by 5% and 3–4%, respectively, with a per degree increase in temperature between 18 and 22 °C during grain filling. Based on this calculation, losses up to 50% in yield potential on exposure to 32–38 °C during the critical grain formation period have been estimated6.
Wheat belongs to the family Poaceae, tribe Triticeae and is placed in the genus Triticum. Triticum comprises diverse types of cultivated and wild species of wheat. The genome size of bread wheat (Triticum aestivum L.), a cultivated allohexaploid species is about 16,000 Mb and contains three different genomes (AABBDD), while durum is a cultivated tetraploid species (AABB) of approximately 12,000 Mb. Origin of wheat passed through a complex pathway involving the crossing of three different genomes followed by their localization in a single nucleus. These genomes though different from each other but are related in their evolutionary pathways. The B genome of the bread wheat and durum wheat has been derived from a closely related diploid species of wild wheat Aegilops speltoides (SS), but this probably happened only once or a few times and involved only a few progenitors. Consequently, the potential genetic diversity of Ae. speltoides have not been represented well in bread and durum wheat germplasm7.
The importance of wild species, particularly, Aegilops species in breeding programs is documented by various studies where they have a role in improvement for tolerance to drought stress8, heat stress9,10 and salinity stress11, resistance to several pests and diseases such as rusts12,13,14 and also have contribution towards traits as complex as yield7. Therefore, besides being a progenitor of bread wheat, Ae. speltoides has established itself as a valuable genetic resource for wheat improvement. Also, the ability of this wild progenitor to adapt to different climatic zones including drought/heat environments, different disease hot spots, and nutrient-poor areas has enhanced its value as a breeding resource.
Heat stress transcription factors (HSFs) regulate core aspects of the heat stress response15 by turning on almost all "heat shock genes" (HSGs) thus, protecting against heat stress. Several heat-inducible genes known as HSGs are up-regulated in response to heat stress and encode heat shock proteins (HSPs) which protect intracellular proteins from denaturation16. The presence of conserved heat shock elements (HSEs) in the promoter region of HSGs initiates transcription in response to heat. Plant HSFs are a complex gene family having an important role in the modulation of transcription during heat stress17. Based on details of their oligomerization domains, plant HSFs have been distinguished into three classes, class A, class B, and class C HSFs15. Only A-type HSFs show transactivation property due to the presence of a C-terminal AHA type activation domain, while B-type HSFs have been proposed as either co-activators or repressors. Multiple copies of HSFs have been reported in plants: 21 in Arabidopsis, 24 in tomato, 25 in pepper, and 56 in wheat18,19,20,21.
Previous studies have highlighted the importance of HSFs in wheat under various abiotic stress conditions, particularly heat stress21,22,23. A seed preferential HSF; TaHSFA2d now annotated as TaHSFA6b was cloned for the first time in wheat23 and its overexpression in Arabidopsis was reported to increase thermotolerance. The induction of HSFA2, HSFA6 in response to heat stress in wheat21, and high expression of TaHSFC2a gene during grain filling and transient induction in the leaves by heat stress have also been reported24. Overexpression of TaHSFC2a-B in transgenic wheat is reported to cause upregulation of thermo-protectant genes-TaHSP70d and TaGalSyn.
The diploid Ae. speltoides (BB), known to be closely related to the species which contributed the B genome to tetraploid and hexaploid wheat. At Punjab Agricultural University (PAU), India, we have a collection of about 160 accessions of Ae. speltoides and preliminary investigations led to the identification of this species as a source of heat stress tolerance as evident by its stay-green trait, normal pollen fertility, and normal seed setting at temperatures as high as 39 °C6. We also developed a number of lines carrying introgressions from different accessions of Ae. speltoides to the durum and hexaploid wheat background under our wheat-wide hybridization program. The present study was designed with an aim to understand the variations in the HSF genes offered by the heat-tolerant Ae. speltoides accession pau 3809 (AS3809) over the two cultivated but heat susceptible cultivars, PBW725 (hexaploid, bread wheat) and PDW274 (tetraploid, durum wheat). This analysis provides fundamental insights into HSFs and their variants available in AS3809 which would be utilized in the wheat improvement programs for heat stress tolerance.
Results
Transcriptomic data filtering and de novo assembly
A comprehensive overview of the transcriptome in three different wheat ploidies yielded 16, 21, and 18 million paired-end reads for AS3809, PBW725, and PDW274, respectively. A summary of statistics of de novo assemblies is given in Table 1. After removing adapter sequences, ambiguous and low-quality reads, the high-quality clean reads (Phred score ≥ 30) led to the assembly of AS3809 with 113,157 trinity transcripts corresponding to 74,946 genes. 121,466 and 109,827 trinity transcripts from PBW725 and PDW274, respectively corresponding to 76,404 and 68,873 genes. N50 values of 1409, 1398, and 1004 bp for AS3809, PBW725, and PDW274 respectively, and average GC content ranging from 47.57 to 48.22%, indicated the good quality of all the assemblies, suitable for further annotation (Table 1).
Assessment of transcriptome assembly completeness
BUSCO analysis revealed that the majority of the Liliopsida core genes were successfully recovered in the AS3809 assembly (Table 2). Specifically, of the 3278 single-copy orthologs searched, 74.3% of core genes were completely recovered in AS3809 as compared to 65.4% and 56.7% in PBW725 and PDW274, respectively. Around 12.1%, 14%, and 16% of genes in AS3809, PBW725, and PDW274, respectively were partially recovered in fragmented form. Only 13.6% of genes were missing from AS3809 assembly as compared to 16.1% and 19.4% missing rate for hexaploid and tetraploid wheat. Given the high quality of the dataset, recovery for both ‘complete-single copy’ and ‘complete-duplicated’, BUSCOs was considerably higher for AS3809 assembly as compared to PBW725 and PDW274. Therefore, the AS3809 transcriptome was used as the standard for extracting transcripts related to heat stress tolerance.
Annotation and Gene Ontology analysis
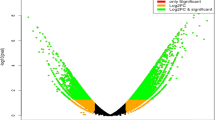
Blast2GO annotation based on the BLASTX results led to functional characterization of 55,838, 50,077, and 49,872 transcripts in AS3809, PBW725, and PDW274, respectively. All the assemblies exhibited a diverse range of GO, suggesting that biological process, molecular function, and cellular component were all well represented (Fig. 1). These three main categories were further divided into 53 GO functional subcategories uniformly across the three transcriptomes. In all the assemblies, genes encoding cellular and metabolic processes along with the response to stimuli were significantly represented in the biological process category while under the molecular function category, the percentage of genes coding for binding and catalytic activities was higher. Within the cellular components, proteins related to cell and cellular parts were expressing more, followed by the cellular membrane and organelles.
Functional terms were also assigned to the trinity transcripts from each wheat assembly using the MapMan Mercator. MapMan employs a basic hierarchical tree structure of terms called "bins" to represent biological contexts and concepts. MapMan annotation resulted in the classification of the transcripts into different bins, each bin representing a different group as photosynthesis, carbohydrate metabolism, glycolysis, gluconeogenesis, oxidative phosphorylation, TCA, cell wall synthesis, lipid and amino acid metabolism, biotic and abiotic stress response, etc. The abiotic stress bin revealed that the majority of the active transcripts were from redox state, respiratory, cell wall, MAPK, defense genes, PR proteins, heat shock proteins, heat shock transcription factors, auxin, hormone signaling, etc. in all three assemblies. The PfamScan categorized transcripts into domains, families, and repeats with all maximum in AS3809 and minimum in PDW274. The annotation statistics obtained for the three assemblies are shown in Table 3.
Identification of orthologous genes
Ortholog transcript detection, based on the OrthoMCL program, demonstrated considerable overlap in transcripts sequences across all three assemblies. Over 39% of the transcripts were identified as putative orthologs between AS3809 and PBW725 assemblies while PDW274 and AS3809 assemblies shared around 37% of the transcripts as putative orthologs. The annotation results from BLAST2GO, MapMan, and PfamScan of the ortholog transcripts were similar suggesting the authenticity of identified ortholog.
Identification of putative heat shock transcription factor (HSF) genes
The three different classes of HSF i.e., A, B, and C were annotated/identified from the transcripts. In total, 37 HSF transcripts were retrieved from AS3809 assembly along with their orthologs from PDW274 and PBW725. Of these, 12 HSFs viz., HSFA1b, HSFA2b, HSFA4b, HSFA3, HSFA5, HSFA6b, HSFA9, HSFB1, HSFB3, HSFB2a, HSFB2b and HSFC1b showing maximum FPKM values were chosen for further analysis. All these 12 HSFs had higher expression in AS3809, followed by PDW274 and the least expression value in PBW725 (Fig. 2). The selected HSF genes have high sequence similarity (95–98%) among three wheat species and possessed HSF DNA binding domain as evident from Pfam analysis. Gene Ontology from Blast2GO and PfamScan annotation of these HSF transcripts are given in Table 4.
In silico expression analysis of HSF genes (tissue-specific and under abiotic stress)
BLASTn search of selected 12 HSF transcripts with the wheatEXP database suggested expression levels of all the HSF genes to be significantly higher in leaf (Z71) tissues. At the basal level, the expression of HSFA6b was found to be the highest, followed by HSFB2b and HSFC1b (Fig. 3). Similarly, at 1 h of heat stress, the expression of HSFA6b was the highest, followed by HSFB3, HSFB1, and HSFB2b, indicating these genes play regulatory functions and be induced in plants within 1 h of heat stress. At 6 h of heat stress, the expression of HSFB2b was found the highest amongst all 12 HSFs. But in the expression database, the expression values recorded under prolonged heat stress (6 h) were less than those observed under 1 h of heat stress.
Protein–protein interaction (PPI) network analysis of the HSF proteins
Protein–protein interaction network data was represented in the form of two distinct variables; node, representing a protein, and an edge, representing the interaction between two proteins. A highly connected network of selected HSFs was observed with 10 nodes representing 10 selected HSFs and 30 interactions in form of edges. This PPI network revealed that there is a complex network of interaction between different HSFs as HSFC1b and HSFA9 interact only with HSFA3 while HSFA1b and HSF4b has two interactions each. HSFA5 and HSFB1were more interactive with five interactions each. But almost all the HSF proteins interactions end to HSFB2a (7 edges) and HSFB2b (6 edges). This reflected that HSFB2a and HSFB2b could be the primary regulators of the heat shock response and their response might have been modulated by other HSFs selected in the present study. HSFA6b on other hand seems to be a mediator, interacting with the other proteins to modulate their functions, rather than directly affecting the heat shock response (Fig. 4).

source and points of arrows show the sources and targets of HSF protein in the network. More arrows at a node mean more number of HSF proteins are interacting with that particular HSF.
Protein–protein interaction (PPI) network analysis exhibiting interaction of HSF proteins with one another. In the network, the nodes correspond to the proteins and the edges represent the interactions. Arrows represent the interaction between one HSF protein with another by degree value. The
Phylogenetic relationship among HSF gene family
To find the evolutionary relationship and potential roles of wheat HSF genes based on known functions of barley and rice HSFs25, an unrooted neighbor-joining (NJ) phylogenetic tree was constructed using Mega 7.0 (Fig. 5). As expected, 12 wheat HSF proteins identified in the present study were clustered closely with respective HSFs from rice and barley, and clearly separated into two main groups. The first group contained the HSF proteins from the B class and was further clustered into two main sub-groups named Ia (comprising of HSFB2b, HSFB3, and HSFB2a) and Ib (comprising of HSFB1). AS3809-HSFB2b was found to be close to Os-HSFB2b while highly diverged from PBW725-HSFB2b. For HSFB2a and HSFB1 proteins, wheat proteins were observed to have close relatedness with each other but were distinct from the barley and rice proteins.
However, the second cluster contained the A and C classes of HSF proteins and was subdivided into two groups, named group IIa and IIb. The IIa cluster contained HSFC1b and HSFA3 proteins and in both these clusters, rice and barley proteins were observed to have less similarity with the wheat proteins. HSFA3 clustered separately from the rest of the class A present in cluster IIb and showed close relatedness with the HSFC1b gene. On the other hand, cluster IIb represented a group of two sub-clusters, having HSFA5 and HSFA4b (ClusterIIb.1), while the second sub-cluster (IIb.2) contained the rest of the four HSF protein categories (HSFA6b, HSFA2b, HSFA9, and HSFA1b).
Validation of the expression of HSF genes
Of the 12 selected HSFs, eight have been validated through qRT-PCR, as four (HSFA1b, HSFA2b, HSFA6b, and HSFB3) failed to amplify in two or more selected wheat species (Fig. 6). After 1 h of heat stress on seedlings, HSFA3, HSFA5, HSFA9, HSFB2a, and HSFC1b exhibited the highest expression in the wild wheat species AS3809 while two HSF genes, HSFA4b and HSFB1 exhibited high expression in cultivated durum PDW274, and hexaploid wheat PBW725 respectively. HSFB2b though amplified in control but did not show expression after heat stress.
Expression analysis of HSF genes in response to control and high-temperature stress in three species of wheat (AS3809, PBW725, and PDW274). Change in transcript abundance of HSF genes in 10 days old seedlings of wheat species of different ploidy levels subjected to heat stress at 35 °C for 1 h. (a) HSFA3 (b) HSFA4b (c) HSFA5 (d) HSFA9 (e) HSFB1 (f) HSFB2a (g) HSFB2b (h) HSFC1b. The expression level at control (AS3809) i.e., 0 h of treatment was normalized as 1.0. The results shown are the means ± SE of three independent experiments.
Discussion
Heat stress is one of the most serious constraints that limit wheat productivity. The mechanism of heat stress tolerance is a complicated phenomenon governed by numerous genes which cause a variety of physiological and biochemical changes. Wild species always fascinate scientists for having a repertoire of exotic genes/alleles which have almost vanished in cultivated wheat due to domestication and breeding8,10,12,13,14. Various studies have highlighted the importance of Ae. speltoides, Ae. tauschii and Ae. geniculata being a valuable genetic resource for enhancing thermotolerance9,26,27. Previous studies6,28 reported Ae. speltoides to be highly heat-tolerant species, which can be utilized to enhance the thermotolerance of wheat27. Therefore, our study was designed to identify the unique transcript from the selected accession AS3809 by comparing it with the cultivated tetraploid and hexaploid wheat and study the expression profiling of selected HSFs among three ploidies.
Transcriptome data analysis of wild and cultivated wheat
De novo transcriptome assembly was performed for all three transcriptomes using Trinity, to avoid any bias arising due to the non-availability of reference genomes of Ae. speltoides and T. durum. Regardless of the huge difference in the genome size (4.9–7 GB)29,30,31 and the number of genes (50,000–100,000)30,32 among the wild diploid, cultivated tetraploid and hexaploid wheat, the total number of expressed transcripts obtained were 113.1 K in AS3809 which is just marginally less than hexaploid wheat PBW725 having 121.4 K and tetraploid PDW274 with 109.8 K transcripts. The disproportionate trends between genome size and gene expression have been reported in polyploid species owing to the momentous number of repetitive sequences in the non-expressing heterochromatin region of higher ploidies as compared to diploid relatives. Furthermore, loss of some genes/functional alleles during artificial selection and domestication seems to play a direct role in the observed trend33,34.
The N50 values obtained for the three transcriptome assemblies were found to be appropriate for downstream analysis35,36. BUSCO analysis-based completeness assessment of the three transcriptomes revealed the highest recovery of conserved single-copy orthologs in the diploid wild followed by hexaploid and tetraploid wheat, indicating good coverage and high quality of AS3809, relative to higher ploidies, demonstrating loss of genes owing to the domestication process as well as polyploidy events37. Recovery reported for the assemblies was enough for identification of single-copy orthologs38 thus the current assemblies of the cultivated as well as wild species of wheat will supplement the published resources for wheat. BLAST2GO and MapMan based comprehensive annotation revealed that GO representations for all three categories of molecular function, biological processes, and cellular component were successfully captured representing GO profiles with the highest proportions of mapped GO terms39.
Identification and validation of HSFs
AS3809 shared 39% and 37% putative orthologs with PBW725 and PDW274 respectively. The transcriptome data have been analyzed for differential expression of putative HSF gene candidates for heat stress tolerance. HSFs play an imperative role in acquired thermotolerance as they are the terminal components of the signal transduction chain mediating the activation of genes responsive to heat stress16. The 12 important HSF genes under study had the highest FPKM in wild wheat indicating higher expression in AS3809 as compared to cultivated durum and hexaploid wheat. These HSFs are primarily involved in stimulating the rapid synthesis and accumulation of heat shock proteins (HSPs), which not only act as molecular chaperones shielding proteins from thermal aggregation but are also involved in various aspects of proteins homeostasis, such as protein translocation and degradation40.
The selected HSFs from the transcriptome data were validated with in-silico expression analysis data from the WheatExp database and with real-time qRT-PCR. Of the 12 important HSF genes with highest expression in AS3809, eight viz., HSFA3, HSFA4b, HSFA5, HSFA9, HSFB1, HSFB2a, HSFB2b and HSFC1b were validated by qRT-PCR, (Fig. 6). Of these 6 genes HSFA3, HSFA5, HSFA9, HSFB2a, HSFB2b, and HSFC1b showed comparative highest expression in the wild AS3809 as proven in comparative transcriptome data. But two important HSFs, HSFA6b and HSFB3 having a very good expression in response to heat stress in the WheatExp database, could not be validated by qRT-PCR. In the WheatEXP database, HSFA6b had higher expression at basal level and after 1 h of heat stress, HSFB1 and HSFB3 after 1 h of heat stress, and HSFC1b having higher expression at a basal level only. All HSF genes in the WheatEXP database revealed that the expression values recorded under prolonged heat stress of 6 h were less than those observed under 1 h of heat stress suggesting the HSF gene family be quickly induced within 1 h of heat stress, activating downstream pathways for thermotolerance as compared to 6 h of heat stress, thus allowing a quick and elevated response system of plants to combat heat stress. HSFC1b and HSFB2b might have a key regulatory role in thermotolerance in AS3809. Higher expression of HSFC1b in transcriptome data and qRT-PCR and the WheatExp database results indicating this to be important HSF in AS3809. HSFB2b could be another important candidate though it expressed only under control conditions in AS3809 and with a decreased expression upon 1 h of heat stress. The early heat-responsive nature of HSFA6b and HSFB2b was observed to be upregulated within 10 min of heat stress onset in barley and HSFA6b also showed considerable tolerance to salinity and drought stresses23,41,42. A decrease in expression of TaHSFB2b under 1.5 h of heat stress21 as compared to control conditions suggests that this gene performs well under non-stress conditions but its transcript level decreases after heat stress. FaHSFC1b gene from Festuca arundinacea in Arabidopsis thaliana plays a positive role in heat stress by upregulating heat-protective genes43. Upregulation of HSFC1 genes in rice (a diploid species) by heat and oxidative stresses or a combination of these stresses44 corroborating with our findings from this experiment.
HSFA3, HSFA5, HSFA9, HSFB2a genes with higher expression in qRT-PCR results were also an important part of the thermotolerance profile of AS3809 though we could not find their expression in the WheatExp database. The data in the WheatExp database is from hexaploid wheat cultivar Chinese spring and TAM107 and the candidate HSFs specific to the S genome of Ae. speltoides could not be represented. Overexpression of homologous DREB2A from Zea mays in Arabidopsis thaliana led to the induction of HS-related genes, including HSFA345. Downregulation of HSFA5 in hexaploid and tetraploid wheat under heat stress has also been reported21. A9 of class A HSFs and subclass B3, B5 of class B HSFs is absent in monocot species like wheat and rice20. Contrastingly in the current investigation, HSFA9 transcript was found in all three assemblies and is also validated by real-time PCR reaction. The results revealed the highest upregulation of this gene in AS3809 followed by PBW725 with a marginal difference. However, the role of HSFA9 in regulating HSP expression during seed development has been demonstrated in Arabidopsis and sunflower46 though no report of seedling expression has been known. The overexpression of wheat TaHSFB2a in transgenic Arabidopsis leads to enhanced thermal and freeze tolerance47.
Two of the candidate HSFs, HSFB1, and HSFA4b exhibited the highest expression under heat stress in cultivated wheat, PDW274, and PBW725, respectively indicating the species-specific expression. Subclass B1 has been reported to be highly heat-inducible and its overexpression activated the expression promoter-driven reporter genes under optimal conditions21.
As per the protein–protein interaction network of wheat HSFs, HSFA6b appears to be the principal source of HSF that interacts with all other HSFs while HSFB2a is the ultimate target, and the rest are intermediates in the network. Thus, HSFA6 is hypothesized to be the primary HSF/early heat-responsive transcription factor that turns on the cascade of interactions with other HSF proteins under heat stress. Our theory is supported by a prior report21 that demonstrated HSFA6 members to play a key regulatory role in wheat by becoming the dominant HSFs during heat stress. It was observed that HSFA3 interacts with HSFA1b. Heteromeric interactions among HSFA1, HSFA2, and HSFA3 factors have been reported7,48 which enhanced target gene activation leading to acquired thermotolerance.
Phylogenetic analysis of HSF proteins of three wheat species with rice and barley revealed that the wheat HSF proteins clustered closely with respective HSFs of rice and barley42. All class A HSFs are grouped in one single major clade along with the C class HSFs. The class C HSF clustered with HSFA3 indicating high similarity between these two protein classes. The class B HSFs also formed a different cluster and indicated a divergence from the type A subgroup. The clustering together of a particular class of HSFs from different species strengthens the notion of conservatory gene function of these orthologs across the species. The anomalous trend of the clustering of HSFA3 with HSFC1 instead of other HSFA clade members has been reported previously42.
Differences in expression profiling of diploid, tetraploid, and hexaploid wheat
Significant differences were observed in the transcriptome expression profiling of allopolyploid wheat species, T. durum, and T. aestivum as compared to one of the wild diploid progenitors, Ae. speltoides. The highest expression of heat stress transcription factor genes was also observed in the diploid wild wheat, in contrast to the cultivated polyploids. Similarly, recent comparative studies49 focused on the expression analysis of gene pairs in the syntenic regions between hexaploid wheat chromosome 3DL and its progenitor 3L arm of Aegilops tauschii demonstrated 60% decreased gene expression in 70% of gene pairs in the hexaploid context of the 3DL genes. Such a reduction in gene expression has been attributed to altered interactions between transcription factors and nucleosomes on the introduction of new genomes leading to alteration in chromosome accessibility. Another study based on analyzing models of homeolog expression patterns has also demonstrated inter-genome interactions to play a key role in altered gene expression50. Thus, polyploidization events bring about changes in the mode of gene action that varies from additive, non-additive and dominant type in addition to the epigenetic and change in transposon activity. The expression changes observed in allopolyploids and the wild progenitor may be due to gene repression, genetic dominance, sub-functionalization, and novel activation in the former relative to the latter. The HSF genes that have been identified and validated in the current study suggest these genes play positive roles in regulating thermotolerance, especially in wild wheat. These genes are of breeding importance as their introgression into cultivars will improve their thermotolerance and thus avoiding the yield penalty due to heat stress, ultimately leading to better productivity.
Conclusion
We generated an important genomic resource of transcriptome representing differential gene expression among three ploidies. The study highlights the conservation of gene expression across the three species as well loss of functional alleles and inter genome interactions during domestication and polyploidy as a major reason behind the reduction of expression in higher ploidies in contrast to their diploid relatives. We report the eight important HSFs in the comparative RNA-Seq analysis of three wheat species of diploid Ae. speltoides, tetraploid T. durum PDW274, and hexaploid T. aestivum PBW725, of which six were validated as potential novel candidates for thermotolerance from Ae. speltoides. Transcript profiling of HSFs under basal and heat stress conditions revealed a good consistency between the expression levels of the eight genes analyzed by qRT-PCR and their transcript levels detected using RNA-Seq in all the three wheat species. Our results encourage the exploitation of novel alleles from Ae. speltoides and other wild relatives for broadening the narrow genetic base of cultivated wheat.
Materials and methods
Plant material
Plant material included diploid wild wheat Ae. speltoides accession pau3809, a heat-tolerant accession, and the two cultivated wheat varieties, tetraploid T. durum cv. PDW274 and hexaploid T. aestivum cv. PBW725 which are susceptible to heat stress. Seeds of Aegilops speltoides accession pau3809 were originally procured from the Weizmann Institute of Science, Rehovot. PDW274 and PBW725 have been developed at PAU, India. These three germplasm lines have been maintained at PAU. Dr. Parveen Chhuneja identified Ae. speltoides accession pau3809 as a heat-tolerant accession and the vouchers specimen of this material has been deposited in the herbarium of National Bureau of Plant Genetic Resources (NBPGR), New Delhi, India. Experimental research and field studies on wild and cultivated varieties, including the collection of plant material, were complied with relevant institutional, national, and international guidelines and legislation. Since the plant material has been maintained by PAU, India, permissions regarding the collection of seed specimens were not required.
RNA extraction and Illumina Sequencing
The plants of the three wheat genotypes were grown in a glasshouse maintained at night/day conditions of 18/22 °C and 80/60% relative humidity with 16 h light (500 μmol m−2 s−1) in pots containing a mixture of sand:soil: peat (3:1:1, v/v/v) at Punjab Agricultural University, Ludhiana. The 10–12 days old seedlings of the three selected wheat lines were taken during the day and immediately snap-frozen in liquid nitrogen and stored at − 80 °C for further use. Seedlings from three different plants were pooled to form one biological replicate. Total RNA from two biological replicates was extracted from the frozen seedling tissue samples using an RNA isolation kit (Qiagen) as per manufacturer protocols. The RNA concentration and purity were determined by Nanodrop™ 1000 Spectrophotometer (Thermo Scientific). Only high-quality RNA samples with OD 260/280 ranging from 1.8 to 2.2 and RIN (RNA integrity number) values ranging from 7.4 to 10.0 were used to construct the RNA-Seq library. Six separate libraries consisting of two biological replicates of the three genotypes were prepared and outsourced for transcriptome sequencing. Indexed TruSeq libraries were prepared for the six RNA samples and 100 bp paired-end sequencing was performed using a HiSeq 2000 platform. The generated raw reads were submitted to the NCBI sequence read archives (SRA) bearing accession number PRJNA767375.
Transcriptomic data filtering and de novo assembly
The quality of reads was assessed using the FastQC toolkit v0.11.951 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The adaptor sequences and low-quality bases (Phred score < 30) were removed from the raw reads using the Trimmomatic tool v0.3852 (http://www.usadellab.org/cms/?page=trimmomatic). De novo transcriptome assembly was performed for all three transcriptomes using Trinity software v2.8.453 (https://github.com/trinityrnaseq/trinityrnaseq). For each of the three de novo assemblies, the transcript abundance was computed by RSEM which estimated the number of RNA-Seq fragments corresponding to each Trinity transcript, including normalized expression values as FPKM (fragments per kilobase of target transcript length per million reads mapped). The assembled transcripts from each assembly were also searched for sequence homology by performing standalone BLASTX with an e-value of 1e − 5 against publicly available NR (NCBI non-redundant protein sequences) database. A schematic representation of the de novo transcriptome reconstruction and analysis pipeline is shown in Fig. 7.
Assessment of transcriptome assembly completeness
The comprehensive and quantitative level of completeness of transcriptome assemblies was assessed by comparing the three assembled transcript sets to a set of highly conserved single-copy orthologs using the BUSCO (Benchmarking Universal Single-Copy Orthologs) v2 pipeline54 (https://busco.ezlab.org/). This compared the assembled transcripts to the predefined set of 3278 Liliopsida single-copy orthologs from the OrthoDB v10 database. The number of complete, duplicated, fragmented, and missing BUSCOs were obtained in each of the three assemblies.
Prediction of protein-coding regions and domains
TransDecoder v5.5.0 (http://transdecoder.github.io/) was used to identify all likely coding regions in the three assemblies, and the single best open reading frame (ORF) per transcript was selected as per the TransDecoder pipeline (-single best orf). The protein sequence file from TransDecoder was used to predict the protein structure domains and families of all transcripts by searching against the Protein family (Pfam) database using the HMM-based tool PfamScan55 (https://www.ebi.ac.uk/Tools/pfa/pfamscan/).
Functional annotation of whole transcriptome assemblies
The identified transcripts in each of the assemblies were annotated based on corresponding homologs identified from the BLASTX program against the NCBI protein database ‘NR’ at an e-value of 1e−5. Based on the NR annotations, the Blast2GO v5.2.5 program (https://www.blast2go.com/) was used to obtain Gene Ontology (GO) annotations for genes in biological process, molecular function, and cellular component category for each assembly separately56. Mercator v3.6 (https://mapman.gabipd.org/app/mercator) was used to assign functional terms to nucleotide sequences using the MapMan ‘BIN’ ontology. MapMan was used for functional analysis and visualization57. The comparative GO terms for molecular function, biological process, and cellular component for each of the three assemblies were plotted using wego v2.058 (https://wego.genomics.cn/).
Identification of orthologous genes
OrthoMCL program59 (https://github.com/stajichlab/OrthoMCL) was used for constructing orthologous groups amongst the three wheat assemblies, using the Markov Cluster algorithm to identify (putative) orthologs and paralogs31. OrthoMCL was conducted for all pair-wise comparisons among the three assemblies. The output of OrthoMCL was used to determine the number of overlapping (shared across species) transcripts across the three assemblies. Further, the transcripts that were annotated/identified as heat stress transcription factors (HSFs) belonging to classes A, B, and C60 were retrieved from transcriptome assembly of AS3809 along with their corresponding orthologs from PBW725 and PDW274. Blast2sequence and multiple sequence alignment with ClustalX were performed to see the extent of homology between HSFs of different wheat species.
In silico expression analysis of genes
The expression patterns of selected HSF transcripts were investigated using wheat transcriptome data from the WheatExp database (https://wheat.pw.usda.gov/WheatExp/). This database comprises RNA-Seq datasets derived from five different tissues (spike, root, leaf, grain, and stem) of hexaploid bread wheat variety Chinese Spring each sampled at three different developmental stages61. Another dataset consists of one-week-old seedlings of hexaploid wheat variety TAM107 treated with high temperature (40 °C) for 1 h and 6 h62. The 12 selected HSF transcripts were BLASTn searched with the wheat transcriptome database with an expected cut-off of 1e − 5.
Protein–protein interaction (PPI) network analysis
The HSF protein sequences were mapped to the STRING (Search Tool for the Retrieval of Interacting Genes) database (http://string-db.org/) to acquire protein–protein interaction (PPI) networks. Active interaction sources, including text mining, experiments, databases, and co-expression as well as species limited to “Aegilops speltoides”. The required confidence score > 0.4 was set as the threshold to identify the PPI pairs among the HSF proteins. Cytoscape v3.6.1 s63 (https://cytoscape.org/) was used to visualize the PPI network.
Phylogenetic analysis of HSF gene family
To access the phylogenetic relationships among previously identified barley and rice HSF genes25 with the HSF genes identified in diploid, tetraploid, and hexaploid wheat and to classify all the members of the family, multiple sequence alignment of protein sequences was done using program ClustalW. Mega 7.0 program64 (https://www.megasoftware.net/) was then used to construct an un-rooted neighbor-hood joining method based phylogenetic tree with 1000 bootstrap replication values with default parameters, by using multiple sequence alignment of the deduced amino acid sequence of diploid, tetraploid, hexaploid wheat along with barley and rice HSF proteins.
Validation of HSF genes by quantitative real-time PCR (qRT-PCR)
The seeds of Ae. speltoides acc. pau3809, T. aestivum cv. PBW725 and T. durum cv. PDW274 were sown in germination trays in a growth chamber maintained at 22 °C/15 °C Day: night temperature, 16:8 h photoperiod. Then heat stress was given to the 10-day old seedlings at 35 °C for 1 h and immediately after stress RNA from control and heat-stressed plants was extracted for performing qRT-PCR. Primers for the 12 selected HSFs were generated using Primer Express software v3.0.1 (https://www.thermofisher.com/order/catalog/product/4363991) following the default parameters and are listed in Supplementary Table 1. Since there was considerable homology (98%) in the active coding region of the HSF genes across the three wheat species, we used the same primers for all three species. TaActin gene65 was used as the internal control. The relative gene expression levels for RT-qPCR data were calculated using the 2−∆∆Ct method66. All reactions were conducted in triplicate.
References
Akter, N. & Rafiqul Islam, M. Heat stress effects and management in wheat. A review. Agron. Sustain. Dev. 37, 37. https://doi.org/10.1007/s13593-017-0443-9 (2017).
Arora, S., Cheema, J., Poland, J., Uauy, C. & Chhuneja, P. Genome-wide association mapping of grain micronutrients concentration in Aegilops tauschii. Front. Plant Sci. 10, 54. https://doi.org/10.3389/fpls.2019.00054 (2019).
Climate change 2007: The Physical Science Basis: Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change (Cambridge University Press, 2007).
Wollenweber, B., Porter, J. R. & Schellberg, J. Lack of interaction between extreme high-temperature events at vegetative and reproductive growth stages in wheat. J. Agron. Crop. Sci. 189, 142–150 (2003).
Gupta, P. K., Balyan, H. S., Gahlaut, V. & Kulwal, P. L. Phenotyping, genetic dissection, and breeding for drought and heat tolerance in common wheat: Status and prospects. in Plant Breeding Reviews (ed. Janick, J.) 85–168 (Wiley, 2012). https://doi.org/10.1002/9781118358566.ch2
Awlachew, Z. T., Singh, R., Kaur, S., Bains, N. S. & Chhuneja, P. Transfer and mapping of the heat tolerance component traits of Aegilops speltoides in tetraploid wheat Triticum durum. Mol. Breed. 36, 78. https://doi.org/10.1007/s11032-016-0499-2 (2016).
Li, J., Wan, H. & Yang, W. Synthetic hexaploid wheat enhances variation and adaptive evolution of bread wheat in breeding processes. J. Syst. Evol. 52, 735–742 (2014).
Farooq, S. Co-existence of salt and drought tolerance in Triticeae. Hereditas 135, 205–210 (2004).
Waines, J. High temperature stress in wild wheats and spring wheats. Funct. Plant Biol. 21, 705–715 (1994).
Jafarzadeh, J. et al. Breeding value of primary synthetic wheat genotypes for grain yield. PLoS ONE 11(9), e0162860. https://doi.org/10.1371/journal.pone.0162860 (2016).
Colmer, T. D., Flowers, T. J. & Munns, R. Use of wild relatives to improve salt tolerance in wheat. J. Exp. Bot. 57, 1059–1078 (2006).
Vikas, V. K. et al. Stem and leaf rust resistance in wild relatives of wheat with D genome (Aegilops spp.). Genet. Resour. Crop Evol. 61, 861–874 (2014).
Huang, M. et al. Genetic analysis of heading date in winter and spring wheat. Euphytica 214, 128. https://doi.org/10.1007/s10681-018-2199-y (2018).
Olivera, P. D., Rouse, M. N. & Jin, Y. Identification of new sources of resistance to wheat stem rust in Aegilops spp. in the tertiary genepool of wheat. Front. Plant Sci. 9, 1719. https://doi.org/10.3389/fpls.2018.01719 (2018).
Von Koskull-Döring, P., Scharf, K.-D. & Nover, L. The diversity of plant heat stress transcription factors. Trends Plant Sci. 12, 452–457 (2007).
Baniwal, S. K. et al. Heat stress response in plants: a complex game with chaperones and more than twenty heat stress transcription factors. J. Biosci. 29, 471–487 (2004).
Nover, L., Bharti, K. & Scharf, K.-D. Arabidopsis and the heat stress transcription factor world: How many heat stress transcription factors do we need?. Cell Stress Chaperones 6(3), 177–189 (2001).
Fragkostefanakis, S., Röth, S., Schleiff, E. & Scharf, K.-D. Prospects of engineering thermotolerance in crops through modulation of heat stress transcription factor and heat shock protein networks: HSFs and HSPs for improvement of crop thermotolerance. Plant Cell Environ. 38, 1881–1895 (2015).
Guo, M. et al. Genome-wide analysis, expression profile of heat shock factor gene family (CaHSFs) and characterisation of CaHsfA2 in pepper (Capsicum annuum L.). BMC Plant Biol. 15, 151. https://doi.org/10.1186/s12870-015-0512-7 (2015).
Scharf, K.-D., Berberich, T., Ebersberger, I. & Nover, L. The plant heat stress transcription factor (Hsf) family: Structure, function and evolution. Biochimica et Biophysica Acta BBA Gene Regul. Mech. 1819, 104–119 (2012).
Xue, G.-P., Sadat, S., Drenth, J. & McIntyre, C. L. The heat shock factor family from Triticum aestivum in response to heat and other major abiotic stresses and their role in regulation of heat shock protein genes. J. Exp. Bot. 65, 539–557 (2014).
Agarwal, P. & Khurana, P. Functional characterization of HSFs from wheat in response to heat and other abiotic stress conditions. Funct. Integr. Genomics 19, 497–513 (2019).
Chauhan, H., Khurana, N., Agarwal, P., Khurana, J. P. & Khurana, P. A seed preferential heat shock transcription factor from wheat provides abiotic stress tolerance and yield enhancement in transgenic Arabidopsis under heat stress environment. PLoS ONE 8(11), e79577. https://doi.org/10.1371/journal.pone.0079577 (2013).
Hu, Q. et al. Meiotic chromosome association 1 interacts with TOP3α and regulates meiotic recombination in rice. Plant Cell 29, 1697–1708 (2017).
Chauhan, H., Khurana, N., Agarwal, P. & Khurana, P. Heat shock factors in rice (Oryza sativa L.): Genome-wide expression analysis during reproductive development and abiotic stress. Mol. Genet. Genomics 286, 171. https://doi.org/10.1007/s00438-011-0638-8 (2011).
Zaharieva, M., Gaulin, E., Havaux, M., Acevedo, E. & Monneveux, P. Drought and heat responses in the wild wheat relative Aegilops geniculata Roth: Potential interest for wheat improvement. Crop Sci. 41, 1321–1329 (2001).
Pradhan, G. P., Prasad, P. V. V., Fritz, A. K., Kirkham, M. B. & Gill, B. S. High temperature tolerance in Aegilops species and its potential transfer to wheat. Crop Sci. 52, 292–304 (2012).
Jakhu, P. et al. Cloning, expression analysis and In silico characterization of HSP101: A potential player conferring heat stress in Aegilops speltoides (Tausch) Gren. Physiol. Mol. Biol. Plants 27, 1205–1218 (2021).
Salse, J. et al. New insights into the origin of the B genome of hexaploid wheat: Evolutionary relationships at the SPA genomic region with the S genome of the diploid relative Aegilops speltoides. BMC Genomics 9, 555. https://doi.org/10.1186/1471-2164-9-555 (2008).
Borrill, P., Adamski, N. & Uauy, C. Genomics as the key to unlocking the polyploid potential of wheat. New Phytol. 208, 1008–1022 (2015).
Ruban, A. S. & Badaeva, E. D. Evolution of the S-genomes in Triticum-Aegilops alliance: Evidences from chromosome analysis. Front. Plant Sci. 9, 1756. https://doi.org/10.3389/fpls.2018.01756 (2018).
El Baidouri, M. et al. Reconciling the evolutionary origin of bread wheat (Triticum aestivum ). New Phytol 213, 1477–1486 (2017).
Appels, R. et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, 6403. https://doi.org/10.1126/science.aar7191 (2018).
Miki, Y. et al. Origin of wheat B-genome chromosomes inferred from RNA sequencing analysis of leaf transcripts from section Sitopsis species of Aegilops. DNA Res. 26, 171–182 (2019).
Liu, W. et al. Transcriptome analysis of wheat grain using RNA-Seq. Front. Agric. Sci. Eng. 1(3), 214–222. https://doi.org/10.15302/J-FASE-2014024 (2014).
Yadav, I. S. et al. Comparative temporal transcriptome profiling of wheat near isogenic line carrying Lr57 under compatible and incompatible interactions. Front. Plant Sci. 7, 1943. https://doi.org/10.3389/fpls.2016.01943 (2016).
Chen, Z. J. & Ni, Z. Mechanisms of genomic rearrangements and gene expression changes in plant polyploids. BioEssays 28, 240–252 (2006).
Kumar Kushwaha, S. et al. Differential gene expression analysis of wheat breeding lines reveal molecular insights in yellow rust resistance under field conditions. Agronomy 10, 1888. https://doi.org/10.3390/agronomy10121888 (2020).
Kumar, R. R. et al. Harnessing next generation sequencing in climate change: RNA-Seq analysis of heat stress-responsive genes in wheat (Triticum aestivum L.). OMICS 19, 632–647 (2015).
Schleiff, E. & Becker, T. Common ground for protein translocation: access control for mitochondria and chloroplasts. Nat. Rev. Mol. Cell. Biol. 12, 48–59 (2011).
Xue, G.-P., Drenth, J. & McIntyre, C. L. TaHsfA6f is a transcriptional activator that regulates a suite of heat stress protection genes in wheat (Triticum aestivum L.) including previously unknown Hsf targets. J. Exp. Bot. 66, 1025–1039 (2015).
Mishra, S. K. et al. Genome-wide identification, phylogeny and expression analysis of HSF gene family in barley during abiotic stress response and reproductive development. Plant Gene 23, 100231. https://doi.org/10.1016/j.plgene.2020.100231 (2020).
Zhuang, L. et al. Characterization and functional analysis of FaHsfC1b from Festuca arundinacea conferring heat tolerance in Arabidopsis. IJMS 19, 2702. https://doi.org/10.3390/ijms19092702 (2018).
Mittal, D., Madhyastha, D. A. & Grover, A. Gene expression analysis in response to low and high temperature and oxidative stresses in rice: Combination of stresses evokes different transcriptional changes as against stresses applied individually. Plant Sci. 197, 102–113 (2012).
Qin, F. et al. Regulation and functional analysis of ZmDREB2A in response to drought and heat stresses in Zea mays L: ZmDREB2A in drought and heat stress response. Plant J. 50, 54–69 (2007).
Kotak, S. et al. Complexity of the heat stress response in plants. Curr. Opin. Plant Biol. 10, 310–316 (2007).
Zhang, S. et al. Overexpression of TaHSF3 in transgenic Arabidopsis enhances tolerance to extreme temperatures. Plant Mol. Biol. Rep. 31, 688–697 (2013).
Li, M., Berendzen, K. W. & Schöffl, F. Promoter specificity and interactions between early and late Arabidopsis heat shock factors. Plant Mol. Biol. 73, 559–567 (2010).
Lu, F.-H. et al. Reduced chromatin accessibility underlies gene expression differences in homologous chromosome arms of diploid Aegilops tauschii and hexaploid wheat. GigaScience 9(giaa070), 1–11. https://doi.org/10.1093/gigascience/giaa070 (2020).
Hu, G. & Wendel, J. F. Cis–trans controls and regulatory novelty accompanying allopolyploidization. New Phytol. 221, 1691–1700 (2019).
Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Haas, B. J. et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512 (2013).
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V. & Zdobnov, E. M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015).
Finn, R. D. et al. Pfam: the protein families database. Nucl. Acids Res. 42, D222–D230 (2014).
Conesa, A. & Götz, S. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genomics 2008, 1–12 (2008).
Thimm, O. et al. mapman: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37, 914–939 (2004).
Ye, J. et al. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 34, W293–W297 (2006).
Li, L. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 13, 2178–2189 (2003).
Doring, P. et al. The role of AHA motifs in the activator function of tomato heat stress transcription factors HsfA1 and HsfA2. Plant Cell 12, 265–278 (2000).
Choulet, F. et al. Structural and functional partitioning of bread wheat chromosome 3B. Science 345, 1249721–1249721 (2014).
Liu, Z. et al. Temporal transcriptome profiling reveals expression partitioning of homeologous genes contributing to heat and drought acclimation in wheat (Triticum aestivum L.). BMC Plant Biol. 15, 152. https://doi.org/10.1186/s12870-015-0511-8 (2015).
Shannon, P. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016).
Risk, J. M. et al. The wheat Lr34 gene provides resistance against multiple fungal pathogens in barley. Plant Biotechnol. J. 11, 847–854 (2013).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001).
Author information
Authors and Affiliations
Contributions
S.K. and P.C.—conceived and designed research, reviewed the manuscript, S.K.—planned the experiments, S.S.—conducted all analysis of transcriptome data, identification of HSF and primer designing, I.S.Y.—helped in analysis of transcriptome data, H.C. and P.S.—designed and conducted the real-time experiment. S.K., S.S., P.M., and A.K.—compiled the work and produced the final draft of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Seni, S., Kaur, S., Malik, P. et al. Transcriptome based identification and validation of heat stress transcription factors in wheat progenitor species Aegilops speltoides. Sci Rep 11, 22049 (2021). https://doi.org/10.1038/s41598-021-01596-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-01596-6
This article is cited by
-
Analyzing the regulatory role of heat shock transcription factors in plant heat stress tolerance: a brief appraisal
Molecular Biology Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.