Abstract
Natural killer cells are modulated through the binding of killer cell immunoglobulin-like receptors (KIRs) with human leukocyte antigen (HLA) class I ligands. This study investigated the association of KIR/HLA pairs with progression to liver cirrhosis, hepatocellular carcinoma (HCC) development, and nucleot(s)ide (NUC) treatment freedom in hepatitis B virus (HBV) infection. KIR, HLA-Bw, and HLA-C were genotyped in 280 Japanese HBV patients for clinical comparisons. No significant associations of KIR/HLA pairs were detected in terms of liver cirrhosis development. The KIR2DS3 positive rate was significantly higher in patients with HCC (n = 39) than in those without (n = 241) [30.8% vs. 14.9%, odds ratio (OR) 2.53, P = 0.015]. The KIR3DL1/HLA-Bw4 pair rate was significantly lower in the NUC freedom group (n = 20) than in the NUC continue group (n = 114) (25.0% vs. 52.6%, OR 0.30, P = 0.042). In conclusion, this study indicated remarkable associations of KIR/HLA with HCC development (KIR2DS3) and freedom from NUC therapy (KIR3DL1/HLA-Bw4) in HBV patients, although the number of cases was insufficient for statistical purposes. Additional multi-center analyses of larger groups are needed to clarify whether KIR/HLA pairs play a role in HBV patient status.
Similar content being viewed by others
Introduction
Hepatitis B virus (HBV) infection is a global health concern, with possible life-threatening liver infection in both acute and chronic disease forms. There are over 250 million estimated HBV carriers in the world, of whom approximately 600,000 die annually from HBV-related liver disease. Chronic HBV infection often leads to liver cirrhosis and eventual hepatocellular carcinoma (HCC)1,2,3,4. Accordingly, it is the second-leading cause of liver cirrhosis and cancer in Japan after hepatitis C virus (HCV)5. Several factors are reportedly involved in the progression to liver cirrhosis and HCC in HBV patients6. In order to prevent disease progression, therapeutic agents including nucleot(s)ide analogues (NUCs) and pegylated interferon are currently available7,8,9,10. However, since it remains difficult to eliminate the covalently closed circular DNA of HBV in the liver with current standard therapies, a life-long commitment to HBV treatment with NUCs and careful HCC surveillance is required, which may also impose a financial burden and potential drug toxicity11. It is of clinical need to identify new factors associated with liver cirrhosis and HCC development towards safe NUC discontinuation.
Natural killer (NK) cells provide rapid responses to viral infection and play a key role in tumor immunosurveillance by directly inducing the death of cancer cells12. Accordingly, the functional impairment of NK cells has been associated with chronic disease onset and cancer development. NK cell function is centrally controlled by activating and inhibitory killer immunoglobulin-like receptors (KIRs), which bind to human leukocyte antigen (HLA) class I molecules13,14,15. We earlier demonstrated that KIR/HLA receptor-ligand combinations were associated with chronic liver disease progression and HCC development in HCV-infected patients16,17. However, no reports have addressed such associations in Japanese patients with HBV to date even though a few studies suggested such associations in other ethnicities18,19.
The present study investigated the association of KIR/HLA pairs with disease progression to liver cirrhosis (Study 1), HCC development (Study 2), and freedom from NUCs (Study 3) in Japanese patients with chronic HBV infection.
Results
Patient characteristics
The cohort’s characteristics are summarized in Table 1. Median age was 62 years, and 52.5% of subjects were male. Regarding HBV markers, HBsAg-positive and HBeAg-positive patients totaled 78.9% and 14.3%, respectively. Clinically, 240 patients were at the chronic hepatitis stage and 40 patients were at the liver cirrhosis stage. Of the 40 patients with liver cirrhosis, 12 patients had no NUC treatment history or indication for NUC therapy since they had already achieved an HBsAg loss and HBV DNA level of 0 LIU/mL but were at the liver cirrhosis stage at the time of enrollment. The remaining 28 patients at the liver cirrhosis stage received NUC treatment in 2018. Thirty-nine patients had experienced HCC by the end of 2018. Of the 134 patients (47.9%) with prior or ongoing NUC treatment, 20 patients had achieved freedom from NUCs by the end of 2018.
Clinical characteristics and KIR/HLA genotyping of patients with HBV-induced cirrhosis (Study 1)
Age, male frequency, FIB-4, APRI, and M2BPGi 1 + or 2 + were significantly higher in patients at the liver cirrhosis stage, while platelet count and HBV DNA were significantly lower (Table 1). To clarify the impact of KIR/HLA pairs on disease progression to liver cirrhosis, KIR and HLA genes were genotyped and their frequencies were compared between patients with and without cirrhosis. No significant differences were detected for any KIR or HLA frequency or KIR/HLA pair frequency between the groups (Table 2).
Clinical characteristics and KIR/HLA genotyping of patients with HBV-induced HCC (Study 2)
Age, male frequency, FIB-4, APRI, and M2BPGi 1 + or 2 + were significantly higher in patients with HCC, whereas platelet count and HBV DNA were significantly lower (Table 1). There were no significant differences in the gene frequencies of HLA-Bw4, HLA-Bw6, HLA-C1, or HLA-C2 in patients with and without HCC. The KIR2DS1 positive rate was significantly higher in the HCC group (53.8% vs. 34.9%; odds ratio [OR] 2.18, P = 0.023), the significance of which disappeared in the KIR2DS1/HLA-C2 pair analysis. The KIR2DS3 positive rate was significantly higher in the HCC group (30.8% vs. 14.9%; OR 2.53, P = 0.015). No significant differences in KIR/HLA proportions were observed between the groups (Table 3).
Independent risk factors of HCC development by logistic regression analysis
We conducted logistic regression analysis to identify the independent risk factors of HCC development. Regression analysis model 1 including all indices confirmed that liver cirrhosis stage (hazard ratio [HR] 13.42, P < 0.001) and KIR2DS3 positivity (HR: 4.15, P = 0.003) were independent risk factors of HCC development (Table 4). Model 2 analysis, which included age ≥ 65 years, male gender, and KIR2DS3, demonstrated age ≥ 65 years (HR 2.87, P = 0.006), male gender (HR 2.93, P = 0.009), and KIR2DS3 positivity (HR 2.90, P = 0.011) as independent risk factors (Table 4).
Clinical characteristics and KIR/HLA genotyping of patients receiving NUC treatment (Study 3)
Twenty of the 134 patients having been treated with NUCs achieved freedom from NUC therapy by the end of 2018. In terms of background characteristics, there were no significant differences between NUC free and NUC continue patients apart from lower frequencies of HBsAg and HBeAg positivity, no progression to cirrhosis, and no development of HCC in the NUC free group (Table 5). The HLA-Bw6 positive rate was significantly lower in the NUC free group than in the NUC continue group (60.0% vs. 89.5%, OR 0.18, P = 0.001). The KIR3DL1/HLA-Bw4 pair was also significantly less frequent in the NUC free group (25.0% vs. 52.6%, OR 0.30, P = 0.042) (Table 6).
Independent risk factors of NUC treatment freedom by logistic regression analysis
Logistic regression analysis was performed to identify the independent risk factors of NUC treatment freedom. Regression analysis model 1 considering all indices confirmed HBsAg positivity (HR 0.07, P < 0.001) and KIR3DL1/HLA-Bw4 positivity (HR 0.25, P = 0.040) as negative independent risk factors of NUC treatment freedom (Table 7). Model 2 analysis, which included age ≥ 65 years, male gender, and KIR3DL1/HLA-Bw4 positivity, demonstrated that KIR3DL1/HLA-Bw4 positivity (HR 0.29, P = 0.026) was a negative independent risk factor of NUC treatment freedom (Table 7).
Discussion
This single-center, cross-sectional study retrospectively examined whether specific KIR/HLA pairs were associated with progression to liver cirrhosis, HCC development, and freedom from NUC treatment in chronic HBV patients. Our results showed that: (1) there was no detectable association of KIR-HLA genotype with liver cirrhosis development, (2) patients with KIR2DS3 had a significantly higher risk of HCC, with no remarkable involvement of any KIR/HLA pair, and (3) patients with the KIR3DL1/HLA-Bw4 pair were significantly more likely to achieve freedom from NUCs. These genotype differences might have affected the clinical course of HBV by stimulating or escaping immune surveillance and other intrahepatic inflammatory processes.
Many factors are involved in the progression to liver cirrhosis in HBV patients, such as the viral factors of HBV DNA, HBsAg level, and HBV genotype, the host factors of gender and age, and such environmental factors as alcohol intake6. Among those, age is a strong predictor of significant fibrosis progression6; indeed, the patients with cirrhosis in the present study were significantly older than those without. Regarding host genetic factors, HLA class II has been widely analyzed and linked to disease progression20. To date, no association studies have considered KIR-HLA combinations in the Japanese. We therefore conducted this investigation to identify associations of such combinations with disease progression to liver cirrhosis. However, no significant relationship was detected among the KIR-HLA combinations tested. In a Gambian population, KIR and HLA class I combinations were also not related to liver cirrhosis19, suggesting that the viral, host, and environmental factors mentioned above might have a stronger influence on liver cirrhosis development in HBV. Longitudinal studies with more subjects are needed to clarify these findings.
Regarding host genetic factors for HCC development in HBV patients, a number of candidate genes specifically for HLA class II regions have been investigated by genetic association studies to evaluate their role in HCC susceptibility20,21. KIR and HLA class I combination was reportedly associated with HCC development in Chinese while its combination was not related to HCC development in Gambians18,19. A genome-wide association study very recently proposed HLA-A*33:03 as a susceptibility allele for HCC in HBV22. The present investigation on the impact of KIRs and HLA-Bw or HLA-C on HCC development detected no remarkable KIR/HLA pairs, although a significant association of KIR2DS3 with HCC onset was found. The ligand to KIR2DS3 has not been identified to date. However, KIR2DS3 has been related to colorectal cancer23, spontaneous HCV clearance failure24, and fatal outcome of Ebola virus infection25. Moreover, KIR2DS3 was seen to be expressed at low level on the NK cell surface with unknown function26, and might therefore represent a genetic marker for a closely related linked gene with biological effects. Further research is required to confirm this hypothesis.
We found no association among KIR-HLA genotypes with liver cirrhosis development, although a significant association of KIR2DS3 with HCC development was detected. In general, HCC occurs after patients progress to a liver cirrhosis disease status as evidenced in the natural course of HCV27, raising the hypothesis that the same KIR/HLA pairs could be risk factors for liver cirrhosis as well as HCC development. However, in addition to liver cirrhosis stage patients, noncirrhotic patients with HBV infection who have such risk factors as a family history of HCC, Asian male over 40 years of age, and Asian female over 50 years of age, can progress to HCC. Indeed, 17 HCC patients at a noncirrhotic stage were included in this study (Table 1), which suggested a risk difference among KIR/HLA genotypes between liver cirrhosis patients and noncirrhotic patients. A larger number of patients is needed for a sufficient sub-group analysis to verify this hypothesis.
Lastly, HBV patients who receive NUCs are expected to continue treatment indefinitely since there is currently no way to completely eliminate the virus from the liver, with discontinuation sometimes leading to virological relapse and hepatic flares. However, some patients are able to control infection without ongoing treatment. We observed that the KIR3DL1/HLA-Bw4 pair frequency was significantly higher in the NUC continue group despite the KIR3DL1 and HLA-Bw4 expression rates being comparable between the groups. This KIR/HLA pair may be a new biomarker for predicting drug continuation or freedom in HBV patients under NUC treatment. Other ligands to KIR3DL1 include HLA-A23, HLA-A24, and HLA-A32 on the HLA-A locus, which have very recently been reported to exhibit a similar biological function to HLA-Bw4, and have thus been termed HLA-ABw428. Associations of the HLA-A genotype with HBV should be addressed.
Based on the above findings, it is possible that particular KIR/HLA pairs play an important role in regulating the innate immune response and tumor surveillance as well as controlling disease susceptibility in HBV-infected patients. The identified KIR/HLA pair presented in this study may represent an effective predictive marker of HCC development after HBV infection, with additional use as a predictor of the efficacy of anti-HBV treatment with NUCs.
There are several limitations to this study. First, as the number of patients with liver cirrhosis, HCC, and NUC freedom were too small for a definitive conclusion, a larger validation analysis with more subjects is needed to confirm our results. Second, this study was cross-sectional and retrospectively designed, and so the criteria for NUC discontinuation were not aligned; patients who achieved NUC discontinuation were defined as belonging to the NUC treatment freedom group as a result of their clinical course. In order to minimize selection bias, a larger number of prospectively enrolled patients is needed to confirm our results regarding an association between NUC discontinuation and the KIR3DL1/HLA-Bw4 pair. Third, this investigation analyzed HLA-Bw4/Bw6 and HLA-C1/C2, but not other HLA class I genotypes. Additional classical HLA class I genotypes, such as HLA-A, and non-classical HLA class I genotypes should be addressed in future studies.
In conclusion, the present study indicated associations of KIR2DS3 with HCC development and the KIR3DL1/HLA-Bw4 pair with freedom from NUCs in HBV patients, although a larger number of cases are required for statistical purposes. Future multi-center analyses are needed to confirm that KIR/HLA pairs play a role in HBV patient status.
Materials and methods
Patients
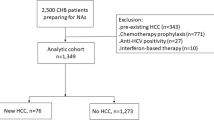
A total of 324 patients of at least 20 years of age who regularly visited Shinshu University Hospital in Matsumoto, Japan, for HBV infection management in the year 2018 (January 1, 2018, to December 31, 2018) were retrospectively targeted. Patients exhibiting other causes of chronic liver disease, such as alcoholic liver disease, non-alcoholic fatty liver disease, primary biliary cholangitis, or autoimmune hepatitis, were not considered. The study participant selection flowchart is depicted in Fig. 1. After the exclusion of 44 cases (2 with acute hepatitis B virus infection and 42 without stored DNA or serum), 280 patients with chronic HBV infection were enrolled in the analysis. The racial background of all patients was uniformly Japanese.
Total bilirubin, alanine aminotransferase, and other relevant biochemical tests were performed using standard methods. HBsAg, HBeAb, and MAC-2 binding protein glycosylation isomer (M2BPGi; Sysmex Co., Kobe, Japan) were measured with a HISCL-5000 (Sysmex Co., Kobe, Japan) system using stocked serum. M2BPGi readings of < 1.00 COI, ≥ 1.00 COI and < 3.00 COI, and ≥ 3.00 COI were judged as negative, 1 + , and 2 + , respectively.
Determination of liver cirrhosis
Patients with liver cirrhosis were judged as those with histologically proven liver cirrhosis and/or the characteristic clinical signs of advanced liver disease in imaging studies in the year 2018. The number of patients who were diagnosed as being in the liver cirrhosis stage primarily by liver biopsy, ultrasonography, computed tomography, and magnetic resonance imaging were 9, 2, 21, and 8, respectively.
Determination of HCC
HCC patients were defined as having HCC in the prior quarter century (i.e., between 1993 and 2018) or complicating HCC in 2018. HCC was diagnosed by imaging characteristics, arterial hypervascularity, and venous or delayed phase washout by contrast-enhanced dynamic computed tomography and/or magnetic resonance imaging when a nodular lesion was detected by ultrasonography or a tumor marker was elevated.
Definition of NUC freedom
Among the 134 patients with prior or ongoing NUC treatment, 20 patients had successfully discontinued therapy by the end of 2018 and were defined as NUC free.
HLA class I and KIR genotyping
Whole-genomic DNA was extracted from whole blood samples from all participants using QuickGene-800 assays (Fujifilm, Tokyo, Japan). HLA-Bw4, HLA-C1, and HLA-C2 genotyping29 as well as KIR genotyping30 were performed using the polymerase chain reaction with sequence-specific primers. Regarding KIR2DS4, full-length KIR2DS4 was typed by the polymerase chain reaction with sequence-specific primers in consideration of its functional difference31. HLA and KIR typing were used to stratify patients into groups according to predicted KIR-ligand interactions and binding affinities. The KIR/HLA pairs of interest were KIR2DL1/2DS1/2DS4-HLA-C2, 2DL2/3/2DS2-HLA-C1, and 3DL1/3DS1-HLA-Bw4. All genotyping was blinded to clinical variables.
Statistical analysis
Statistical analysis and data visualization were carried out using StatFlex ver. 7.0.11 software (Artech Co., Ltd., Osaka, Japan). Continuous variables were compared using the Mann–Whitney U test. Categorical variables were evaluated by Pearson’s chi-squared test, Fisher’s exact test, or Yates' continuity correction, as appropriate. Multivariate analysis was performed by means of regression analysis with a stepwise method after categorizing continuous variables to minimize interference. A P-value of < 0.05 was considered statistically significant.
Ethical statement
This investigation was conducted according to the guidelines of the Declaration of Helsinki and approved by the Ethics Committee of Shinshu University School of Medicine (protocol code 302 approved on October 10, 2010, and 527 approved on August 10, 2015).
Informed consent
Informed consent was obtained from all participants involved in this study.
Abbreviations
- HBV:
-
Hepatitis B virus
- HCC:
-
Hepatocellular carcinoma
- HCV:
-
Hepatitis C virus
- NUC:
-
Nucleot(s)ide analogue
- cccDNA:
-
Covalently closed circular DNA
- NK:
-
Natural killer
- KIR:
-
Killer immunoglobulin-like receptor
- HLA:
-
Human leukocyte antigen
- M2BPGi:
-
MAC-2 binding protein glycosylation isomer
- OR:
-
Odds ratio
- HR:
-
Hazard ratio
References
Beasley, R. P., Hwang, L. Y., Lin, C. C. & Chien, C. S. Hepatocellular carcinoma and hepatitis B virus: A prospective study of 22 707 men in Taiwan. Lancet 2, 1129–1133 (1981).
Fattovich, G., Bortolotti, F. & Donato, F. Natural history of chronic hepatitis B: Special emphasis on disease progression and prognostic factors. J. Hepatol. 48, 335–352 (2008).
Ganem, D. & Prince, A. M. Hepatitis B virus infection–natural history and clinical consequences. N. Engl. J. Med. 350, 1118–1129 (2004).
Yim, H. J. & Lok, A. S. Natural history of chronic hepatitis B virus infection: What we knew in 1981 and what we know in 2005. Hepatology 43, S173-181 (2006).
Tateishi, R. et al. A nationwide survey on non-B, non-C hepatocellular carcinoma in Japan: 2011–2015 update. J. Gastroenterol. 54, 367–376 (2019).
de Jongh, F. E. et al. Survival and prognostic indicators in hepatitis B surface antigen-positive cirrhosis of the liver. Gastroenterology 103, 1630–1635 (1992).
Hosaka, T. et al. Long-term entecavir treatment reduces hepatocellular carcinoma incidence in patients with hepatitis B virus infection. Hepatology 58, 98–107 (2013).
Liaw, Y. F. et al. Lamivudine for patients with chronic hepatitis B and advanced liver disease. N. Engl. J. Med. 351, 1521–1531 (2004).
Matsumoto, A. et al. Efficacy of lamivudine for preventing hepatocellular carcinoma in chronic hepatitis B: A multicenter retrospective study of 2795 patients. Hepatol. Res. 32, 173–184 (2005).
Lampertico, P. et al. Long-term suppression of hepatitis B e antigen-negative chronic hepatitis B by 24-month interferon therapy. Hepatology 37, 756–763 (2003).
Gill, U. S. et al. Assessment of bone mineral density in tenofovir-treated patients with chronic hepatitis B: Can the fracture risk assessment tool identify those at greatest risk?. J. Infect. Dis. 211, 374–382 (2015).
Guillerey, C., Huntington, N. D. & Smyth, M. J. Targeting natural killer cells in cancer immunotherapy. Nat. Immunol. 17, 1025–1036 (2016).
Cella, M., Longo, A., Ferrara, G. B., Strominger, J. L. & Colonna, M. NK3-specific natural killer cells are selectively inhibited by Bw4-positive HLA alleles with isoleucine 80. J. Exp. Med. 180, 1235–1242 (1994).
Colonna, M., Borsellino, G., Falco, M., Ferrara, G. B. & Strominger, J. L. HLA-C is the inhibitory ligand that determines dominant resistance to lysis by NK1- and NK2-specific natural killer cells. Proc. Natl. Acad. Sci. U S A 90, 12000–12004 (1993).
Colonna, M., Brooks, E. G., Falco, M., Ferrara, G. B. & Strominger, J. L. Generation of allospecific natural killer cells by stimulation across a polymorphism of HLA-C. Science 260, 1121–1124 (1993).
Saito, H. et al. KIR2DL2 combined with HLA-C1 confers risk of hepatitis C virus-related hepatocellular carcinoma in younger patients. Oncotarget 9, 19650–19661 (2018).
Umemura, T. et al. KIR/HLA genotypes confer susceptibility and progression in patients with autoimmune hepatitis. JHEP Rep. 1, 353–360 (2019).
Pan, N. et al. KIR and HLA loci are associated with hepatocellular carcinoma development in patients with hepatitis B virus infection: A case-control study. PLoS ONE 6, e25682 (2011).
Yindom, L. M. et al. KIR content genotypes associate with carriage of hepatitis B surface antigen, e antigen and HBV viral load in Gambians. PLoS ONE 12, e0188307 (2017).
Nishida, N. et al. New susceptibility and resistance HLA-DP alleles to HBV-related diseases identified by a trans-ethnic association study in Asia. PLoS ONE 9, e86449 (2014).
Jiang, D. K. et al. Genetic variants in STAT4 and HLA-DQ genes confer risk of hepatitis B virus-related hepatocellular carcinoma. Nat. Genet. 45, 72–75 (2013).
Sawai, H. et al. Genome-wide association study identified new susceptible genetic variants in HLA class I region for hepatitis B virus-related hepatocellular carcinoma. Sci. Rep. 8, 7958 (2018).
Diaz-Peña, R. et al. Analysis of killer immunoglobulin-like receptor genes in colorectal cancer. Cells 9, 514 (2020).
Dring, M. M. et al. Innate immune genes synergize to predict increased risk of chronic disease in hepatitis C virus infection. Proc. Natl. Acad. Sci. U S A 108, 5736–5741 (2011).
Wauquier, N., Padilla, C., Becquart, P., Leroy, E. & Vieillard, V. Association of KIR2DS1 and KIR2DS3 with fatal outcome in Ebola virus infection. Immunogenetics 62, 767–771 (2010).
VandenBussche, C. J., Mulrooney, T. J., Frazier, W. R., Dakshanamurthy, S. & Hurley, C. K. Dramatically reduced surface expression of NK cell receptor KIR2DS3 is attributed to multiple residues throughout the molecule. Genes Immun. 10, 162–173 (2009).
Kiyosawa, K. et al. Interrelationship of blood transfusion, non-A, non-B hepatitis and hepatocellular carcinoma: Analysis by detection of antibody to hepatitis C virus. Hepatology 12, 671–675 (1990).
Saunders, P. M. et al. The role of the HLA class I α2 helix in determining ligand hierarchy for the killer cell ig-like receptor 3DL1. J. Immunol. 206, 849–860 (2021).
Tajik, N., Shahsavar, F., Nasiri, M. & Radjabzadeh, M. F. Compound KIR-HLA genotype analyses in the Iranian population by a novel PCR-SSP assay. Int. J. Immunogenet. 37, 159–168 (2010).
Vilches, C., Castaño, J., Gómez-Lozano, N. & Estefanía, E. Facilitation of KIR genotyping by a PCR-SSP method that amplifies short DNA fragments. Tissue Antigens 70, 415–422 (2007).
Graef, T. et al. KIR2DS4 is a product of gene conversion with KIR3DL2 that introduced specificity for HLA-A*11 while diminishing avidity for HLA-C. J. Exp. Med. 206, 2557–2572 (2009).
Acknowledgements
We thank Yuki Akahane and Asami Yamazaki for their technical assistance and Trevor Ralph as the Senior Editor of Impact Language Services for his English editorial assistance.
Funding
The authors sincerely appreciate the research support provided in part by the Japan Society for the Promotion of Science (JSPS) KAKENHI Grant-in-Aid for Scientific Research (C) (18K08000) (20K08282), Grant-in-Aid for Early-Career Scientists (20K17019), and Japan Agency for Medical Research and Development (AMED) (JP21fk0210084).
Author information
Authors and Affiliations
Contributions
S.J. is responsible for conceptualization, original draft preparation, formal analysis, data curation, and methodology. M.O. are responsible for sample analysis. S.J., H.K., S.W., Y.Y., A.S., T.Y., E.T., and T.U. are responsible for data curation. S.J. and T.U. are responsible for supervision.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Joshita, S., Ota, M., Kobayashi, H. et al. Association analysis of KIR/HLA genotype with liver cirrhosis, hepatocellular carcinoma, and NUC freedom in chronic hepatitis B patients. Sci Rep 11, 21424 (2021). https://doi.org/10.1038/s41598-021-01014-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-01014-x
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.