Abstract
Stephanandra incisa is a wild-type shrub with beautiful leaves and white flowers and is commonly used as a garden decoration accessory. However, the limited availability of genomic data of S. incisa has restricted its breeding process. Here, we identified EST-SSR markers using de novo transcriptome sequencing. In this study, a transcriptome database containing 35,251 unigenes, having an average length of 985 bp, was obtained from S. incisa. From these unigene sequences, we identified 5,555 EST-SSRs, with a distribution density of one SSR per 1.60 kb. Dinucleotides (52.96%) were the most detected SSRs, followed by trinucleotides (34.64%). From the EST-SSR loci, we randomly selected 100 sites for designing primer and used the DNA of 60 samples to verify the polymorphism. The average value of the effective number of alleles (Ne), Shannon’s information index (I), and expective heterozygosity (He) was 1.969, 0.728, and 0.434, respectively. The polymorphism information content (PIC) value was in the range of 0.108 to 0.669, averaging 0.406, which represented a middle polymorphism level. Cluster analysis of S. incisa were also performed based on the obtained EST-SSR data in our work. As shown by structure analysis, 60 individuals could be classified into two groups. Thus, the identification of these novel EST-SSR markers provided valuable sequence information for analyzing the population structure, genetic diversity, and genetic resource assessment of S. incisa and other related species.
Similar content being viewed by others
Introduction
Stephanandra incisa is a wild-type shrub belonging to the genus Stephanandra and is widely distributed in eastern Asia1. Due to its peculiar leaf shape, and white flowers in clusters, it is commonly used as an ornament for garden decoration. Additionally, its wood is expensive and is used for construction purposes2,3. Studies on S. incisa have primarily focused on its distribution, system evolution, cultivation, and breeding4,5. However, limited knowledge on the molecular markers of S. incisa, especially codominant markers have restricted further research efforts to understand the genetic and molecular biological processes of this species.
Molecular markers are efficient tools that facilitate genetic diversity analysis and selection of the germplasm6,7. One of the most common molecular markers include simple sequence repeats (SSR) due to their polymorphism, high stability, cross-transferable nature, and simplicity of analysis8,9. They are randomly distributed across the genome in several plant species. The SSRs are generally classified into expressed sequence tag-SSRs (EST-SSRs) and genomic-SSRs (gSSRs) based on the original sequences. EST-SSRs are more efficient as well as time-and cost-effective compared with g-SSRs10. Additionally, EST-SSRs are derived from mRNA transcript sequences, are more evolved, and exhibit more transferability between species11,12. Until now, several EST-SSRs have been identified, and used in breeding system research, construction of genetic linkage map, and genetic diversity analysis to evaluate EST-SSRs polymorphism in various plant species13,14,15. However, there are no published reports on the identification of EST-SSRs in S. incisa.
High throughput RNA-seq is a next-generation sequencing (NGS) technology, which provides large-scale genomic and transcriptomic information in the fields of conservation genetics and evolution16,17. NGS technology is also used for generating genome-based, functional data for non-model plants, such as Rhododendron latoucheae, and numerous EST sequences for gene annotation and target discovery analysis as well as comparative genomics18,19. Previously, genomic analysis was done using conventional sequencing methods; however, it had several disadvantages, such as high cost, labor-intensive, and time-consuming. The newly developed NGS technology, especially de novo transcriptome sequencing, is capable of generating large volumes of information to be used for detecting molecular markers, novel gene discovery, and identification of polymorphic loci20,21. This technique can be used for the analysis of model as well as non-model plants22. The high throughput and cost-efficient features highlight the accuracy and efficiency of the de novo transcriptome sequencing, which has made it the method of choice in life sciences research23,24. It has used to identify several EST sequences in several plants, such as Pinus bungeana, Lemna gibba, Curcuma alismatifolia, Styrax japonicus, and Sophora japonica6,14,25,26,27.
Here, the Illumina sequencing platform was used to construct the first transcriptome of S. incisa to (1) assemble and annotate the transcriptome data of S. incisa, (2) develop and identify several EST-SSRs markers for S. incisa, and (3) analyze polymorphism of these EST-SSRs markers among Laoshan (LS) and Anshan (AS) populations to further develop and utilize S. incisa EST resources and (4) analyze genetic identity based on the obtained EST-SSR data. This information will contribute to further the genetic diversity research and assessment of S. incisa.
Results
Illumina sequencing and de novo assembly
Of the 40,053,100 raw reads generated for S. incisa, 40,010,736 clean reads containing 97.98% Q20 bases were generated post strict filtration and quality control. The reads contained 5,968,100,025 nucleotides (nt), with 48.72% GC content. We identified 35,251 unigenes having an average length of 985 bp and with an N50 and N60 value of 7,212 and 23,437 bp, respectively. The unigenes were 201 to 11,634 bp long with a total length of 34,743,756 bp (Table 1). In the final unigene collection, 14,652 unigenes (41.56%) were 200 to 499 bp long, 7,881 unigenes (22.36%) were 500 to 999 bp long, 4978 unigenes (14.12%) were 1000 to 1499 bp long, 3507 unigenes (9.95%) were 1500 to 1999 bp long, and 4233 unigenes (12.01%) were longer than 2000 bp. The results showed that with an increase in the length of the unigenes sequence, there was a decrease in the number of assembled unigenes (Fig. 1).
Functional annotation and classification
We compared the unigenes obtained by transcriptome sequencing with the Swiss-Prot, Nr, KOG, and KEGG databases (Fig. 2). A total of 35,251 unigenes were annotated. The results showed that 26,604 (Nr: 75.47%), 18,140 (Swissprot: 51.46%), 14,676 (KOG: 41.63%), and 11,067 (KEGG: 31.42%) unigenes were functionally annotated. Among these, 8830 (25.42%) unigenes were annotated in both the KEGG and the KOG databases. However, only 2246 (6.37%) were annotated in the KEGG database. Additionally, 14,550 (41.28%) unigenes were annotated in both the KOG and the Nr databases (Fig. 2).
The results of the comparative analysis of the homology of the S. incisa transcriptome unigenes in the Nr database found highest homology with Prunus mume unigene. We found that 11,565 unigenes were annotated to Prunus mume, 3561 unigenes to Malus domestica, and 3189 unigenes to Pyrus x bretschneideri. It exhibited low homology with Medicago truncatula and Arabidopsis thaliana (Figure S1 and Table S1).
GO annotated unigenes were classified into the following three main categories: molecular function, cellular component, and biological processes. Unigenes in these three biological function categories included 50 level-2 categories. Amongst the biological processes, the largest group was comprised of unigenes of metabolic process (6195; 11.83%), followed by cellular process (5,517; 10.54%). Amongst the cellular components, the largest group was comprised of unigenes of cell (3401; 6.50%) and cell parts (3401; 6.50%), followed by organelle (2531; 4.83%) and membranes (1918; 3.66%). Amongst the molecular function, the largest group was comprised of unigenes of catalytic activity (10.90%, 5706), followed by binding (4362; 8.33%) (Fig. 3 and Table S2).
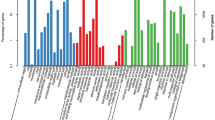
For the KOG classification, 14,676 (41.63%) annotated unigenes were classified into 25 molecular families. The dominant group was comprised of general function prediction only (3775; 16.79%), followed by signal transduction mechanisms (2,593; 11.53%), protein turnover, post-translational modification, chaperones (2540; 11.29%), and ribosomal structure, translation, and biogenesis (1274; 5.66%). The smallest group was comprised of cell motility (17; 0.076%). Other classifications were more average in proportion, mostly containing approximately 1000 unigenes (Fig. 4A and Table S3).
(A) KOG functional classification of S. incisa unigenes (The y-axis indicates the number of unigenes in a specific functional cluster. The x-axis indicates the KOG functional classification). (B) KEGG metabolic pathway of S. incisa (the y-axis is the name of the KEGG metabolic pathway, and the x-axis is the ratio of the number of genes. https://www.genome.jp/dbget-bin/www_bget?pathway+map01100).
Next, 11,067 unigenes were classified into five categories, including 133 KEGG pathways. The largest category was comprised of metabolism (8453; 76.38%), followed by the genetic information processing (2408; 21.76%), environmental information processing (524, 4.73%), cellular processes (465, 4.20%), and organismal systems (432, 3.90%) (Fig. 4B and Table S3).
Frequency and distribution of SSRs in the unigenes
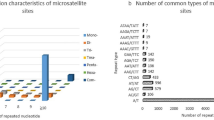
Of the 4519 unigenes, we identified 5555 potential EST-SSRs, amongst which 846 unigenes had greater than one EST-SSR locus, and 495 were identified as compound microsatellites (Table 1). The distribution density of SSRs in the S. incisa unigenes was one SSR per 1.60 kb (5555 SSRs in 34.74 Mb) at a frequency of 41.14%. The most common SSRs were di-nucleotide SSRs (2,942, 52.96%), followed by tri-nucleotide (1924, 34.64%), and hexa-nucleotide (313, 5.63%). The remaining SSRs accounted for 6.76% of the total SSRs. Additionally, the most common repeats of the EST-SSR were six tandem repeats (1317, 23.71%), followed by five tandem repeats (1137, 20.47%), seven tandem repeats (774, 13.93%), four tandem repeats (475, 8.55%), eight tandem repeats (454, 8.17%), nine tandem repeats (415, 7.47%), ten tandem repeats (365, 6.57%). The remaining tandem repeats accounted for < 12% of the EST-SSR (Fig. 5 and Table S4).
We observed a significant motif-type bias in S. incisa (Fig. 6). The most common motif in the di-nucleotide repeats was AG/CT (2739, 49.31%), followed by AC/GT (147, 2.65%), and AT/AT (51, 0.92%). The most common motif in the tri-nucleotide repeats was AAG/CTT, followed by AGC/CTG (300, 5.40%), ACC/GGT (270, 4.86%), ATC/ATG (224, 4.03%), AGG/CCT (213, 3.83%), AAC/GTT (169, 3.04%), CCG/CGG (90, 1.62%), ACG/CGT (79, 1.42%), and ACT/AGT (33, 0.59%). These 15 repeat motifs accounted for 88.91% of the total motifs, while the remaining types accounted for 11.09% (Fig. 6 and Table S5).
Development and validation of novel EST-SSRs
We used Primer5 software to design primer pairs that meet the required criteria. We randomly selected and synthesized 100 primer pairs, and initially tested the polymorphism of SSR primers. 29 primer pairs with polymorphism were separated by agarose gel (Figure S2). Finally, amplicons and amplicon polymorphisms were assessed using DNA from 60 samples. The polymorphisms of loci were detected by capillary electrophoresis (Fig. 7).
Partial capillary electrophoresis diagram of 60 samples (Genemarker2.2.0; https://genemarker.software.informer.com/2.2/).
Polymorphism of SSR loci
The results showed that there were 29 polymorphic loci in 100 loci. Additionally, we detected 90 alleles in these two populations, at an average of 3.1 alleles/locus. S21 was the locus with the largest number of alleles (6), with the values for Ne, He, and PIC as 2.933, 0.665, and 0.592, respectively. The value of Ne lied in the range of 1.130 and 3.569 with an average of 1.969. S8 has the highest value of I (1.324) and S3 had the lowest value of I (0.230) with an average value of 0.728. The value of He ranged from 0.070 to 0.723 with a mean of 0.434. The value of PIC varied from 0.108 to 0.669 with an average of 0.406. For overall PIC, nine loci had values > 0.50 (highly informative), while S01, S03, S04, S06, S13, and S19 had values < 0.25 (minimally informative), and the remaining 14 loci had values between 0.25 and 0.50 (moderately informative; Table S6).
Cluster analysis
All the amplified alleles were used for cluster analysis of the 60 individuals via the UPGMA method. A UPGMA dendrogram showed that all the individuals were divided into two clusters, in which cluster 1 contained A27, LS06, LS07, LS23 and cluster 2 contained all other individuals (Figure S3). Clearly, a high level of genetic similarity was displayed, with similarity coefficients distributed in a narrow range of 0.75–1.0. The results indicated low diversity in the screened germplasm and narrow genetic.
Discussion
SSR markers are commonly used for genetic diversity analysis, constructing cultivar DNA fingerprinting, marking assisted breeding, and studying genome-wide association28. Transcriptome sequencing has been efficiently used to develop SSR markers in several plant species. It can also be used in non-model plants and plays a crucial role in the discovery of novel genes, gene expression analysis, and for the identification of molecular markers29,30,31. S. incisa is a non-model plant with high ornamental and medicinal value. However, there are no published reports on the transcriptome sequence of S. incisa. In this study, the transcriptome of S. incisa was reported using Illumina sequencing technology. We identified 40,053,100 paired-end raw reads and 40,010,736 high-quality clean reads with a Q20 level of 97.98%, to ensure the quality of sequencing. The assembled unigenes were shorter than Saccharina japonica (44,362,190) but longer than Chinese Hawthorn (28,888,844)32,33. The average length of these unigenes was longer than that of Pinus bungeana (922 bp) and Dalbergia odorifera (676 bp)27,34. Next, these assembled unigenes were successfully annotated to registered public databases, including Swiss-Prot, Nr, KEGG, and KOG. These annotations provide useful information, which could be used for the genetic diversity analysis of S. incisa in the future. The result of the annotation showed that S. incisa was closely related to Prunus, Housefly, and Pyrus x bretschneideri, all belonging to the family Rosaceae. These results were consistent with the previous taxonomic studies. In the GO classification results, metabolic processes and catalytic activity were classified as the largest groups among the three functional categories. The prediction cluster occupied an important position in the KOG classification, consistent with previous results, followed by signal transduction mechanisms and protein turnover, posttranslational modification, chaperones categories. As for KEGG pathways database, 11,076 (31.42%) unigenes had high matches and were classified to five categories, including 133 KEGG pathways. Thus, these results provided valuable information to be used to study genetic diversity and improved adaptation of S. incisa.
Molecular markers can provide adequate genetic diversity information for plants35,36,37. EST-SSR markers have been an effective tool for analyzing gene structure, linkage mapping, and QLT analysis38. Thus, the identification of EST-SSR markers of S. incisa could promote its molecular breeding process, especially in mapping and anchoring parental maps. However, there exist no published reports on the molecular markers in S. incisa. Here, we identified 5555 EST-SSRs, among which, dinucleotide repeat sequences and trinucleotide repeat sequences accounted for a large proportion. The most common dinucleotide repeat in S. incisa was AG/CT, followed by AC/GT, and AT/AT. Although the functional significance of SSRs in plant transcript regions is unclear, the AG/CT motif, a homopurine-homopyrimidine stretch frequently found in the 5′ untranslated region, reportedly plays a vital role in regulating the gene expression and nucleic acid metabolism in plants39. The most common trinucleotide repeat was AAG/CTT, followed by AGC/CTG, and ACC/GGT. These results agreed with the results of previous research and showed that the AAG may be the most important motif in dicotyledonous plants40. Additionally, the results agreed with the previous studies on the Rosacea, such as Prunus mume41. Thus, the frequency of SSRs was closely linked to the size of the database, mining tool used, the genome structure, and the species differences42.
The location site of EST-SSR markers in the genes facilitates their use for detecting valuable genetic diversity that is possibly associated with valuable breeding traits. The EST-SSRs are considered appropriate for designing specific primers due to the high quality amplicons43. In this study, we randomly selected and synthesized 100 primer pairs, followed by the identification of 29 polymorphic genic SSR markers and 90 alleles among the two S. incisa populations. Additionally, the average values of Ne, I, and He were 1.969, 0.728, and 0.434, respectively. These results indicated that S. incisa has a medium level of genetic diversity. The PIC value is used to assess the level of genetic information44. Additionally, it is used employed to assess the polymorphic level of the markers and classified into high-level (< 0.5), moderate-level (0.5 < PIC < 0.25), and low-level (< 0.25) categories45. The value of PIC ranged from 0.108 to 0.669, which represented a middle polymorphism level. The level of polymorphism could be attributed to several factors, such as the difference in quantity in the sample. Here, polymorphism was detected based on the identified EST-SSR-containing sequences, which might be a valuable resource for the identification of genetic markers for future research on S. incisa. An admixture model-based approach was implemented to evaluate the population structure, and suggested two clusters were the best for the 60 S. incisa samples. The analysis of neighbor-joining confirmed this result. Based on the cluster analysis results, no obvious correlation of genotypes of the S. incisa to their geographical locations.
These polymorphic microsatellite markers would provide an important reference for developing polymorphic molecular markers for S. incisa and could be applied in population genetics analysis, linkage mapping, and marker-assisted selective breeding in the future. Thus, the identified EST-SSR makers were effective for the genetic analysis of the S. incisa populations. Additionally, these results could act as a new valuable resource for genomic studies on S. incisa. However, only 60 individuals from two groups were genetically analyzed. Thus, further research with an extended sample size needs to be conducted for the analysis of genetic diversity and structure of the species.
Conclusions
This is the first study to analyze the transcriptome of S. incisa. A total of 35,251 unigenes, with an average length of 985 bp, were obtained from S. incisa to create a transcriptome database, from which 5555 EST-SSRs were identified. All unigenes were annotated in the Nr (75.47%), Swiss-Prot (51.46%), KOG (41.63%), and KEGG (31.42%) databases. From these EST-SSR loci, we randomly selected 100 sites for designing primer and used the DNA of 60 samples to verify the polymorphism. Diversity parameters showed that these primer pairs had a middle polymorphism level among the two S. incisa populations. Cluster analysis indicated that 60 individuals could be classified into two groups. This is the first report to identify EST-SSR markers in S. incisa, which could be used for further genetic research and breeding approaches in S. incisa.
Methods
Plant materials and RNA isolation
We collected grown leaves of S. incisa plants from Laoshan to conduct transcriptomics analysis. The leaves were flash frozen in liquid nitrogen and stored at − 80 °C until further use. The TRIzol reagent was used for extracting total RNA following the specified protocol (Invitrogen, USA). For confirming polymorphism, 60 individual samples from the Laoshan (LS) and Anshan (AS) populations were selected, and each of the sampled individuals was kept more than 150 m apart to minimize the genetic relationships among the sample trees (Fig. 8). The fresh and healthy leaves were stored in silica gel. The total genomic DNAs were extracted using CTAB46. The extracted RNA and DNA were quantified using Thermo Scientific Nanodrop2000 spectrophotometer. Agarose gel electrophoresis was used to assess the integrity of the quantified DNA and RNA. The high-quality RNA samples were used to prepare the complementary cDNA library.
Geographic distribution of the two investigated S. incisa populations sampled from China. ArcMap10.5 (http://www.esri.com/) was used to generate the map.
Construction of cDNA library and illumina sequencing
Following the Illumina sequencing platform manufacturer’s instructions, first, the first and the second strand were synthesized. Next, the cDNA was purified, ends were repaired, ligated to the adapter, followed by library enrichment. The cDNA quality and concentration were evaluated using Qseq100 DNA Analyzer (Bioptic Inc, China). Finally, the Illumina HiSeq 2000 platform was used to sequence the cDNA library.
Sequence assembly and data analysis
The raw sequencing data can be filtered by removing the adapter contaminants, reads containing a high number of poly-N, and read data with < 20 bp. The SeqPrep program can be used to screen high-quality clean read data for the de novo assembly, empty reads, and reads with Q < 20, followed by the calculation of the GC content, sequence replication levels, along with the Q20 and Q30 values of the obtained clean data47. Thus, we filtered the raw sequencing reads (reads with unknown base ‘N’, low-quality reads with the adapter, along with other low-quality sequences) to obtain the clean reads, which were assembled using Trinity software (version 2.4.0, https://github.com/trinityrnaseq/trinityrnaseq/issues/270) to get the consensus sequences. The software combined the reads with overlapping nucleic acid sequences into contigs. The sequences that could not extend at both ends of contigs were classified as unigenes48. These assembled unigenes were used for annotation analysis. These unigenes were compared with public databases (E < 1e−5), such as NCBI non-redundant (Nr, http://www.ncbi.nlm.nih.gov/)49, Swiss-Prot (http://www.expasy.ch/sprot/)50, KOG (http://www.ncbi.nlm.nih.gov/COG), and KEGG (http://www.genome.jp/kegg/) via BLASTX (version 2.2.26, ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/) to get information on classification and functional annotation51. From the NCBI Nr database, Blast2GO was used to attain the gene ontology (GO) annotation information of unigenes to identify their molecular functions, cellular components, and biological processes52. For KEGG pathway analysis, BLASTX software was used to perform the required operations to construct KEGG database53.
Identification of EST-SSRs loci and primer design
The SSR loci contain di/tri/tetra/penta/hexa nucleotide sequences with at least 8/6/4/3/3 repeats, respectively. The Microsatellite identification tool (MISA, http://www.gramene.org/db/markers/ssrtool) was used to identify EST-SSRs54. Primer 5.0 (http://www.premierbiosoft.com/primerdesign/) was used to design the primers based on the following parameters: annealing temperature (55–60 °C), GC content (40–60%), primer lengths (18–22 bp), and target PCR product size (100–300 bp)55. All primers were synthesized by the Beijing Ruibiotech Company.
Validation and application of SSR markers
The CTAB method was used to extract total genomic DNA56. The PCR reactions were carried out in a final volume of 20 µL, which contained 2 × Mix (10 µL), reverse primer (0.15 µL), forward primer (0.15 µL), DNA (1 µL) and dd H2O up to 20 µL. PCR amplification was carried out at 95 ℃ for 5 min, followed by 35 denaturation cycles at 95 ℃ for 30 s, annealing at 52 ℃ for 30 s, and then extension at 72 ℃ for 30 s. A final extension at 72℃ and 10 min was also carried out. All the PCR products were verified through capillary electrophoresis. Electrophoretograms of capillary electrophoresis were read and analyzed through Gene Marker V2.2.0 (https://genemarker.software.informer.com/2.2/) software.
Statistical analysis
The number of observed alleles (Na), expected heterozygosity (He), the number of effective alleles (Ne), and Shannon’s information index (I)57 were calculated using the POPGENE V1.32 (https://www.softpedia.com/get/Science-CAD/Popgene-Population-Genetic-Analysis.shtml) software. The Polymorphism information content (PIC) was calculated using the POWER-MARKER v3.25 (http://statgen.ncsu.edu/powermarker/index.html)45. Unweighted pair group (UPGMA) in NTSYS 2.1 (https://en.freedownloadmanager.org/Windows-PC/NTSYSpc.html) was used to cluster the populations of S. incisa, and a dendrogram was generated from this clustering.
Data availability
The dataset is available from the NCBI Short Read Archive (SRA) with accession number SRR12158738.
References
Huang, Y., Zhang, J., Liu, Q., Wang, K. & Liu, Q. Research on shade tolerance of Stephanandra incisa. Chin. Agric. Sci. Bull. 16, 77–81 (2012).
Li, B. & Gong, W. Resources of wild woody medicinal plants in Qingdao Laoshan Coastal Zone. J. Oceanogr. Huanghai Bohai Seas 18, 56–65 (2000).
Fang, J. & Cheng, Z. Preliminary study on wild woody ornamental plant resources of rosaceae exploitation and utilization. Resour. Dev. Market 24, 452–454 (2008).
Oh, S. H. Neillia includes Stephanandra (Rosaceae). Novon J. Bot. Nomenclature 16, 91–95 (2006).
Oh, S. H. & Potter, D. Phylogenetic utility of the second intron of LEAFY in Neillia and Stephanandra (Rosaceae) and implications for the origin of Stephanandra. Mol. Phylogenet. Evol. 29, 203–215. https://doi.org/10.1016/s1055-7903(03)00093-9 (2003).
Li, W. et al. De novo transcriptomic analysis and development of EST-SSRs for Styrax japonicus. Forests 9 (2018).
Al-Ashkar, I., Alderfasi, A. & Ben Romdhane, W. Morphological and genetic diversity within salt tolerance detection in eighteen wheat genotypes. Plants (Basel) 9, https://doi.org/10.3390/plants9030287 (2020).
Shu, W. et al. Analysis of genetic diversity and population structure in Sophora japonica Linn. in China with newly developed SSR markers. Plant Mol. Biol. Rep. 37, 87–97 (2019).
Sharma, H. et al. Development of polymorphic EST-SSR markers and their applicability in genetic diversity evaluation in Rhododendron arboreum. Mol. Biol. Rep. 1–11 (2020).
Vasek, J. et al. New EST-SSR markers for individual genotyping of opium poppy cultivars (Papaver somniferum L.). Plants (Basel) 9, https://doi.org/10.3390/plants9010010 (2019).
Wang, X. et al. Development of EST-SSR markers and their application in an analysis of the genetic diversity of the endangered species Magnolia sinostellata. Mol. Genet. Genomics 294, 135–147 (2019).
Zhang, J., Liu, T. & Rui, F. Development of EST-SSR markers derived from transcriptome of Saccharina japonica and their application in genetic diversity analysis. J. Appl. Phycol. 30, 2101–2109 (2018).
He, D. et al. Development of SSR markers in Paeonia based on De novo transcriptomic assemblies. PLoS ONE 15, e0227794 (2020).
Lu, Y. et al. Development of EST-SSR markers and their application in the analysis of the genetic diversity of Sophora japonica Linn. Trees https://doi.org/10.1007/s00468-020-01985-w (2020).
Banjanac, T. et al. Phenotypic and genetic variation of an interspecific Centaurium Hybrid (Gentianaceae) and its parental species. Plants (Basel) 8, https://doi.org/10.3390/plants8070224 (2019).
Köse, M. A., Çetinsağ, N. & Gürcan, K. De novo transcriptome assembly and SSR marker development in apricot (Prunus armeniaca). Turk. J. Agric. For. 41, 305–315 (2017).
Fan, J., Lou, Y., Shi, H., Chen, L. & Cao, L. Transcriptomic analysis of dark-induced senescence in bermudagrass (Cynodon dactylon). Plants (Basel) 8, https://doi.org/10.3390/plants8120614 (2019).
Wu, T. et al. The first Illumina-based de novo transcriptome sequencing and analysis of pumpkin (Cucurbita moschata Duch.) and SSR marker development. Mol. Breed. 34, 1437–1447, https://doi.org/10.1007/s11032-014-0128-x (2014).
Zhang, L., Wan, X., Xu, J., Lin, L. & Qi, J. D. novo assembly of kenaf (Hibiscus cannabinus) transcriptome using illumina sequencing for gene discovery and marker identification. Mol. Breed. 35, 192. https://doi.org/10.1007/s11032-015-0388-0 (2015).
Rai, H. S. et al. Transcriptome characterization and detection of gene expression differences in aspen (Populus tremuloides). Tree Genet. Genomes 9, 1031–1041. https://doi.org/10.1007/s11295-013-0615-y (2013).
Yoo, M. J., Wendel, J. F. & Bomblies, K. Comparative evolutionary and developmental dynamics of the cotton (Gossypium hirsutum) fiber transcriptome. PLoS Genet. 10, e1004073 (2014).
Gonzálezballester, D. et al. RNA-seq analysis of sulfur-deprived Chlamydomonas cells reveals aspects of acclimation critical for cell survival. Plant Cell 23, 1679–1681 (2011).
Zhou, Q. et al. Development and cross-species transferability of EST-SSR markers in Siberian wildrye (Elymus sibiricus L.) using Illumina sequencing. Sci. Rep. 6, 20549 (2016).
Davey, J. W. et al. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat. Rev. Genet. 12, 499–510. https://doi.org/10.1038/nrg3012 (2011).
Sima et al. De novo assembly of transcriptomes, mining, and development of novel EST-SSR markers in Curcuma alismatifolia (Zingiberaceae family) through Illumina sequencing. Sci. Rep. (2019).
Fu, L., Ding, Z., Kumpeangkeaw, A., Tan, D. & Zhang, J. De novo assembly, transcriptome characterization, and simple sequence repeat marker development in duckweed Lemna gibba. Physiol. Mol. Biol. Plants 26 (2019).
Cai, Q. et al. De novo sequencing and assembly analysis of transcriptome in Pinus bungeana Zucc. ex Endl. Forests 9, 156 (2018).
Carneiro, V. M. L., Luciane, S., Lima, D. A. & Freitas, M. C. D. Microsatellite markers: What they mean and why they are so useful. Genet. Mol. Biol. 39, 312–328 (2016).
An, M., Deng, M., Zheng, S. & Song, Y. De novo transcriptome assembly and development of SSR markers of oaks Quercus austrocochinchinensis and Q-kerrii (Fagaceae). Tree Genet. Genomes 103 (2016).
Dai, F. et al. De novo assembly, gene annotation, and marker development of mulberry (Morus atropurpurea) transcriptome. Tree Genet. Genomes 11, 26–26 (2015).
Yan, J. et al. De novo transcriptome sequencing and gene expression profiling of spinach (Spinacia oleracea L.) leaves under heat stress. Sci. Rep. 6, 19473 (2016).
Zhang, J., Liu, T. & Rui, F. Development of EST-SSR markers derived from transcriptome of Saccharina japonica and their application in genetic diversity analysis. J. Appl. Phycol. (2017).
Ma, S., Dong, W., Lyu, T. & Lyu, Y. An RNA sequencing transcriptome analysis and development of EST-SSR markers in Chinese Hawthorn through illumina sequencing. Forests 10, 82 (2019).
Liu, F. et al. De novo transcriptome analysis of Dalbergia odorifera and transferability of SSR markers developed from the transcriptome. Forests 10 (2019).
Sanabam, R., Singh, N. S., Sahoo, D. & Devi, H. S. Genetic relationship of rough lemon landraces and under-utilised citrus genotypes from North-East India revealed by SSR and RAPD markers. Trees (2018).
Mohammad-Panah, N., Shabanian, N., Khadivi, A., Rahmani, M. S. & Emami, A. Genetic structure of gall oak (Quercus infectoria) characterized by nuclear and chloroplast SSR markers. Tree Genet. Genomes 13, 1–12 (2017).
Chhajer, S., Jukanti, A. K., Bhatt, R. K. & Kalia, R. K. Genetic diversity studies in endangered desert teak [Tecomella undulata (Sm) Seem] using arbitrary (RAPD), semi-arbitrary (ISSR) and sequence based (nuclear rDNA) markers. Trees (2018).
Pinto, M. V., Poornima, H. S., Sivaprasad, V. & Naik, V. G. A new set of mulberry-specific SSR markers for application in cultivar identification and DUS testing. J. Genet. (2018).
Martienssen, R. A. & Colot, V. DNA methylation and epigenetic inheritance in plants and filamentous fungi. Science (New York, N.Y.) 293, 1070–1074, https://doi.org/10.1126/science.293.5532.1070 (2001).
Cavagnaro, P. F. et al. Genome-wide characterization of simple sequence repeats in cucumber (Cucumis sativus L.). BMC Genomics 11, 569, https://doi.org/10.1186/1471-2164-11-569 (2010).
Gong, Q. et al. On the characteristics of EST-derived SSR marker development of Prunus mume and its transferability to Prunus cerasifera. J. Beijing Univ. Agric. (2017).
Biswas, M. K., Chai, L., Mayer, C. & Xu, Q. Exploiting BAC-end sequences for the mining, characterization and utility of new short sequences repeat (SSR) markers in Citrus. Mol. Biol. Rep. (2012).
Jia, X., Deng, Y., Sun, X., Liang, L. & Su, J. D. novo assembly of the transcriptome of Neottopteris nidus using Illumina paired-end sequencing and development of EST-SSR markers. Mol. Breed. 36, 94 (2016).
Wu, J., Cai, C., Cheng, F., Cui, H. & Zhou, H. Characterisation and development of EST-SSR markers in tree peony using transcriptome sequences. Mol. Breed. 34, 1853–1866 (2014).
Botstein, D., White, R. L., Skolnick, M. & Davis, R. W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 32, 314–331 (1980).
Yang, W. CTAB DNA extraction protocol of P. pruinosa. https://doi.org/10.17504/PROTOCOLS.IO.ICDCAS6 (2017).
Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. https://doi.org/10.1038/nbt.1883 (2011).
Yue, Z., Xue, Z., Wang, Y. & Shen, S. De novo assembly of transcriptome and development of novel EST-SSR markers in Rhododendron rex Lévl. through illumina sequencing. Front. Plant Sci. 8, 1664 (2017).
Pruitt, K. D., Tatiana, T. & Maglott, D. R. NCBI reference sequence (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 35 (2005).
UniProt Consortium, T. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 46, 2699–2699. https://doi.org/10.1093/nar/gky092 (2018).
Camacho, C., Coulouris, G., Avagyan, V., Ma, N. & Papadopoulos, J. BLAST+: architecture and applications. BMC Bioinform. 10, 1–9 (2009).
Conesa, A. et al. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676 (2005).
Kanehisa, M., Goto, S., Kawashima, S., Okuno, Y. & Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 32, D277-280. https://doi.org/10.1093/nar/gkh063 (2004).
Huang, L., Yang, X., Sun, P., Tong, W. & Hu, S. The first illumina-based de novo transcriptome sequencing and analysis of safflower flowers. PLoS ONE 7, e38653. https://doi.org/10.1371/journal.pone.0038653 (2012).
Xin-Yu, Z. & Yan-Ning, G. To design PCR primers with oligo 6 and Primer Premier 5. Bioinformatiocs (2004).
Doyle, J. J. T. & Doyle, J. L. Isolation of plant DNA from fresh tissue. Focus 12 (1990).
Srivastava, V. et al. OnPLS integration of transcriptomic, proteomic and metabolomic data shows multi-level oxidative stress responses in the cambium of transgenic hipI-superoxide dismutase Populus plants. BMC Genomics 14, 893. https://doi.org/10.1186/1471-2164-14-893 (2013).
Acknowledgements
This work was supported by Shandong Provincial Major Agricultural Application Technology Innovation Project (LCNZ2016-36); High-level Science Foundation of Qingdao Agricultural University [663/1114306]. We thank the National Forest Genetic Resources Platform (NFGRP) for providing the Stephanandra incisa resources.
Author information
Authors and Affiliations
Contributions
K.W. conceived and designed the experiments; Z.W. and C.Z. performed the experiments, conducted the data analysis, and wrote the manuscript; W.L. revised the manuscript; X.J. contributed the plant material; and Y.L. analyzed the data.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, C., Wu, Z., Jiang, X. et al. De novo transcriptomic analysis and identification of EST-SSR markers in Stephanandra incisa. Sci Rep 11, 1059 (2021). https://doi.org/10.1038/s41598-020-80329-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-80329-7
This article is cited by
-
Development of EST-SSR Markers and Population Genetic Analysis of Hemsleya zhejiangensis, an Endangered Species Endemic to Eastern China
Plant Molecular Biology Reporter (2024)
-
Development of unigene-derived SSR markers from RNA-seq data of Uraria lagopodioides (Fabaceae) and their application in the genus Uraria Desv. (Fabaceae)
BMC Plant Biology (2023)
-
Genetic diversity, population structure, and DNA fingerprinting of Ailanthus altissima var. erythrocarpa based on EST-SSR markers
Scientific Reports (2023)
-
Development of EST-SSR Markers and Genetic Diversity Analysis Among Wild Pistachio Species
Erwerbs-Obstbau (2023)
-
Analysis of the genetic diversity and population structure of Monochasma savatieri Franch. ex Maxim using novel EST-SSR markers
BMC Genomics (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.