Abstract
The leaf curl disease of Jatropha caused by geminiviruses results in heavy economic losses. In the present study, we report the identification of a new strain of a Jatropha leaf curl Gujarat virus (JLCuGV), which encodes six ORFs with each one having RNA silencing suppressor activity. Therefore, three artificial microRNAs (amiRNAs; C1/C4, C2/C3 and V1/V2) were designed employing overlapping regions, each targeting two ORFs of JLCuGV genomic DNA and transformed in tobacco. The C1/C4 and C2/C3 amiRNA transgenics were resistant while V1/V2 amiRNA transgenics were tolerant against JLCuGV. The relative level of amiRNA inversely related to viral load indicating a correlation with disease resistance. The assessment of photosynthetic parameters suggests that the transgenics perform significantly better in response to JLCuGV infiltration as compared to wild type (WT). The metabolite contents were not altered remarkably in amiRNA transgenics, but sugar metabolism and tricarboxylic acid (TCA) cycle showed noticeable changes in WT on virus infiltration. The overall higher methylation and demethylation observed in amiRNA transgenics correlated with decreased JLCuGV accumulation. This study demonstrates that amiRNA transgenics showed enhanced resistance to JLCuGV while efficiently maintaining normalcy in their photosynthesis and metabolic pathways as well as homeostasis in the methylation patterns.
Similar content being viewed by others
Introduction
Viruses have emerged as major plant pathogens causing severe losses to agricultural production worldwide1. Geminiviridae is the second largest family of plant viruses2, possessing the potential to exploit a broad range of plants as its host. The geminiviruses are characterized by geminate virus particles that are categorized into nine genera based on their genome organization, insect vector, host range and phylogeny reconstruction, viz., Becurtovirus, Begomovirus, Curtovirus, Eragrovirus, Mastrevirus, Topocuvirus, Turncurtovirus, Capulavirus and Grablovirus. Based on the number of DNA molecules in the viral genome, they can be: monopartite (one DNA molecule) or bipartite (two DNA molecules: DNA-A and DNA-B)3. The viral genome in monopartite viruses and the DNA-A component of bipartite viruses are homologous and have a similar organization that includes the V1/AV1, C1/AC1, C2/AC2, C3/AC3, and C4/AC4 ORFs3.
The geminiviruses infect a wide range of economically important crops like cassava, cotton, grain legumes and tomato, causing significant financial losses across the world. These economic losses have been estimated to about US$ 1300–2300 million for cassava in Africa4, US$ 5 billion for cotton in Pakistan, US$ 300 million for grain legumes in India5 and the US$ 140 million in Florida for tomato alone6. The DNA viral mutation rates roughly range between 10–8 and 10–6 substitutions per nucleotide per cell infection7. This high rate of recombination and mutation leads to their great diversity worldwide8, thereby increasing their host range and geographical distribution. Thus, geminiviruses are emerging as widely spread and diverse plant DNA viruses posing severe threat to crop production.
Jatropha, belonging to family Euphorbiaceae, has various medicinal properties9. It has also gained importance as a biodiesel producing crop as Jatropha seeds contain 27–40% oil10. The oil yield is affected by attacks of various pathogens such as viruses, bacteria, fungi and nematodes. The leaf curl disease in Jatropha, caused by the whitefly transmitted geminiviruses, is the most devastating. In India, it causes a reduction of up to 80% in seed yield11 and 40% in oil production12. The symptoms of the disease include mosaic and curling of leaves and malformed fruits. To date, both monopartite and bipartite geminiviruses have been detected in Jatropha from India, Kenya, Nigeria, Dominic Republican, Jamaica and the United States of America (USA)13.
For generating resistance to geminiviruses, various molecular strategies based on RNA silencing and gene editing have been reported including antisense RNAs14, pathogen-derived resistance15, expression of double-stranded (ds) RNA homologues16 and CRISPR/Cas9 system17. The RNA silencing mechanism in plants forms the first line of defence by targeting the viral mRNA or the RNA genome18. Interestingly, viruses have evolved to acquire a variety of unrelated RNA silencing suppressors (RSSs) that affect different yet multiple steps in the silencing pathway19 to favour virus replication. Several reports have demonstrated that different geminiviral proteins encode RSS activity20,21,22 and may supplement each other to overcome host RNA silencing response. A molecular strategy to target the virus-encoded suppressors is expected to boost plant tolerance to infection. The artificial miRNA (amiRNA) provide a highly specific alternative to silence the viral transcripts for targeting the RSS for developing disease resistance23.
Therefore, in this study, we adopted the strategy of identifying and inactivating the virus-encoded RSSs to boost Jatropha plant resistance against geminiviruses. For this purpose, amiRNAs were designed from genomic regions producing overlapping ORFs to target multiple viral transcripts simultaneously.
Results
Characterization of geminivirus infecting Jatropha curcas
The geminivirus infected leaves were collected from J. curcas CP9 accession (IC 565735), from CSIR-CSMCRI Jatropha plantation fields, Neswad (Gujarat, India). The infected plants showed severe to moderate leaf curling with > 70% disease incidence.
For identification of the virus, the infected leaves were analyzed for the presence of DNA-A, DNA-B and DNA-β. Only DNA-A was amplified, and no amplification was observed for DNA-B and DNA-β. This led us to infer that the viral species infecting Jatropha CP9 accession was probably monopartite. For confirmation, the complete genome was amplified by RCA (rolling circle amplification) and sequenced. Sequence analysis confirmed the presence of only one genomic DNA (KM411359, 2758 bp) in the isolated virus. Based on sequence homology to other begomoviruses, it was identified to be a novel isolate and named as Jatropha leaf curl Gujarat virus (JLCuGV). Its genomic DNA possessed two ORFs in the sense strand (AV1 and AV2) and four ORFs in complementary strand (AC1, AC2, AC3 and AC4; see Supplementary Fig. S1).
JLCuGV genes function as RNA silencing suppressors
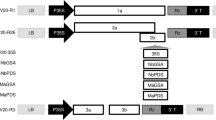
For identification of potential RSS in the JLCuGV, the six ORFs of the genome (AV1, AV2, AC1, AC2, AC3 and AC4) were individually cloned in pCAMBIA-1301 (Fig. 1a) and were independently agroinfiltrated in the in-house-developed stable green fluorescent protein (GFP)-silenced tobacco lines. The presence of strong green fluorescence indicated a reversal of GFP silencing. The assay was performed using the well-characterized RSS, FHVB2 (Flock house virus B2 gene), as a positive control24 and empty vector and mock (only buffer) infiltration as a negative control [Fig. 1b(iv)]. The infiltrated leaf patches were observed under UV at regular intervals of 7, 11 and 15 dpi (days post-inoculation; Fig. 1b(i–iii)). The GFP fluorescence was obtained with all six JLCuGV genes indicating that all the ORFs contained inherent RSS activity. This was confirmed by checking for the presence of GFP transcripts in the corresponding infiltrated regions (Fig. 1c). The cDNA generated from the leaves infiltrated with FHVB2 or individual JLCuGV ORFs showed a ~ 300 bp band for GFP transcript, but no band amplified from the mock control. However, detailed molecular analysis is required to characterize the strength, duration and complementation of the RSS activity exhibited by individual ORFs.
Identification of RNA silencing suppressor activity in the ORFs of JLCuGV genomic DNA. (a) The schematic representation to show the cloning strategy of individual JLCuGV ORFs in pCAMBIA-1301. (b) Pictures of leaves showing restoration of GFP fluorescence on agroinfiltration of AV1, AV2, AC1, AC2, AC3 and AC4 after (i) 7 dpi (ii) 11 dpi and (iii) 15 dpi. Flock house virus B2 (B2) gene was used as a positive control in each case. (iv) control panel showing the leaves infiltrated with Flock house virus B2 (B2) gene as a positive control and empty vector (EV) and Mock (only buffer) as a negative control at 15 dpi. (c) Amplification of gfp gene (upper panel) and actin gene (loading control; lower panel) from the cDNA isolated from leaves agroinfiltrated independently with each JLCuGV ORFs (AV1, AV2, AC1, AC2, AC3 and AC4), B2: Flock house virus B2 gene (positive control), C: Control mock (only buffer infiltrated leaf).
Tobacco transgenic plants expressing amiRNAs display resistance against JLCuGV
Confirmation of the transgenics
For developing an effective antiviral strategy, amiRNA constructs were designed to target the RSS. In the genomic region, the ORFs arise from overlapping regions (C1/C4, C2/C3 and V1/V2); so, these regions were selected as targets for the designing of amiRNAs. Three amiRNAs (C1/C4, C2/C3 and V1/V2) were designed in silico (Table 1). The amiRNA constructs were generated by overlapping PCRs and cloned in pCAMBIA-1301 backbone containing the gus gene under constitutive promoters (Supplementary Fig. S2). The constructs were transformed in Nicotiana benthamiana plants by Agrobacterium tumefaciens. The transgenic leaves of T1 plants showed complete or partial blue colouration for GUS assay, indicating positive transgenic lines (L61, L64, L67, L73, L79, L95 of C1/C4 amiRNA; L41, L42, L44, L48, L51, L58 of C2/C3 amiRNA; L13, L14, L15, L17, L19 and L21 of V1/V2 amiRNA; Supplementary Fig. S3a). Molecular analysis using gene-specific PCR primers confirmed the presence of gus (1.2 kb), hptII (1.2 kb) and amiRNA (259 bp) genes (Supplementary Fig. S3b). Three putative single-copy transgenic lines of each amiRNA construct (L61, L64 and L67 of C1/C4; L41, L42 and L44 of C2/C3; L13, L14 and L15 of V1/V2 amiRNA) were selected for further analysis (Supplementary Fig. S3c).
Transgenics show resistance against JLCuGV
To analyze the tobacco plants producing amiRNAs for resistance to virus infection, the agroinfectious clone of JLCuGV (Supplementary Fig. S4) was used for infiltration. The agroinfiltration in WT and VA (vector alone) plants served as control. All the WT (total 3 out of 3 inoculated plants) and VA (total 3 out of 3 inoculated plants) plants showed curl-mosaic symptoms after 21 dpi, and the symptoms increased with an increase in time (Fig. 2a,b). Visible symptoms were not observed in the C1/C4 (L61, L64, L67) and C2/C3 (L41, L42, L44) amiRNA transgenic plants (9 out of 9 plants of each C1/C4 and C2/C3 transgenics; Fig. 2c,d), but 3 out of 9 plants expressing amiRNA V1/V2 (L13, L14, L15) showed weak symptoms on leaves (Fig. 2e).
Characterization of the N. benthamiana transgenics overexpressing the amiRNA against C1/C4, C2/C3 and V1/V2 ORFs, respectively. Representative pictures to show viral symptoms in (a) WT, (b) VA, (c) C1/C4, (d) C2/C3 and (e) V1/V2 amiRNA transgenics after 28 dpi. Arrow indicates yellow mosaic/ leaf distortion symptoms. (f) PCR amplification of genomic DNA (upper panel) and actin gene as a control (lower panel) in agroinfiltrated WT, VA and transgenics for detection of JLCuGV in infected samples (at 28 dpi). (g) HindIII digested RCA products of the agroinfiltrated C1/C4, C2/C3, V1/V2 transgenics, WT and VA for detection of JLCuGV in infected samples (at 28 dpi). (h) The qRT-PCR based estimation of viral load in the infiltrated plants of WT, VA and V1/V2 transgenics after 14, 21, 28 and 35 dpi. (i) Relative expression of amiRNA in transgenics. Values are represented as means ± SD (n = 3) and marked with different alphabets (from ‘a’ to ‘e’) to indicate a significant difference as determined by Tukey test at P ≤ 0.05 probability.
At 28 dpi, the genomic DNAs from all these plants were checked for the presence of viral DNA by PCR (450 bp) using specific primers (Fig. 2f) and reconfirmed by checking for genomic DNA amplicon (2.7 kb) obtained by HindIII digestion of RCA product (Fig. 2g). The actin gene was amplified as loading control (Fig. 2f,g). The DNA bands were observed in virus-infected WT and VA plants, but no amplification was seen in case of C2/C3 and C1/C4 amiRNA transgenics. In virus-infected V1/V2 amiRNA transgenic plants, a faint band was obtained.
The WT, VA and V1/V2 amiRNA transgenics, which were found positive for JLCuGV infection, were further checked for the viral load using qRT-PCR for the quantitative determination of infection severity. The viral load of WT and VA was 4959 and 4999 copies/ng, respectively at 14 dpi, which increased by 1.93- and 1.89-fold respectively at 35 dpi (Fig. 2h). However, the viral load of V1/V2 amiRNA transgenics (5364 copies/ng at 14 dpi) decreased by 0.88-fold at 35 dpi (Fig. 2h).
For determining the correlation between the expressed amiRNA levels in transgenics with the JLCGV infection, the quantitative estimation of amiRNA was performed in transgenics using stem-loop qRT-PCR. The level of amiRNA increased from 1.3-fold (C1/C4), 1.15-fold (C2/C3) and 0.97-fold (V1/V2) at 14 dpi to 5.4-fold (C1/C4), 5.4-fold (C2/C3), 2.88-fold (V1/V2) at 35 dpi in amiRNA transgenics (Fig. 2i).
Methylation profiling of transgenics expressing amiRNA
The genomic DNA from T1 amiRNA transgenics of C1/C4 (L61, L67), C2/C3 (L42, L43), V1/V2 (L13, L15), VA and WT plants were used for MSAP (methylation-sensitive amplified polymorphism) analysis with 20 primers, which were earlier characterized to be best polymorphic MSAP primers in tobacco (More et al.25). Among 20 primers, 15 primers (Supplementary Table S1) showed consistent and clear profiles. Out of 2520 bands in total, 1528 bands showed methylation. The global methylation events were higher in transgenics as compared to WT and VA. The C1/C4 amiRNA transgenic exhibited very high methylation polymorphism (L67 = 78.73%, L61 = 73.97%), full methylation (L67 = 56.83%, L61 = 40%) and hemi-methylation (L61 = 33.97%, L67 = 21.90%). C2/C3 amiRNA transgenics also showed > 60% methylation polymorphism (L43 = 67.94%, L42 = 61.90%), > 40% full methylation (L42 = 40.63%, L43 = 40.63%) and > 20% hemi-methylation (L43 = 27.30%, L42 = 21.27%). The V1/V2 amiRNA transgenics showed lower values than C1/C4 and C2/C3 amiRNA transgenics for methylation polymorphism (L13 = 54.60%, L15 = 52.06%), full methylation (L15 = 37.46%, L13 = 36.83%) and hemi-methylation (L13 = 17.78%, L15 = 14.60%). The lowest values for methylation patterns (full methylation: 36% and hemi-methylation: 9%), were observed in WT (Table 2).
For analyzing the changes in methylation patterns of transgenics in comparison to WT, the total bands were further classified as “A to P” (16 combinations; Table 3) based on their banding pattern. The banding pattern was classified into three types: no change (A to D), methylation (E to J) and demethylation (K to P; Table 3). The pairwise comparison between transgenics and WT revealed maximum “no change” and minimum “no change” in VA (65.03%) and C1/C4 amiRNA transgenics L67 (28.76%), respectively (Table 3). The L43 of C2/C3 amiRNA transgenics showed maximum change in methylation (14.51%) pattern compared to WT, whereas the lowest change in methylation pattern (9.59%) was observed in VA. The demethylation percentage was found to be maximum in L67 (45.08%) of C1/C4 amiRNA transgenics, while the lowest demethylation in VA (13.47%) in comparison to WT (Table 3).
Improved photosynthesis parameters in amiRNA expressing transgenics on JLCuGV infiltration
For understanding the effect of virus infection on the photosynthesis process, the WT, VA and amiRNA transgenics (C1/C4, C2/C3 and V1/V2) were infected (in 3 replicates) with JLCuGV and analyzed for various photosynthetic parameters.
Gas exchange related photosynthetic parameters
The photosynthesis rate decreased significantly on JLCuGV infection in WT and V1/V2 transgenics, while no significant change was observed in C1/C4 and C2/C3 transgenics. In WT, the photosynthesis rate showed a significant decrease of 2.2- and 3.9-fold after 28 dpi and 35 dpi, respectively, as compared to control plants. The virus infiltrated transgenics showed a significantly better photosynthesis rate than the WT at all time points. Among the transgenics, the agroinfiltrated V1/V2 transgenics showed 1.34- and 1.55-fold decrease in photosynthesis rate after 28 dpi and 35 dpi, respectively, as compared to the control plant (Fig. 3a). In C1/C4, C2/C3 and V1/V2 transgenics, the transpiration rate increased significantly by 1.76-, 1.12- and 1.76-fold, respectively, at 28 dpi. The water use efficiency (WUE) and stomatal conductance increased significantly by 4.53-fold (C1/C4 transgenics), 3.11-fold (C2/C3 transgenics) and 2.28-fold (V1/V2 transgenics) by 35 dpi and 1.92-fold (C1/C4 transgenics), 1.12-fold (C2/C3 transgenics) and 1.93-fold (C1/C4 transgenics) at 28 dpi, respectively, as compared to WT (Fig. 3b–d). The internal CO2 (Ci) and the ratio of intercellular to ambient CO2 (Ci/Ca) increased in virus infiltrated WT and V1/V2 plants, but decreased in C2/C3 and C1/C4 transgenics, as compared to their respective uninfected control plants (Fig. 3e,f). At 35 dpi, the virus infiltrated WT plants showed increased Ci and Ci/Ca values as compared to C1/C4 (1.93- and 1.87-fold, respectively), C2/C3 (1.46- and 1.56-fold, respectively), V1/V2 (1.17- and 1.17-fold, respectively).
Analysis of gas exchange related photosynthesis parameters of WT, C1/C4, C2/C3 and V1/V2 amiRNA plants with and without JLCuGV agroinfiltration (a) Photosynthesis rate, (b) Transpiration rate, (c) Water use efficiency, (d) Stomatal conductance, (e) Ci, (f) Ci/Ca ratio, (g) ETR and (h) ΦCO2. Values are represented as means ± SD (n = 3) and marked with different alphabets to indicate a significant difference as determined by Tukey test at P ≤ 0.05 probability.
On the contrary, the electron transport rate (ETR) and CO2 assimilation (ɸCO2) decreased in WT and V1/V2 transgenics and increased in C1/C4 and C2/C3 transgenics after virus infiltration. Upon virus infiltration (35 dpi), ETR and ɸCO2 increased significantly in C1/C4 (4.12- and 4.47-fold, respectively), C2/C3 (3.46- and 3.63-fold, respectively) and V1/V2 transgenics (1.82- and 2.39-fold, respectively) than WT (Fig. 3g,h).
Chlorophyll fluorescence related photosynthetic parameters
The efficiency of photosystem II (Fv/Fm) was similar in both WT and transgenics with and without virus infiltration (Fig. 4a). The quantum yield of PSII electron transport (ɸPSII) and photochemical quenching (qP) increased significantly, while 1-qP significantly decreased in C1/C4, C2/C3 and V1/V2 amiRNA transgenics than WT after virus infiltration at all time points. ɸPSII increased in virus infiltrated C1/C4, C2/C3 and V1/V2 transgenics by 4.12-, 3.11- and 1.82-fold, respectively than WT at 35 dpi (Fig. 4b). Similarly, at 35 dpi, qP also increased on virus infiltration in C1/C4, C2/C3 and V1/V2 amiRNA transgenics by 4.65-, 3.53- and 2.22-fold, respectively, as compared to WT (Fig. 4c). The comparison of the virus infiltrated plant with its control plant showed that ɸPSII and qP decreased in WT and V1/V2 transgenics on virus infiltration, whereas it increased in C1/C4 and C2/C3 amiRNA transgenics. The 1-qP decreased significantly in virus infiltrated C1/C4, C2/C3 and V1/V2 amiRNA transgenics by 1.33-, 1.21- and 1.09-fold, respectively, as compared to WT at 35 dpi (Fig. 4d).
Analysis of chlorophyll fluorescence parameters of WT, C1/C4, C2/C3 and V1/V2 amiRNA plants with and without JLCuGV agroinfiltration. (a) ΦPSII, (b) Fv/Fm ratio (c) qP and (d) 1-qP. Values are represented as means ± SD (n = 3) and marked with different alphabets to indicate a significant difference as determined by Tukey test at P ≤ 0.05 probability.
Metabolite profiling of amiRNA expressing transgenics
Metabolite analysis was performed in both WT and amiRNA transgenics with and without JLCuGV agroinfiltration. Twenty-one common metabolites were detected in both healthy and virus infiltrated tissue (Supplementary Table S2). The identified metabolites were categorized into eight different groups like alkenes, amino acids, carboxylic acids, fatty acids, fatty alcohols, polyols, polyphenols and sugars. Sugars formed the largest group, followed by carboxylic acids and polyols. In WT, the concentration of sugar was more on geminivirus infiltration (68%) than the healthy tissue (54%), while the carboxylic acids were more in healthy (19%) than infected tissue (6%; Fig. 5a). In C1/C4 transgenics, healthy and virus infiltrated tissue showed less change in concentration of sugars (21% and 28%, respectively), carboxylic acids (36% and 34%, respectively) and polyols (38% and 33%, respectively; Fig. 5b). Also, in C2/C3 transgenics, healthy and virus infiltrated tissue showed less change in concentration of sugars (39% and 41%, respectively), carboxylic acids (27% and 21%, respectively) and polyols (32% and 30%, respectively; Fig. 5c). While in V1/V2 transgenics, the concentration of sugar was more on geminivirus infiltration (56%) than the healthy tissue (40%), while the carboxylic acids were higher in healthy (25%) than infected tissue (11%; Fig. 5d). The other compounds like polyphenols, fatty acids, fatty alcohols, alkenes and amino acids were less than 6% in both WT and transgenics with healthy and infected tissue.
Comparison of different metabolite responses in (a) WT, (b) C1/C4, (c) C2/C3 and (d) V1/V2 amiRNA transgenics with and without (control) JLCuGV agroinfiltration, (e) heat map of metabolomics with row represents the metabolite and column represents the sample. The metabolite concentrations are shown in different colours.
In amiRNA transgenics, except V1/V2 transgenics, no significant changes were observed in the concentration of metabolites in healthy (control) and virus infiltrated tissues. In WT, C2/C3 and V1/V2 transgenics, fructose and glucose content increased, with a maximum increase of 8.76- and 8.10-fold, respectively, in WT on infiltration. The C1/C4, however, showed a decrease in fructose and glucose content on virus infiltration. The sucrose content increased in transgenics (1.19–1.26-fold), whereas decreased by 1.95-fold in WT with virus infiltration (Table 4). The malic and quinic acid content increased in C1/C4 and C2/C3 transgenics, whereas decreased in WT and V1/V2 transgenics on virus infiltration. The quinic acid content showed a noticeable increase of 3.15-fold and a decrease of 4.29-fold in C2/C3 and V1/V2 transgenics, respectively. The pyruvic acid showed a reduction in both WT and transgenics, with a remarkable decrease of 17.7-fold in WT on virus infiltration (Table 4). In WT, C2/C3 and V1/V2 transgenics, an increase in myo-inositol content (1.23–2.22-fold) was observed. The glycerol content decreased in both WT and transgenics, with a maximum decrease of 4.77-fold in C1/C4 transgenics on virus infiltration (Table 4).
Out of all the detected metabolites in the healthy tissue of WT, sucrose (45.09%) was detected in the highest amount, followed by myo-inositol (14.30%) and malic acid (13.52%), while in C1/C4 and C2/C3 transgenics, malic acid (35.06% and 25.17%, respectively) was detected in the highest amount, followed by myo-inositol (29.29% and 23.55%, respectively; Fig. 5e). In virus infiltrated WT tissue, fructose was detected in highest amount (29.85%), followed by glucose (25.30%) and myo-inositol (18.45%; Fig. 5e, Supplementary Table S2), while in virus infiltrated C1/C4 transgenics, malic acid was detected in highest amount (32.34%), followed by myo-inositol (28.55%) and sucrose (26.27%; Fig. 5e, Supplementary Table S2).
Discussion
This study shows that each of the 6 ORFs of the newly isolated JLCuGV genomic DNA (AC1, AC2, AC3, AC4, AV1 and AV2), independently show strong RSS activity. The reversal of GFP expression in GFP silenced tobacco lines on agroinfiltration of AC1, AC2, AC3, AC4, AV1 and AV2 ORFs, independently, confirmed the strong RSS activity in all the six ORFs. The positive amplification of GFP gene from the cDNA generated from infiltrated leaves, gave the molecular confirmation of RSS activity exhibited by JLCuGV ORFs. The RSS activity acquired by individual JLCuGV ORFs seems to have increased its virulence. In the case of geminiviruses, ORFs like AV2, AC1, AC2, and AC4 were reported as RSSs26,27. In Bhendi yellow vein mosaic virus, strong RSS activity detected in AC4 and βC1, while low RSS activity is detected for AC228. In the Tomato leaf curl New Delhi virus, the RSS activity was identified in AC2, AV2 and AC4 proteins22. It was shown that the RSS activity of AC4 and AV2 appeared during early infection and supplemented the RSS activity of AC2 to sustain the response for a longer duration22. Here, we report for the first time that all six ORFs of JLCuGV genomic DNA have acquired a strong RSS activity, although detailed molecular analysis is required to characterize the strength, duration and complementation of the RSS activity exhibited by individual ORFs.
As a strategy to boost plant resistance to virus infection, we designed amiRNAs targeting overlapping regions between ORFs C1/C4, C2/C3 and V1/V2 to silence their transcripts and hence inhibit their RSS activity. The N. benthamiana transgenics independently expressing C1/C4 and C2/C3 amiRNAs showed resistance against JLCuGV while transgenics expressing V1/V2 amiRNAs showed tolerance to JLCuGV. The WT and VA plants showed high viral load with severe disease symptoms, whereas no viral load and no symptoms were detected in C1/C4 and C2/C3 transgenics. This indicated that JLCuGV resistance in C1/C4 and C2/C3 transgenics could be due to the expression of amiRNA. Several other reports are available that have shown that amiRNAs are effective in bringing out the degradation or silencing of the target viral genes23,29.
Interestingly, amiRNA levels were lower in V1/V2 amiRNA transgenics as compared to the C1/C4 and C2/C3 transgenics. The reason needs to be investigated though, in all probability, it may be due to the allelic states of amiRNA precursor, based on the integration of T-DNA into the plant genome, which may have altered the level of amiRNA expression29. The lower amiRNA expression level in V1/V2 amiRNA transgenics correlates with the development of mild symptoms and limited tolerance to virus infection. It was observed that the viral load in V1/V2 amiRNA transgenics was much lower than that observed in WT or VA plants. This shows the important role of miRNAs in providing protection from virus infection and the manifestation of pathogen symptoms30. Earlier, Vu et al.29 have shown an inverse correlation between the expression levels of amiRNA and the development of disease symptoms.
DNA methylation plays a major role in protecting plants under both biotic and abiotic stresses. Several reports state that plants can methylate the invading virus genome to suppress their growth30. The changes in methylation in Nicotiana tabacum genome were observed after the Cucumber mosaic virus (CMV) infection31. The predominance of CHH hypomethylation has emerged as a major defence response against viral infection31. The transgenics C1/C4 and C2/C3 amiRNA exhibited higher methylation across the genome after virus infection as compared to WT, VA and V1/V2 amiRNA transgenics. The higher methylation and demethylation in amiRNA transgenics, corelated with decreased JLCuGV accumulation. This suggested that the homeostasis in methylation pattern in C1/C4 and C2/C3 amiRNA transgenics helped to block the JLCuGV accumulation, leading to resistance against JLCuGV. It has been reported that geminiviruses counteract plant defence strategy by inhibiting DNA methylation. The geminiviral AC2 genes act as RSS by interacting and inhibiting adenosine kinase (ADK), an essential gene of the methylation pathway32. ADK plays a role in the production of the methyl group donor S-adenosyl methionine (SAM) and defects in ADK lead to a loss in methylation in plants33. As C2/C3 amiRNA transgenics target the AC2 gene, the viral counter defence to suppress ADK was not functional, and the plants could successfully follow the methylation pathway to inhibit the virus growth.
Virus infection usually affects the process of photosynthesis34,35, such that decreased photosynthesis is correlated with increased disease severity and increased virus multiplication36,37. JLCuGV infiltrated WT showed severe symptoms and significantly decreased photosynthesis as compared to amiRNA transgenics (C1/C4 and C2/C3). The V1/V2 transgenics showed mild symptoms and relatively less reduction of photosynthesis parameters as compared to WT. The decrease in photosynthesis rate has been previously reported in geminivirus-infected leaves of Eupatorium makinoi35 and mustard after turnip mosaic virus infection37. Naidu et al.38 suggested that a reduced level of chlorophyll-a may be responsible for the reduced rate of photosynthesis.
The transpiration rate, WUE and stomatal conductance were significantly higher in virus-infected transgenics compared to WT. The reduced transpiration rate and stomatal conductance were also observed in mustard after geminivirus infection37. The Ci and Ci/Ca were significantly higher in WT, suggesting that the CO2 taken by the plants was not utilized during photosynthesis and got accumulated in the cells, increasing the Ci and Ci/Ca ratio. It also correlates with the reduced rate of photosynthesis observed in WT. Similar results were reported with tobamovirus infection for reduced stomatal conductance, transpiration rate and Ci in pepper39. The ɸCO2 was significantly higher in transgenics, suggesting efficient CO2 assimilation, which correlated with both increased photosynthesis rate and reduced Ci of transgenics. The reduced ɸCO2 is reported for pea on Pea enation mosaic virus (family Luteoviridae) infection40. The higher ETR seen in amiRNA transgenics suggests better redox homeostasis. The high ETR was found associated with improved reactive oxygen species (ROS) homeostasis during salinity41. Although Guo et al.37 report no change for ETR during geminivirus infection, a significant reduction in ETR was observed in grapevine on virus infection42.
Fv/Fm was maintained at a healthy 0.8 level31 in both WT and transgenics even after virus infiltration. Similarly, no change in Fv/Fm was reported for geminivirus infected mustard37, and Pepper mild mottle virus (PMMoV) infected pepper39. ɸPSII was significantly higher in transgenics, suggesting proper functioning of PSII as it is a measure of PSII performance. In WT, ɸPSII was significantly reduced, and reduced ɸPSII is reported to cause the accumulation of reduced bound quinone, which causes damage to the primary electron acceptor plastoquinone of PSII43. Therefore, reduced ɸPSII can be correlated to the reduction of ETR observed in WT during infection. The qP indicates the efficient conversion of light into chemical energy37 and significantly improved qP in transgenics can be attributed to the maintenance of photochemical quenching followed by increased photosynthesis rate in transgenics. The overexcitation of the photochemical system after turnip mosaic virus infection caused reduced qP in mustard37.
In WT, the sugar content was significantly increased and intermediates of the tricarboxylic acid (TCA) cycle (malic acid, pyruvic acid and citric acid) reduced after virus infiltration. This suggests that sugar metabolism is affected and the normal functioning of the TCA cycle is disturbed in WT after JLCuGV infection. In transgenics, no remarkable changes are observed, indicating better maintenance of metabolic pathways on virus infiltration. The increase in sugar content of WT is correlated with the virus infection in Tobacco mosaic virus-infected tobacco44, Tobacco rattle virus-infected Arabidopsis45 and CMV infected beet plants46. Similarly, the decrease in carboxylic acids of WT is correlated with virus infection in Papaver somniferum on geminivirus Ageratum enation virus (AEV) infection47. The decrease in sucrose concentration is reported in Potato virus Y (PVY)-inoculated potato leaves, while the increase in sucrose content reported in Jatropha infected by geminivirus48. In WT, a decrease in sucrose and an increase in fructose and glucose content after virus infiltration might be due to activation of invertase enzyme which converts sucrose into fructose and glucose49,50. An increase in glucose concentration is also reported in tomato infected with Tomato mosaic virus50 and PVY-inoculated potato leaves51.
In conclusion, the study reports that a new isolate of JLCuGV was responsible for the leaf curl disease in Jatropha CP9 accession. All six ORFs of JLCuGV genomic DNA exhibited RSS activity in-planta. Three amiRNAs were designed against C1/C4, C2/C3 and V1/V2 transcripts by targeting the overlapping regions. Transgenics expressing these amiRNAs showed a resistance (C1/C4 and C2/C3) and tolerance (V1/V2) response to viral infection. A direct correlation of the viral load and disease symptoms with relative levels of amiRNA was observed. The photosynthetic parameters were significantly better in amiRNA transgenics as compared to WT on virus infiltration, suggesting that both light and dark centres of photosynthesis are maintained due to reduced viral load. Likewise, the metabolite profiles were not much altered in amiRNA transgenics, while sugar metabolism is disturbed in WT on virus infiltration, again suggesting resistance of the amiRNA transgenics to the virus. MSAP analysis revealed an increase in methylation and demethylation profiling in amiRNA transgenics as compared to VA and WT, suggesting that methylation pathways get activated in amiRNA transgenics, and increased level of methylation profiling is correlated with a degree of virus resistance in amiRNA transgenics. All the results indicated that the amiRNA transgenics were better adapted to resist/tolerate JLCuGV infection. Importantly, as amiRNAs designed in this study share homology to related geminiviruses that infect other crops like tomato, pepper and papaya, so these have the potential to be employed for developing geminiviral resistance in other susceptible economically important crops.
Materials and methods
Plant material
The J. curcas leaves CP9 accession showing mosaic and leaf curl symptoms were collected from Nesvad, Jatropha experimental fields of Plant Omics division, CSIR-CSMCRI, Gujarat. Collections were made during July and September 2014. The collected samples were stored at − 80 °C until use.
Detection and identification of begomoviruses
The genomic DNA of the collected samples was isolated by modified CTAB method51. The detection and characterization of begomoviral DNA components (DNA-A, DNA-B, DNA- β) from the samples was performed by PCR with degenerate primers (Supplementary Table S3) using 100 ng of genomic DNA. The complete genome of the begomovirus was amplified from the genomic DNA (100 ng) by RCA using a TempliPhi Amplification Kit (GE Life Sciences, UK). The RCA products were digested with HindIII to get the complete begomoviral genome (2.7 kb), cloned in pBluescriptII KS(+) vector and sequenced.
Cloning of JLCuGV ORFs independently in pCAMBIA-1301
The individual ORFs of JLCuGV genome (AV1, AV2, AC1, AC2, AC3 and AC4) were cloned in pRT101, thereafter, the entire cassette with CaMV35S promoter-ORF- PolyA was cloned in pCAMBIA-1301 vector using PstI (AV1, AV2 and AC1) or HindIII (AC2, AC3 and AC4; Fig. 1a). The cloned ORF constructs were then mobilized in A. tumefaciens LBA4404 strain for agroinfiltration.
Agroinfiltration of JLCuGV ORF constructs
The Agrobacterium cultures (OD600 = 1; 1 ml per plant) of individual JLCuGV ORF constructs were injected into leaves of stable N. tabacum cv. Xanthi GFP silenced lines into separate patches in each leaf for identification of RSS activity. Agroinfiltration was performed by a method described by Hamilton et al.52. The Agrobacterium cultures (OD600 = 1; 1 ml per plant) were injected into leaves of the N. benthamiana (WT and transgenics) at 4–6 leaf stage using a 2 ml syringe. The plants were kept in an insect-free chamber at a constant temperature (25–28 °C) under 14–16 h lighting.
Detection and confirmation of RSS activity
The agroinfiltrated leaves of the GFP silenced lines were observed under UV light for visualization of green fluorescence at 7, 11 and 15 dpi. The ORFs having inherent RSS activity were expected to restore the GFP expression by suppressing the RNA silencing machinery.
Total RNA was isolated from the individual regions of leaves52 agroinfiltrated independently with single JLCuGV ORF (AV1, AV2, AC1, AC2, AC3 and AC4) and FHVB2 construct, a well-known RSS used as control. The cDNA was prepared using 5 μg of total RNA using SuperScript IV First-Strand synthesis system (Invitrogen), as per manufacturer’s instructions. cDNA (1 μl), from each sample was used for amplification of gfp gene (~ 300 bp) with specific primers (Supplementary Table S3). The cDNA from mock (only buffer infiltrated) leaves was used as a negative control as it lacked the GFP expression. The samples showing amplification of the gfp transcript confirmed the restoration of GFP expression in the agroinfiltrated region of the leaf, thereby confirming the inherent RSS activity of the particular ORF.
In silico designing and construction of amiRNA plasmids for tobacco transformation
The common overlapping sequences between the two genes (AV1–AV2, AC1–AC4 and AC2–AC3) were used for designing amiRNA (Table 1, Supplementary Fig. S5a). The amiRNA design was carried out by using the WMD3 (Web MicroRNA Designer 3) online tool (http://wmd3.weigelworld.org). While designing of amiRNA, we also tried to take the sequences, which can be used to target the begomovirus infecting tomato, papaya, pepper, ageratum etc. (Supplementary Fig. S5b–d), so that same amiRNA construct can also be used against other begomoviruses.
The three designed amiRNAs (Table 1) were amplified by four sets of primers (Supplementary Table S3) via overlapping PCR on the pNW55 vector backbone for obtaining pre-miRNA as given in Sharma et al.53. The pre-amiRNA fragment was then cloned in pRT101, using XhoI and BamHI sites for directional cloning, under CaMV-35S promoter. Then the entire cassettes of C1/C4, C2/C3 and V1/V2 amiRNA with CaMV-35S promoter were cloned in pCAMBIA-1301 using HindIII site. The schematic representation of the amiRNA gene construct in pCAMBIA-1301 showed in Supplementary Fig. S2.
Tobacco transformation
Three amiRNA constructs (C1/C4, C2/C3 and V1/V2 amiRNA) cloned in pCAMBIA-1301 vector and vector backbone alone (VA) were mobilized into the A. tumefaciens strain LBA4404. The transformed agrobacterium was used for the transformation of N. benthamiana leaf discs, according to Clemente54.
Molecular validation of transgenic plants expressing the amiRNA
The transgenic plants (T1) were checked by GUS assay to confirm the transgene expression. These were later confirmed for gene integration by PCR. The PCR was carried out using the primers specific for amiRNA gene, gus gene and hygromycin (hptII) gene (Supplementary Table S3). The positive lines were then checked for copy number by quantitative Real-Time PCR (qRT-PCR).
Construction of infectious clone for agroinfiltration
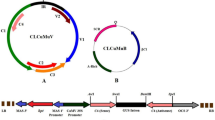
The construction of infectious clone was performed in two steps. In the first step, the RCA product was double digested with BamHI and HindIII, then the digested product (0.65 kb) was cloned in pCAMBIA-1301 to get a recombinant vector pCAM-JLCuGV-Partial. In the second step, the full-length genome of RCA product generated by HindIII was ligated with pCAM-JLCuGV-Partial, to get a complete infectious clone pCAM-JLCuGV-Inf. The orientation of the inserts was confirmed with sequencing. The positive clones that contain tandemly repeated viral genomes having two ori (origin of replication) sites were selected. The infectious clone was transformed in A. tumefaciens LBA4404 strain. For causing infection, the agroinfectious clone of JLCuGV (Supplementary Fig. S4) was infiltrated in leaves of WT and transgenic lines of each amiRNA (L61, L64, L67 of C1/C4 amiRNA; L41, L42, L44 of C2/C3 amiRNA and L13, L14, L15 of V1/V2 amiRNA) in three replicates.
Molecular confirmation of JLCuGV replicating in agroinfiltrated tobacco plants
The genomic DNA was isolated from the tobacco plants55 agroinfiltrated with JLCuGV infectious clone. The presence of an infectious clone of JLCuGV was checked by PCR with specific degenerate primers (Supplementary Table S3) using 100 ng of each genomic DNA as a template. The actin gene was amplified as a loading control using NtActF and NtActR primers (Supplementary Table S3). The complete begomoviral genome was amplified by RCA using a TempliPhi Amplification Kit (GE Life Sciences, UK). The amplified RCA product was digested with HindIII to get 2.7 kb genomic DNA.
Determination of viral load
Geminivirus copy number in the agroinfiltrated plant was determined as reported in Legarrea et al.56. The pCAMBIA-1301 containing JLCuGV genomic DNA plasmid was linearized with BglII restriction enzyme. The quantified plasmid DNA was used to calculate the number of copies based on the formula:
where 650 denotes weight (Da) of a base pair.
The standard curve was prepared by serial dilution of 109 to 101 plasmid of pCAMBIA-1301 containing JLCuGV genomic DNA. The qRT-PCR reaction performed using the AV2 gene primers (Supplementary Table S3). The copy number was quantified using the Ct (cycle threshold) value plotted against the standard curve formula. Three lines of each transgenic event were used for the analysis.
Small RNA isolation
The total RNA was isolated from transgenic lines by Tris-SDS buffer57 and then the small RNA was isolated by using different concentrations of polyethylene glycol (PEG) according to Singh and Jha58. The quality of the purified RNA was examined by 1% agarose gel electrophoresis and the absorbance ratio (A260/A280 and A260/A230) using Epoch Microplate Spectrophotometer (BioTek, USA).
The quantitative estimation of amiRNA
The cDNA was synthesized using small RNA as template and specifically designed stem-loop primers (Supplementary Table S3) by SuperScript IV First-Strand Synthesis System (Invitrogen). The quantitative estimation of amiRNA was carried out using cDNA with stem-loop primers based qRT-PCR as given in Czimmerer et al.59 and actin gene was used as an internal control.
Gas exchange and chlorophyll fluorescence measurement
Photosynthetic gas exchange parameters were measured at photosynthetic photon flux density (PPFD) of 1000 μmol m−2 s−1 by the open infrared gas analyzer (IRGA, Model LI-6400XT, LI-COR). All other conditions for gas exchange and chlorophyll fluorescence measurement were kept the same as reported in Shukla et al.60. Photosynthetic gas exchange parameters and chlorophyll fluorescence parameters were recorded in WT and amiRNA transgenics (L61, L64, L67 of C1/C4, L41, L42, L44 of C2/C3 amiRNA and L13, L14, L15 of V1/V2) at 21, 28 and 35 dpi.
Gas chromatography–mass spectrometry (GC–MS) analysis
The sample preparation and derivatization by N-methyl-N-trimethylsilyl trifluoroacetamide (MSFTA) was carried out as given in Lisec et al.61. The derivatized samples were then transferred into glass vials suitable for GC–MS analysis. The GC–MS analysis was carried out in a SHIMADZU prominent instrument (SHIMADZU Corporation, Kyoto, Japan) equipped with EI mass spectra and autosampler. The injection volume for each sample was 1.0 μl throughout the analysis. Gas chromatographic separation was performed using the silica capillary SH-Rtx-5 column (30 m × 0.25 mm)62. Injection temperature was 230 °C, the interface was set to 250 °C and the ion source was adjusted to 200 °C. Helium flow was 1 mL min−1. After a 5-min solvent delay time at 70 °C, the oven temperature was increased at 5 °C min−1 to 310 °C, 1 min isocratic, cool-down to 70 °C, followed by an additional 5-min delay. The ion source temperature was 200 °C and mass ions in the range of 45–600 m/z were scanned at a rate of 1.0 scans s−1.
MSAP analysis
MSAP analysis was performed as reported earlier25. For MSAP analysis, WT, VA and two representative lines of C1/C4 (L61, L67), V1/V2 (L13, L15) and C2/C3 (L42, L43) amiRNA transgenics were selected. The DNA was isolated from each plant with and without virus agroinfiltration after 35 dpi.
Statistical analysis
The mean values and standard deviations were calculated of three independent biological replicates and the bars in figures represent the mean ± SD. Analysis of variance was calculated using the Tukey test by InfoStat software (version 2018) at P ≤ 0.05 to determine the significance of the difference in viral load of WT, VA and V1/V2 amiRNA transgenics and relative expression of amiRNA in the C1/C4, C2/C3 and V1/V2 amiRNA transgenics at 14, 21, 28 and 35 dpi. Similarly, the analysis of variance was also calculated in the photosynthesis parameters between the means of control and virus infiltrated plants (WT, C1/C4, C2/C3 and V1/V2 amiRNA transgenics) at 21, 28 and 35 dpi. Different alphabets indicated mean values of treatments that were significantly different from each other.
References
Rojas, M. R. & Gilbertson, R. L. Emerging plant viruses: a diversity of mechanisms and opportunities. In Plant Virus Evolution (ed. Roossinck, M. J.) 27–51 (Springer, Berlin Heidelberg, 2008). https://doi.org/10.1007/978-3-540-75763-4_3.
Yadava, P., Suyal, G. & Mukherjee, S. Begomovirus DNA replication and pathogenicity. Curr. Sci. 98, 360–368 (2010).
Stanley, J. et al. Family geminiviridae. In Virus taxonomy: eighth report of the International Committee on Taxonomy of Viruses. pp. 301–326 (2005).
Thresh, J. M., Otim-Nape, G. W. & Fargette, D. The components and deployment of resistance to cassava mosaic virus disease. Integr. Pest Manag. Rev. 3, 209–224 (1998).
Varma, A., Dhar, A. K. & Mandal, B. MYMV transmission and control in India. Mungbean yellow mosaic disease (1992).
Moffat, A. S. Geminiviruses emerge as serious crop threat. Science (80-) 286, 1835 (1999).
Sanjuán, R. & Domingo-Calap, P. Mechanisms of viral mutation. Cell. Mol. Life Sci. 73, 4433–4448 (2016).
Martin, D. P. et al. Recombination in eukaryotic single stranded DNA viruses. Viruses 3, 1699–1738 (2011).
Pullaiah, T. & Bahadur, B. Economic and medicinal importance of Jatrophas. In Jatropha, Challenges for a New Energy Crop (eds Bahadur, B. et al.) 187–217 (Springer, Berlin, 2013).
Achten, W. M. J. et al. Jatropha biodiesel fueling sustainability?. Biofuels Bioprod. Biorefin. 1, 283–291 (2007).
Kancharla, N. et al. De Novo sequencing and hybrid assembly of the biofuel crop Jatropha curcas L.: identification of quantitative trait loci for geminivirus resistance. Genes (Basel) 10, 69 (2019).
Narayana, D. S. A. et al. Natural occurrence of Jatropha mosaic virus disease in India. Curr. Sci. 91, 584–586 (2006).
More, P., Agarwal, P. & Agarwal, P. K. Geminiviruses: Molecular biodiversity and global distribution in Jatropha. Physiol. Mol. Plant Pathol. 108, 101439 (2019).
Lucioli, A. et al. Tomato Yellow Leaf Curl Sardinia virus rep-derived resistance to homologous and heterologous geminiviruses occurs by different mechanisms and is overcome if virus-mediated transgene silencing is activated. J. Virol. 77, 6785–6798 (2003).
Baulcombe, D. C. Mechanisms of pathogen-derived resistance to viruses in transgenic plants. Plant Cell 8, 1833–1844 (1996).
Ye, J. et al. Engineering geminivirus resistance in Jatropha curcus. Biotechnol. Biofuels 7, 149 (2014).
Yin, K. et al. Engineer complete resistance to Cotton Leaf Curl Multan virus by the CRISPR/Cas9 system in Nicotiana benthamiana. Phytopathol. Res. 1, 9 (2019).
Vaucheret, H., Béclin, C. & Fagard, M. Post-transcriptional gene silencing in plants. J. Cell Sci. 114, 3083–3091 (2001).
Burgyán, J. & Havelda, Z. Viral suppressors of RNA silencing. Trends Plant Sci. 16, 265–272 (2011).
Padmanabhan, C., Zhang, X. & Jin, H. Host small RNAs are big contributors to plant innate immunity. Curr. Opin. Plant Biol. 12, 465–472 (2009).
Ruiz-Ferrer, V. & Voinnet, O. Roles of plant small RNAs in biotic stress responses. Annu. Rev. Plant Biol. 60, 485–510 (2009).
Sundaresan, G., Das, S. S., Tripathi, A., Mukherjee, S. K. & Sanan-Mishra, N. Evaluating the strength of RNA silencing suppressor proteins encoded by two geminiviruses using assay based on reversal of GFP silencing. Australas. Plant Pathol. https://doi.org/10.1007/s13313-019-00678-4 (2020).
Niu, Q.-W. et al. Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat. Biotechnol. 24, 1420–1428 (2006).
Karjee, S. et al. The 7a accessory protein of severe acute respiratory syndrome coronavirus acts as an RNA silencing suppressor. J. Virol. 84, 10395–10401 (2010).
More, P., Agarwal, P., Mastan, S. G. & Agarwal, P. K. MSAP marker based DNA methylation study in Salicornia brachiata DREB2A transgenic tobacco. Plant Gene 6, 77–81 (2016).
Rodríguez-Negrete, E. et al. Geminivirus Rep protein interferes with the plant DNA methylation machinery and suppresses transcriptional gene silencing. New Phytol. 199, 464–475 (2013).
Sharma, P. & Ikegami, M. RNA-silencing suppressors of geminiviruses. J. Gen. Plant Pathol. 74, 189–202 (2008).
Gopal, P. et al. Differential roles of C4 and βC1 in mediating suppression of post-transcriptional gene silencing: evidence for transactivation by the C2 of Bhendi yellow vein mosaic virus, a monopartite begomovirus. Virus Res. 123, 9–18 (2007).
Vu, T. V., Roy Choudhury, N. & Mukherjee, S. K. Transgenic tomato plants expressing artificial microRNAs for silencing the pre-coat and coat proteins of a begomovirus, Tomato leaf curl New Delhi virus, show tolerance to virus infection. Virus Res. 172, 35–45 (2013).
Paprotka, T., Deuschle, K., Metzler, V. & Jeske, H. Conformation-selective methylation of geminivirus DNA. J. Virol. 85, 12001–12012 (2011).
Baker, N. R. & Rosenqvist, E. Applications of chlorophyll fluorescence can improve crop production strategies: an examination of future possibilities. J. Exp. Bot. 55, 1607–1621 (2004).
Wang, H., Buckley, K. J., Yang, X., Buchmann, R. C. & Bisaro, D. M. Adenosine kinase inhibition and suppression of RNA silencing by geminivirus AL2 and L2 proteins. J. Virol. 79, 7410–7418 (2005).
Moffatt, B. A. et al. Adenosine kinase deficiency is associated with developmental abnormalities and reduced transmethylation. Plant Physiol. 128, 812–821 (2002).
Funayama, S., Sonoike, K. & Terashima, I. Photosynthetic properties of leaves of Eupatorium makinoi infected by a geminivirus. Photosynth. Res. 53, 253–261 (1997).
Funayama, S. & Terashima, I. Effects of geminivirus infection and growth irradiance on the vegetative growth and photosynthetic production of Eupatorium makinoi. New Phytol. 142, 483–494 (1999).
Bassi, M., Appiano, A., Barbieri, N. & D’Agostino, G. Chloroplast alterations induced by tomato bushy stunt virus inDatura leaves. Protoplasma 126, 233–235 (1985).
Guo, D.-P. et al. Photosynthetic rate and chlorophyll fluorescence in leaves of stem mustard (Brassica juncea var. tsatsai) after turnip mosaic virus infection. Plant Sci. 168, 57–63 (2005).
Naidu, R. A., Krishnan, M., Nayudu, M. V. & Gnanam, A. Studies on peanut green mosaic virus infected peanut (Arachis hypogaea L.) leaves. II. Chlorophyll-protein complexes and polypeptide composition of thylakoid membranes. Physiol. Plant Pathol. 25, 191–198 (1984).
Janeczko, A. et al. Comparative studies of compatible and incompatible pepper–Tobamovirus interactions and the evaluation of effects of 24-epibrassinolide. Photosynthetica 56, 763–775 (2018).
Kyseláková, H. et al. Photosynthetic alterations of pea leaves infected systemically by pea enation mosaic virus: a coordinated decrease in efficiencies of CO2 assimilation and photosystem II photochemistry. Plant Physiol. Biochem. 49, 1279–1289 (2011).
Agarwal, P., Dabi, M., Sapara, K. K., Joshi, P. S. & Agarwal, P. K. Ectopic expression of JcWRKY transcription factor confers salinity tolerance via salicylic acid signaling. Front. Plant Sci. 7, 1541 (2016).
Sampol, B., Bota, J., Riera, D., Medrano, H. & Flexas, J. Analysis of the virus-induced inhibition of photosynthesis in malmsey grapevines. New Phytol. 160, 403–412 (2003).
Baker, N. R. Chlorophyll fluorescence: a probe of photosynthesis in vivo. Annu. Rev. Plant Biol. 59, 89–113 (2008).
Bazzini, A. A. et al. Metabolic and miRNa profiling of TMV infected plants reveals biphasic temporal changes. PLoS ONE 6, e28466 (2011).
Fernández-Calvino, L. et al. Virus-induced alterations in primary metabolism modulate susceptibility to Tobacco rattle virus in arabidopsis. Plant Physiol. 166, 1821–1838 (2014).
Xu, P. et al. Virus infection improves drought tolerance. New Phytol. 180, 911–921 (2008).
Srivastava, A. et al. Ageratum enation virus infection induces programmed cell death and alters metabolite biosynthesis in Papaver somniferum. Front. Plant Sci. 8, 1172 (2017).
Sidhu, O. P. et al. Metabolic and histopathological alterations of Jatropha mosaic begomovirus-infected Jatropha curcas L. by HR-MAS NMR spectroscopy and magnetic resonance imaging. Planta 232, 85–93 (2010).
Wynd, F. L. Metabolic phenomena associated with virus infection in plants. Bot. Rev. 9, 395 (1943).
López-Gresa, M. P. et al. Metabolic fingerprinting of tomato mosaic virus infected Solanum lycopersicum. J. Plant Physiol. 169, 1586–1596 (2012).
Kogovšek, P. et al. Primary metabolism, phenylpropanoids and antioxidant pathways are regulated in potato as a response to Potato virus y infection. PLoS ONE 11, e0146135 (2016).
Hamilton, A., Voinnet, O., Chappell, L. & Baulcombe, D. Two classes of short interfering RNA in RNA silencing. EMBO J. 21, 4671–4679 (2002).
Sharma, N., Panchal, S. & Sanan-Mishra, N. Protocol for artificial microRNA mediated over-expression of miR820 in indica rice. Am. J. Plant Sci. 6, 1951 (2015).
Clemente, T. Nicotiana (Nicotiana tobaccum, Nicotiana benthamiana). In Agrobacterium Protocols (ed. Wang, K.) 143–154 (Humana Press, Totowa, 2006). https://doi.org/10.1385/1-59745-130-4:143.
Doyle, J. & Doyle, J. Isolation of plant DNA from fresh tissue. Focus (Madison) 12, 13–15 (1990).
Legarrea, S., Barman, A., Marchant, W., Diffie, S. & Srinivasan, R. Temporal effects of a begomovirus infection and host plant resistance on the preference and development of an insect vector, Bemisia tabaci, and implications for epidemics. PLoS ONE 10, e0142114 (2015).
Agarwal, P., Dabi, M., Das, M., Patel, K. & Agarwal, P. K. An economical and efficient protocol for total RNA isolation from Jatropha curcas. Int. J. Environ. Stud. 72, 624–630 (2015).
Singh, D. & Jha, B. The isolation and identification of salt-responsive novel microRNAs from Salicornia brachiata, an extreme halophyte. Plant Biotechnol. Rep. 8, 325–336 (2014).
Czimmerer, Z. et al. A versatile method to design stem-loop primer-based quantitative PCR assays for detecting small regulatory RNA molecules. PLoS ONE 8, e55168 (2013).
Shukla, P. S., Agarwal, P., Gupta, K. & Agarwal, P. K. Molecular characterization of an MYB transcription factor from a succulent halophyte involved in stress tolerance. AoB Plants 7, plv054 (2015).
Lisec, J., Schauer, N., Kopka, J., Willmitzer, L. & Fernie, A. R. Gas chromatography mass spectrometry–based metabolite profiling in plants. Nat. Protoc. 1, 387–396 (2006).
Halket, J. M. et al. Chemical derivatization and mass spectral libraries in metabolic profiling by GC/MS and LC/MS/MS. J. Exp. Bot. 56, 219–243 (2005).
Acknowledgements
CSIR-CSMCRI Communication No.-PRIS 5/2020. The authors are thankful to the Department of Science and Technology (DST) and Council of Scientific and Industrial Research (CSIR), New Delhi, India. PA acknowledges the financial support from DST-WOS-A scheme. NSM acknowledges a research grant from DBT for developing the assay for suppressor analysis. PM is supported by the DST-INSPIRE SRF fellowship.
Author information
Authors and Affiliations
Contributions
P.M. performed major experiments. P.A. helped in designing and executing experiments. A.A. and N.S.M. involved in the identification of RNA silencing suppressors. P.A. and P.K.A. conceived and coordinated the experiments. All authors have contributed to M.S. writing and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
More, P., Agarwal, P., Anand, A. et al. Artificial miRNA mediated resistance in tobacco against Jatropha leaf curl Gujarat virus by targeting RNA silencing suppressors. Sci Rep 11, 890 (2021). https://doi.org/10.1038/s41598-020-79134-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-79134-z
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.