Abstract
Lipid composition in breast cancer, a central marker of disease progression, can be non-invasively quantified using 2D MRS method of double quantum filtered correlation spectroscopy (DQF-COSY). The low signal to noise ratio (SNR), arising from signal retention of only 25% and depleted lipids within tumour, demands improvement approaches beyond signal averaging for clinically viable applications. We therefore adapted and examined combination algorithms, designed for 1D MRS, for 2D MRS with both internal and external references. Lipid composition spectra were acquired from 17 breast tumour specimens, 15 healthy female volunteers and 25 patients with breast cancer on a clinical 3 T MRI scanner. Whitened singular value decomposition (WSVD) with internal reference yielded maximal SNR with an improvement of 53.3% (40.3–106.9%) in specimens, 84.4 ± 40.6% in volunteers, 96.9 ± 54.2% in peritumoural adipose tissue and 52.4% (25.1–108.0%) in tumours in vivo. Non-uniformity, as variance of improvement across peaks, was low at 21.1% (13.7–28.1%) in specimens, 5.5% (4.2–7.2%) in volunteers, 6.1% (5.0–9.0%) in peritumoural tissue, and 20.7% (17.4–31.7%) in tumours in vivo. The bias (slope) in improvement ranged from − 1.08 to 0.21%/ppm along the diagonal directions. WSVD is therefore the optimal algorithm for lipid composition spectra with highest SNR uniformly across peaks, reducing acquisition time by up to 70% in patients, enabling clinical applications.
Similar content being viewed by others
Introduction
Lipid composition is a central marker for the pathogenesis of breast cancer1, 2, the most commonly diagnosed cancer among women3. Conventional magnetic resonance spectroscopy (MRS) of stimulated echo acquisition mode (STEAM) with short echo time can detect lipid spectral peaks in the breast non-invasively on standard clinical scanners4, and further enhancement in specificity is valuable for clinical applications. Spectral editing methods of double quantum filtering (DQF), effectively suppress background signals, but only target a single metabolite, such as polyunsaturated fatty acids (PUFA) in 1D MRS5. The two dimensional (2D) MRS method of correlation spectroscopy (COSY)6 resolves lipid composition on a 2D map, but suffers from the dominant water signal and wide peak spread7. DQF-COSY, combining the strength of spectral editing and 2D MRS, allows unobscured identification of individual lipid resonances through sharp peak appearance and suppression of water contamination signals8. However, both the signal retention of only 25% in DQF-COSY7 and depleted lipids within breast tumours59 contribute to low signal to noise ratio (SNR), posing a challenge for accurate quantification. Since DQF-COSY collects a series of 1D spectra demanding a long acquisition time (typical scan time of 15–20 min)10, SNR improvement approaches beyond signal averaging are required for clinically viable applications.
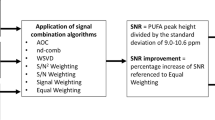
Phased-array coils have been widely adopted in routine clinical practice, with signal combination algorithms developed to enhance SNR and reduce acquisition time11,12. Adaptively Optimised Combination (AOC)13, amongst current combination algorithms developed for 1D MRS (Table 1)13,14,15,16, is the optimal approach for spectra acquired in the brain using conventional MRS13 and PUFA spectra acquired in the breast using spectral editing MRS17. The SNR of a single spectral peak has been adopted as the common assessment criteria in the comparison of combination algorithms. However, lipid composition in 2D MRS is determined utilising multiple spectral peaks across the 2D map, demanding an algorithm with uniform improvement. In contrast to spectral editing MRS, DQF-COSY retains the presence of dominant metabolites, at reduced amplitude, for the estimation of sensitivities and phases of coil elements, potentially eliminating the need to acquire an additional reference spectrum.
We hypothesise that AOC is the optimal algorithm to provide maximal SNR improvement uniformly across the 2D lipid composition spectrum in breast cancer. We therefore adapted current algorithms (Table 1), with a particular focus on noise decorrelated algorithms, for 2D MRS and applied on lipid composition spectra acquired using DQF-COSY. The combination algorithms were evaluated on spectra acquired from breast tumour specimens, healthy female volunteers and patients with breast cancer, with data from the tumour and the peritumoural adipose tissue (Fig. 1a). Each algorithm was implemented twice with weighting coefficients derived from the spectrum without water suppression (external reference, denoted by subscript “e”) and first signal of the DQF-COSY acquisition (internal reference, denoted by subscript “i”) (Fig. 1b). The non-uniformity of SNR improvement across spectral peaks (Table 2) and the direction of non-uniformity was additionally evaluated. The non-uniformity was defined as the coefficient of variance of SNR improvement across spectral peaks. The direction of non-uniformity was quantified as the slopes along the diagonal (bias along the frequency axes) and the off-diagonal (bias along the encoding axes) on a plane regressed to the SNR improvement at the spectral location of each peak.
Diagram of study design and data processing. (a) Study design. Combination algorithms were evaluated on DQF-COSY spectra acquired from ex vivo and in vivo experiments by comparing the SNR. (b) Processing steps. Combination algorithms were applied on DQF-COSY spectra after signal averaging, apodisation and zero filling. AOC = adaptively optimised combination, nd-comb = noise decorrelated combination, WSVD = whitened singular value decomposition.
Results
SNR among algorithms
For tumour specimens, the SNR from WSVDi (median 81.9, interquartile range 57.2–136.4) was comparable against WSVDe, but significantly higher than the other approaches (Table 3, Fig. 2a). For healthy volunteers, WSVDi yielded the highest SNR (833.6 ± 308.6) with statistical significance (Table 3, Fig. 2b). For peritumoural adipose tissue in patients, the SNR from WSVDi (584.6 ± 294.7) was comparable against AOCe, but significantly higher than the other approaches (Table 3, Fig. 2c). For tumours in vivo, the SNR from WSVDi (49.8, 32.6–102.8) was significantly higher (p = 0.028) than nd-combi (43.1, 27.5–99.3), but comparable against other approaches (Table 3, Fig. 2d). Linear algorithms gave lower SNR compared with the noise decorrelated algorithms, and are presented only for information purposes (Table 3). WSVDi improved the SNR by 53.3% (40.3–106.9%) in specimens, 84.4 ± 40.6% in volunteers, 96.9 ± 54.2% in peritumoural adipose tissue and 52.4% (25.1–108.0%) in tumours in vivo, reducing the acquisition time by 50–70% in tumour and adipose tissue respectively. The combined 2D spectra using WSVDi from a specimen, volunteer, peritumoural tissue, and tumour in vivo are shown in Fig. 3.
Comparison of the combination algorithms on DQF-COSY spectra using external and internal references. SNR of DQF-COSY spectra from (a) breast tumour specimens, (b) healthy volunteers, (c) peritumoural adipose tissue, and (d) tumours in vivo, using AOC, nd-comb, and WSVD. Median and interquartile range are shown for non-normally distributed data while mean and standard deviation are shown for normally distributed data. External and internal references were derived from the unsuppressed water spectrum and the spectrum of first signal (increment) in DQF-COSY respectively.
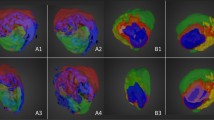
DQF-COSY combined spectra. DQF-COSY combined spectra using WSVDi from (a) a specimen of invasive carcinoma, grade III, (b) the left breast of a healthy volunteer, (c) peritumoural adipose tissue from a patient with invasive carcinoma, grade III, and (d) an invasive carcinoma, grade III from a patient.
Non-uniformity of SNR improvement across spectrum
For tumour specimens, WSVDi had significantly the lowest non-uniformity (21.1%, 13.7–28.1%, p < 0.05) (Table 4, Fig. 4a), with higher improvement towards low frequencies along diagonal direction (slope of − 3.79%/ppm) and mixing encoding (t1/F1) along off-diagonal direction (slope of 4.90%/ppm) (Table 4, Fig. 5a,b). For healthy volunteers, WSVDi had significantly lower non-uniformity (5.5%, 4.2–7.2%) compared against nd-combi (6.6%, 4.6–12.9%, p < 0.001), but no significant differences against the other algorithms (Table 4, Fig. 4b). Higher improvement was found towards high frequencies along diagonal direction (slope of 0.21%/ppm) and reading encoding (t2/F2) along off-diagonal direction (slope of − 0.18%/ppm) (Table 4, Fig. 5a,c). For peritumoural adipose tissue, WSVDi had significantly lower non-uniformity (6.1%, 5.0–9.0%) than AOCi (8.2%, 5.6–10.7%), nd-combi (7.9%, 6.4–11.4%) and nd-combe (8.3%, 6.9–9.9%), but no significant difference from the other algorithms (Table 4, Fig. 4c). Higher improvement was found towards low frequencies along diagonal direction (slope of − 1.08%/ppm) and reading encoding (t2/F2) along off-diagonal direction (slope of − 0.30%/ppm) (Table 4, Fig. 5a,d). For tumours in vivo, there were no significant differences in non-uniformity from WSVDi (20.7%, 17.4–31.7%) against the other algorithms (Table 4, Fig. 4d), with higher improvement towards low frequencies along diagonal direction (slope of − 1.1%/ppm) and mixing encoding (t1/F1) along off-diagonal direction (slope of 2.68%/ppm) (Table 4, Fig. 5a,e).
Comparison of non-uniformity of SNR improvement across the 2D spectrum in DQF-COSY. The non-uniformity of SNR improvement using the noise decorrelated algorithms of AOC, nd-comb, and WSVD is shown for DQF-COSY spectra in (a) breast tumour specimens, (b) healthy volunteers, (c) peritumoural adipose tissue, and (d) tumours in vivo. The non-uniformity of improvement was defined as the coefficient of variance of SNR improvement across all spectral peaks in a 2D spectrum. The error bars show the median and interquartile range. Results using both internal and external references are shown. Data with negative SNR improvement were excluded (1 dataset from a healthy volunteer using AOCi, 1 dataset from a healthy volunteer using nd-combi, and 1 dataset from a tumour in vivo using nd-combi).
SNR improvement distribution in a 3D plane for DQF-COSY. (a) The SNR improvement obtained from WSVDi is plotted at the spectral location of each peak in a 3D scatter plot. Colour maps showing the direction and degree of non-uniformity of SNR improvement are shown for WSVDi for (b) tumour specimens, (c) healthy volunteers, (d) peritumoural adipose tissue, and (e) tumours in vivo. The dots show the location of diagonal and cross peaks. The colour bar shows the SNR improvement.
Discussion
In this work, current combination algorithms, designed for 1D MRS, were adapted and evaluated for lipid composition spectra from breast acquired using 2D MRS, with a particular focus on noise decorrelation algorithms. WSVDi was identified as the most effective signal combination approach in 2D MRS, instead of AOC, the optimal algorithm for 1D MRS13,17. WSVDi provided maximal SNR improvement in patients (97% in peritumoural adipose tissue, 52% in tumour) and low non-uniformity of 6% and 21% respectively. WSVDi, eliminating the need for acquiring an additional reference spectrum (typically scan time of 2 min), reduces scan time by 50–70% from 17 to 8 min in tumour and 5 min in peritumoural adipose tissue.
Noise decorrelation algorithms outperformed all linear algorithms substantially through the cancellation of correlated noise, as found in 1D MRS studies13,17. WSVD performance was not degraded in low SNR spectra acquired from the tumours, in contrast to 1D spectra reported in previous studies17,18. In high SNR spectra from adipose tissue, WSVD and AOC yielded comparable SNR and outperformed nd-comb, as observed in 1D PUFA spectra17. Among the linear algorithms, S/N2 Weighting had the best SNR performance, in line with the 1D MRS studies14,17.
External reference methods (WSVDe, AOCe and nd-combe) showed comparable SNR ex vivo, with the performance of nd-combe degraded in vivo due to the larger variation in coil weightings associated with the voxel location away from the isocentre14,15,19. For internal reference, WSVDi outperformed AOCi and nd-combi. External reference, with higher SNR than internal reference, is expected to provide more accurate weighting coefficients and in turn higher SNR of combined spectra20, as observed in AOC and nd-comb. However, WSVD is less sensitive to the SNR of the reference spectrum as observed in this work, with weighting coefficients generated from the entire spectrum16 instead of a dominant peak, as in AOC13 and nd-comb15.
WSVDi, in addition to providing maximal SNR, eliminates the need for the acquisition of a reference spectrum, reducing scan time by approximately 2 min. WSVDi improved the SNR by 97% in peritumoural adipose tissue and 52% in tumours in vivo, allowing the acquisition of a lipid composition spectrum in less than 8 min in patients instead of 17 min (15 min for lipid composition spectrum and 2 min for reference spectrum).
WSVDe and WSVDi showed better or comparable non-uniformity against other external and internal reference algorithms, with similar performance between WSVDe and WSVDi. WSVDi had non-uniformity of 21% in specimens, 5% in volunteers, 6% in peritumoural adipose tissue and 21% in tumours in vivo, with the variation in non-uniformity reflecting the effects of noise on SNR improvement. The slope along the diagonals further confirmed the observation of non-uniformity, as higher slope (bias in SNR improvement) was associated with lower SNR (observed in spectra from tumours) and lower slope was associated with higher SNR (observed in spectra from adipose tissue). The magnitude of the slop was small in adipose tissue (0.21%/ppm and 0.18%/ppm in healthy volunteers and 1.08%/ppm and 0.30%/ppm in peritumoural adipose tissue), indicating negligible changes of SNR improvement of 0.92% and 0.79% in healthy volunteers and 4.8% and 1.32% in peritumoural adipose tissue at a maximal frequency gap between peaks of interest (0.9 ppm to 5.3 ppm). However, the slopes found in tumours were noticeably higher with higher improvement towards low frequencies and mixing encoding directions, likely due to the signal elevation closer to contamination water signal stripe along the mixing encoding direction (Fig. 3d). Hence, WSVDi provides minimal non-uniformity for SNR improvement across lipid composition spectra.
DQF-COSY8, similar to STEAM21, is composed of three 90° RF pulses, allowing a short echo time and a minimal chemical shift displacement. DQF-COSY, different from STEAM, modulates the evolution time \({\mathrm{t}}_{1}\) for 2D spectral encoding and incorporates quantum coherence pathway selection gradients to suppress background signal, allowing enhanced specificity at the expense of a portion of SNR8. DQF-COSY directly resolves monounsaturated fatty acids (MUFA) and PUFA through J-coupling sensitivity8, while STEAM instead demands a mathematical model22. All lipid peaks could be well detected using conventional 1D MRS under reasonable water suppression23, while DQF-COSY may have a big advantage for the detection of lipid peaks at 4.1 ppm and 4.25 ppm under challenging conditions for water suppression. Hence, DQF-COSY, supported by its intrinsic minimal chemical shift displacement, high specificity and SNR enhancement from a phased-array combination approach may help studying small lesions or area of interest.
The extensive experiments on ex vivo tumour specimens, healthy participants and patients (over 70 datasets from clinical population) encompassed a wide range of physiological environments encountered in a clinical setting. The acquisition voxel was adjusted to the size of the tumour in diseased breast and standardised to 2 \(\times\) 2 \(\times\) 2 cm3 in healthy breast, allowing investigation in both low and high SNR conditions. The comparison among algorithms was comprehensive, covering both internal and external references, with outcome measures extended beyond conventional SNR to non-uniformity. Both standard clinical hardware (scanner and coil) and routine patient imaging procedures were adopted to ensure the immediate clinical translation of the research findings. This study was limited to a single scanner and two different coils, and multi-centre studies on scanners and coils from a range of vendors are required before wider clinical adoption. Patients with invasive breast carcinoma were studied in this work to reduce the confounding factor of experimental setup, however larger patient cohorts with other phenotypes of breast cancer should be investigated in the future.
WSVD, the most effective combination algorithm for 2D MRS, can enhance the sensitivity of inconspicuous lipid constituents found in tumours and accelerate the acquisition in adipose tissue. The extension of single voxel 2D MRS into chemical shift imaging (CSI) of 2D MRS allows the investigation of spatial distribution of lipid composition in tumour but is limited by the voxel size and a demanding acquisition time proportional to the number of voxels acquired. WSVD can potentially allow smaller voxel sizes and reduce acquisition time through trading the enhanced SNR, allowing the investigation of spatially heterogeneous response to neoadjuvant chemotherapy in breast cancer24. CSI of 2D MRS, with further support from compressed sensing, can achieve direct lipid composition mapping of the entire breast, for the early detection and prevention of breast cancer, without the need of a mathematical model of lipid amplitudes based on the empirical assumptions made in the Dixon method25. However, further investigation is needed to consider the spatial variability in the coil sensitivity and the combination of WSVD with compressed sensing for CSI of 2D MRS.
In conclusion, WSVDi, instead of AOC, is the optimal approach for processing lipid composition spectra acquired using 2D MRS on phased-array coils from the breast. WSVDi not only provides maximal SNR improvement without the need of additional reference spectra, but also delivers consistent improvement across lipids with high uniformity. With improved SNR, the acquisition can be achieved at a clinically viable time of 8 min (instead of 17 min), enabling the routine clinical assessment of lipid composition.
Methods
A total of 72 lipid composition spectra were acquired using DQF-COSY from excised human breast tumour specimens, healthy female volunteers and patients with breast cancer (Fig. 1a). The ex vivo study was approved by the North West—Greater Manchester East Research Ethics Committee (REC reference: 16/NW/0032). The in vivo studies were approved by the North of Scotland Research Ethics Service (REC reference: 16/NS/0077) and the London—Central Research Ethics Service (REC reference: 17/LO/1777). All experiments were conducted in accordance with the Declaration of Helsinki guidelines and all participants provided written informed consent prior to the study. All scans were performed on a 3 T clinical MRI scanner (Achieva TX, Philips Healthcare, Best, Netherlands) using a body coil for uniform transmission.
Ex vivo study
Seventeen female patients (mean age 61 years, age range 42–78 years) with invasive carcinoma (eight grade II and nine grade III), without prior hormonal therapy or chemotherapy and a tumour size greater than 10 mm in diameter were enrolled. The freshly excised whole tumour at surgery was immediately scanned before formalin treatment using a 32-element phased-array receiver coil for signal detection. Clinical standard T1-weighted and T2-weighted anatomical images were acquired for voxel localisation. 2D spectra of lipid composition were acquired using DQF-COSY8 with repetition time (TR) of 552 ms, initial echo time (TE) of 25 ms, a t1 increment of 1 ms, 256 increments (mixing encoding t1 time domain axis, F1 frequency domain axis), 256 sampling points (reading encoding t2 time domain axis, F2 frequency domain axis), 4 repeats per increment, spectral bandwidth of 1000 Hz, and DQF gradients of 30/40/100 ms mT/m. Reference spectra without water suppression were acquired using single voxel PRESS sequence26 with TR/TE of 1250/144 ms, 1024 data points, spectral bandwidth of 2000 Hz and 16 averages. The voxel was positioned to cover the whole tumour, with a voxel volume ranging from 2.7 to 16.5 cm3 according to tumour size.
In vivo study
Fifteen healthy female volunteers (mean age 66 years, age range 58–76 years) without previous breast cancer or family history of breast cancer participated in the study. Fifteen patients (mean age 63 years, age range 53–71 years, seven grade II and eight grade III) and a further ten patients (mean age 52 years, age range 36–63 years, one grade II and nine grade III) with invasive carcinoma were enrolled into the study for the acquisition of lipid composition spectra from peritumoural adipose tissue and tumour respectively. Patients with a tumour size greater than 10 mm, without prior chemotherapy or hormonal therapy, and no conditions contraindicative to MRS were eligible. All participants were scanned in the prone position as clinical routine practice using a 16-element phased-array breast receiver coil for signal detection. Standard sagittal T1-weighted anatomical images, axial T2-weighted anatomical images and diffusion weighted images were acquired for voxel localisation. 2D spectra of lipid composition were acquired using DQF-COSY8 with TR of 552 ms, initial TE of 25 ms, a t1 increment of 1 ms, 256 increments (mixing encoding t1 time domain axis, F1 frequency domain axis), 256 sampling points (reading encoding t2 time domain axis, F2 frequency domain axis), 2 repeats per increment, spectral bandwidth of 1000 Hz, and DQF gradients of 30/40/100 ms mT/m. Reference spectra without water suppression were acquired using single voxel PRESS sequence26 with TR/TE of 1250/144 ms, 1024 data points, spectral bandwidth of 2000 Hz and 16 averages. In healthy volunteers, data were acquired from both breasts and the voxel size was set to 2 \(\times\) 2 \(\times\) 2 cm3 containing primarily the adipose tissue. In patients, the voxel covering the tumour had a volume ranging from 2.2 to 21 cm3 for tumours in vivo (10 patients), while a voxel of 2 \(\times\) 2 \(\times\) 2 cm3 was positioned at 1 cm from the tumour for peritumoural adipose tissue (15 patients).
Data processing
All the algorithms were developed in MATLAB (MathWorks, Natick, MA, USA) with the processing flowchart shown in Fig. 1b. The raw data were averaged across repeated acquisitions before signal combination. The averaged signal, organised as a 2D map based on t1 and t2 time domain axes for each coil element, was subsequently apodised using squared sine bell along both time domain axes and zero filled to 512 \(\times\) 512 points. The reference spectrum without water suppression was used as external reference with the maximum peak (either water or lipid) as target metabolite17 while the first t1 increment of the DQF-COSY acquisition was used as internal reference with the maximum peak in frequency domain (either residual water or lipid) as the target metabolite. Both external (denoted by subscript “e”) and internal (denoted by subscript “i”) weighting coefficients, containing weights and phase, were computed using external and internal references respectively for each dataset and for each algorithm. The weighting coefficient derived for a coil element was applied to the apodised and zero filled signals at the corresponding coil element. The combined 2D time domain signal was the summation across all the coil elements, and the combined 2D spectrum was subsequently derived using 2D Fast Fourier transform.
The SNR of a spectral peak (Table 2) was computed as the peak height in the magnitude spectral map divided by the standard deviation of the real part of the noise in the square region covering (F1: 6.9–8.4, F2: 6.0–7.5) ppm27, with the overall SNR quantified at (1.3, 1.3) ppm and SNR improvement referenced to Equal Weighting algorithm27. The non-uniformity of SNR improvement across the 2D spectrum was subsequently derived as the coefficient of variance (standard deviation divided by the mean) of the SNR improvement across all the spectral peaks19. A 3D scatter plot of SNR improvement at the spectral location of the peak was then created (Fig. 5a). A plane was subsequently regressed onto the 3D scatter plot to derive the direction and degree of bias in frequency along the diagonal (low at (0.9, 0.9) ppm to high at (5.3, 5.3) ppm) and in encoding along the off diagonal (reading t2/F2 at (0.9, 5.3) ppm to mixing t1/F1 at (5.3, 0.9) ppm).
Statistical analysis
Statistical analysis was performed in SPSS (Release 24.0, SPSS Inc., Chicago, USA). Shapiro–Wilk test was performed on the SNR and non-uniformity to assess if the distribution was normal. The SNR across noise decorrelated algorithms was compared using a one-way ANOVA with repeated measures and a Wilcoxon signed-rank test for normally and non-normally distributed data respectively. The SNR of the linear algorithms was also reported for reference purposes. Data with negative SNR improvement for the calculation of the non-uniformity were excluded (1 healthy volunteer using AOCi, 1 healthy volunteer using nd-combi, and 1 tumour in vivo using nd-combi). A p-value < 0.05 was considered statistically significant.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Freed, M. et al. Evaluation of breast lipid composition in patients with benign tissue and cancer by using multiple gradient-echo MR imaging. Radiology 281, 43–53 (2016).
Bree, E. D. et al. Adipose tissue fatty acid composition in Greek patients with breast cancer versus those with benign breast tumors. Anticancer Res. 33, 1667–1672 (2013).
Bray, F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 (2018).
Coum, A. et al. In vivo MR spectroscopy of human breast tissue: Quantification of fatty acid composition at a clinical field strength (3 T). Magn. Reson. Mater. Phys. Biol. Med. 29, 1–4 (2016).
He, Q. et al. In vivo MR spectroscopic imaging of polyunsaturated fatty acids (PUFA) in healthy and cancerous breast tissues by selective multiple-quantum coherence transfer (Sel-MQC): A preliminary study. Magn. Reson. Med. 58, 1079–1085 (2007).
Thomas, M. A., Binesh, N., Yue, K. & DeBruhl, N. Volume-localized two-dimensional correlated magnetic resonance spectroscopy of human breast cancer. J. Magn. Reson. Imaging 14, 181–186 (2001).
Cavanagh, J., Fairbrother, W. J., III, A. G. P. & Rance, M. Principles and Practice: Protein NMR Spectroscopy. (2007).
Prescot, A. P. et al. Localized COSY and DQF-COSY1H-MRS sequences for investigating human tibial bone marrow in vivo and initial application to patients with acute leukemia. J. Magn. Reson. Imaging 22, 541–548 (2005).
Agarwal, K. et al. Study of lipid metabolism by estimating the fat fraction in different breast tissues and in various breast tumor sub-types by in vivo 1H MR spectroscopy. Magn. Reson. Imaging 49, 116–122 (2018).
Thomas, M. A. et al. Two-dimensional MR spectroscopic characterization of breast cancer in vivo. Technol. Cancer Res. Treat. 4, 99–106 (2005).
Roemer, P. B., Edelstein, W. A., Hayes, C. E., Souza, S. P. & Mueller, O. M. The NMR phased array. Magn. Reson. Med. 16, 192–225 (1990).
Wright, S. M. & Wald, L. L. Theory and application of array coils in MR spectroscopy. NMR Biomed. 10, 394–410 (1997).
Fang, L., Wu, M., Ke, H., Kumar, A. & Yang, S. Adaptively optimized combination (AOC) of magnetic resonance spectroscopy data from phased array coils. Magn. Reson. Med. 75, 2235–2244 (2015).
Hall, E. L., Stephenson, M. C., Price, D. & Morris, P. G. Methodology for improved detection of low concentration metabolites in MRS: Optimised combination of signals from multi-element coil arrays. NeuroImage 86, 35–42 (2014).
Martini, N. et al. Noise correlations and SNR in phased-array MRS. NMR Biomed. 23, 66–73 (2010).
Rodgers, C. T. & Robson, M. D. Receive array magnetic resonance spectroscopy: Whitened singular value decomposition (WSVD) gives optimal Bayesian solution. Magn. Reson. Med. 63, 1–11 (2017).
Mallikourti, V. et al. Optimal phased-array signal combination for polyunsaturated fatty acids measurement in breast cancer using multiple quantum coherence MR spectroscopy at 3T. Sci. Rep. 9, 9259 (2019).
Wu, M., Fang, L., Ray, C. E. Jr., Kumar, A. & Yang, S. Adaptively optimized combination (AOC) of phased-array MR spectroscopy data in the presence of correlated noise: Compared with noise-decorrelated or whitened methods. Magn. Reson. Med. 30, 672–712 (2016).
Abdoli, A. & Maudsley, A. A. Phased-array combination for MR spectroscopic imaging using a water reference. Magn. Reson. Med. 76, 733–741 (2015).
Dong, Z. & Peterson, B. The rapid and automatic combination of proton MRSI data using multi-channel coils without water suppression. Magn. Reson. Imaging 25, 1148–1154 (2007).
Frahm, J. et al. Localized high-resolution proton NMR spectroscopy using stimulated echoes: Initial applications to human brain in vivo. Magn. Reson. Med. 9, 79–93 (1989).
Dimitrov, I. E. et al. In vivo determination of human breast fat composition by 1H magnetic resonance spectroscopy at 7 T. Magn. Reson. Med. 67, 20–26 (2011).
Thakur, S. B. et al. Quantitative in vivo proton MR spectroscopic assessment of lipid metabolism: Value for breast cancer diagnosis and prognosis. J. Magn. Reson. Imaging 50, 239–249 (2019).
Kim, Y. J. et al. Histogram analysis of apparent diffusion coefficients after neoadjuvant chemotherapy in breast cancer. Jpn. J. Radiol. 34, 657–666 (2016).
Ma, J. Dixon techniques for water and fat imaging. J. Magn. Reson. Imaging 28, 543–558 (2008).
Bottomley, P. A. Spatial localization in NMR spectroscopy in vivo. Ann. N. Y. Acad. Sci. 508, 333–348 (1987).
Wilson, N. E., Burns, B. L., Iqbal, Z. & Thomas, M. A. Correlated spectroscopic imaging of calf muscle in three spatial dimensions using group sparse reconstruction of undersampled single and multichannel data. Magn. Reson. Med. 74, 1199–1208 (2015).
Acknowledgements
The author would like to thank Dr Matthew Clemence (Philips Healthcare Clinical Science, UK) for clinical scientist support, Ms Bolanle Brikinns, Ms Louisa Pirie, Ms Linda Lett, and Ms Kate Shaw, for patient recruitment support, Ms Dawn Younie for logistic support, Mr Roger Bourne and Ms Mairi Fuller for providing access to the patients as well as Mrs Beverly MacLennan, Mrs Nicola Crouch, Mr Mike Hendry, and Ms Laura Reid for radiographer support.
Funding
This project was funded by Friends of Aberdeen and North Centre for Haematology, Oncology and Radiotherapy (ANCHOR), Tenovus Scotland, and NHS Grampian Endowment. Vasiliki Mallikourti’s PhD study is supported by The Princess Royal Tenovus Scotland Medical Research Scholarship.
Author information
Authors and Affiliations
Contributions
V.M. implemented signal combination algorithms, conducted recruitment, performed in vivo data acquisition, conducted data analysis, interpreted the results and drafted the manuscript. S.C. managed the study paperwork, conducted recruitment, performed ex vivo and in vivo data acquisition, interpreted results, and reviewed the manuscript. T.G. performed radiological assessment, conducted patient recruitment, secured the funding, interpreted results and reviewed the manuscript. N.S. managed the study paperwork, conducted patient recruitment, interpreted results and reviewed the manuscript. Y.M. performed the surgical intervention, performed patient recruitment, secured the funding, interpreted results and reviewed the manuscript. T.M. conducted patient recruitment, interpreted results and reviewed the manuscript. R.S. secured the funding, conducted patient recruitment, interpreted results and reviewed the manuscript. S.H. secured the funding, interpreted results and reviewed the manuscript. J.H. secured the funding, designed the study, coordinated the experiments, interpreted the results and drafted the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mallikourti, V., Cheung, S.M., Gagliardi, T. et al. Phased-array combination of 2D MRS for lipid composition quantification in patients with breast cancer. Sci Rep 10, 20041 (2020). https://doi.org/10.1038/s41598-020-74397-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-74397-y
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.