Abstract
Desorption electrospray ionisation mass spectrometry (DESI-MS) can image hundreds of molecules in a 2D tissue section, making it an ideal tool for mapping tumour heterogeneity. Tumour lipid metabolism has gained increasing attention over the past decade; and here, lipid heterogeneity has been visualised in a glioblastoma xenograft tumour using 3D DESI-MS imaging. The use of an automatic slide loader automates 3D imaging for high sample-throughput. Glioblastomas are highly aggressive primary brain tumours, which display heterogeneous characteristics and are resistant to chemotherapy and radiotherapy. It is therefore important to understand biochemical contributions to their heterogeneity, which may be contributing to treatment resistance. Adjacent sections to those used for DESI-MS imaging were used for H&E staining and immunofluorescence to identify different histological regions, and areas of hypoxia. Comparing DESI-MS imaging with biological staining allowed association of different lipid species with hypoxic and viable tissue within the tumour, and hence mapping of molecularly different tumour regions in 3D space. This work highlights that lipids are playing an important role in the heterogeneity of this xenograft tumour model, and DESI-MS imaging can be used for lipid 3D imaging in an automated fashion to reveal heterogeneity, which is not apparent in H&E stains alone.
Similar content being viewed by others
Introduction
The revised 4th edition of the classification edited by the World Health Organisation (WHO) recognises more than 120 tumour entities with distinctive morphology, location, age distribution and biologic behaviour. Tumour entities are graded with a ‘malignancy scale’ that is intended to predict their natural biological behaviour. Gliomas account for the commonest primary brain tumours in adults. They are graded from I to IV based on histological features. The identification of molecular and genetic signatures such as mutations in the isocitrate dehydrogenase 1 and 2 (IDH1/2) genes, ATRX and hTERT, CDKN2A/B deletions, EGFR gene amplifications and co-deletions of chromosomes 1p19q has improved the diagnostic accuracy and stratification of patients with glioma. Glioblastoma (GBM) (WHO grade IV) is the most aggressive among gliomas with a patients’ mean survival of 15 months from onset1. GBM can occur as a primary tumour or can result from the progression of a lower grade astrocytoma. Primary (de novo) GBM is the most common type accounting approximately for 90% of cases. Glioblastomas are highly heterogeneous tumours. Regional genetic, epigenetic and metabolic differences determine intratumoural heterogeneity. Intratumour heterogeneity is a result of evolutionary changes that occur following dynamic remodelling of the tumour microenvironment and the effects of treatment2,3. These genetic alterations can contribute towards resistance to therapy4. Among metabolic changes in GBM, lipid levels including free fatty acids are altered compared to normal tissue5,6 as the result of hypoperfusion and subsequent hypoxia7,8. Fatty acid synthase (FASN) is responsible for de novo synthesis of fatty acids, and is upregulated in gliomas9, amongst other cancer types10,11, with an increase in expression being associated with malignant gliomas12.
Gliomas are diagnosed and monitored with magnetic resonance imaging (MRI). Molecular imaging modalities such as positron emission tomography (PET) can also contribute to the management of patients with glioma. Both MRI and PET have progressed substantially in investigating the heterogeneity of glial tumours as they can image whole tumours in-vivo13,14,15 but they may fail to detect subtle but relevant structural or functional features due to their low spatial resolution.
Mass spectrometry imaging is an ex-vivo molecular imaging method that offers advantages over PET and MRI because it can image hundreds of molecules at a spatial resolution below 200 μm. Desorption electrospray ionisation mass spectrometry (DESI-MS) is an ambient mass spectrometry technique which has been used for molecular imaging since 200416,17,18. Although DESI-MS is an ex-vivo technique, it causes minimal degradation to the sample, and so staining is possible on the same section post-DESI-MS imaging, making it an ideal tool for imaging brain tumours. Unlike in-vivo techniques such as PET or MRI, MS imaging only captures 2D images. However, MS imaging can acquire serial tissue sections, and stack as 3D images. Matrix-assisted laser desorption ionisation (MALDI-MS) imaging is the most commonly used MS imaging technique and has already been used for 3D imaging19 including the mapping of drug distribution in tumours20. Eberlin et al. reported 3D DESI-MS imaging of a mouse brain21. 3D MS imaging is a lengthy process and involves human input to set up and acquire new images. Here, for the first time, an automated 3D DESI-MS imaging method has been used to visualise tumour heterogeneity.
A robotic slide loader that can autonomously mount and change slides on the DESI-MS instrument allows rapid acquisition of a set of 2D images that are automatically processed and aligned to render a 3D image. This enables straightforward and rapid generation of 3D DESI-MS images. DESI-MS is an excellent tool for the detection of lipids, and has been used in numerous lipidomic studies in cancer22,23,24. In gliomas, DESI-MS lipid fingerprints could distinguish between healthy and tumour tissue and between different glioma grades25,26.
In this study we performed 3D DESI-MS imaging using a heterotopic xenograft glioblastoma model to investigate lipid heterogeneity and immunofluorescence to find correlations between the MS lipid heterogeneity data and the downstream marker of hypoxic signalling, carbonic anhydrase-9 (CA-9).
Results and discussion
Fifteen sections were taken every 120 μm from each GBM xenograft and underwent DESI-MS imaging in both negative and positive ion mode. The 2D data were combined and reconstructed to generate 3D DESI-MS images for both polarities. Multivariate analysis (MVA) was performed on the 3D data set to reveal areas of the tumour which had different regions according to their lipid profiles (Fig. 1). 3D DESI-MS imaging was therefore able to visualise areas of distinct lipid heterogeneity within the tumour.
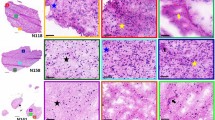
2D images of representative peaks from the distinct tumour areas as separated by the MVA is shown in Fig. 2. A peak appearing in negative ion mode at m/z 885 has been tentatively identified as PI (38:4) based on exact mass and what has previously been reported in the literature, with Eberlin et al. also reporting m/z 885 (PI 38:4) in viable GBM25. Sections adjacent to those used for DESI-MS were stained with CA-9 (to visualise hypoxic regions) and H&E, these were interpreted by a neuropathologist. Molecularly different regions visualised in the DESI-MS images showed different cellularity, with hypoxic regions appearing less cellular than the remaining tumour.
Comparing DESI-MS images with CA-9 staining (Figs. 3 and 4) revealed that the peaks at m/z 400 and m/z 428 (positive ion mode) co-localise with areas of lower cellularity (shown as blue in Fig. 3). The peaks at m/z 400 and m/z 428 co-localised with hypoxia but only in some hypoxic regions forming a thin band within the areas of CA-9 staining. The m/z 400 and 428 band was overlaid with CA-9 images to visualise exactly where in the hypoxic area the m/z 400/428 band was forming (Fig. 5). The m/z 400/428 band was found to localise in the midst of CA-9 staining; rather than forming a boundary between hypoxic and normoxic tissue. These findings suggest that the lipids at m/z 400 and 428 may only be upregulated in very specific oxygen tensions. Furthermore, the 3D positive ion mode DESI-MS image was able to visualise this metabolically unique area, allowing three dimensional interrogation of this heterogeneity.
Peaks observed at m/z 428 and m/z 400 have been identified as stearoylcarnitine and palmitoylcarnitine respectively, based on the Lipid Maps database27 and what has been previously reported in hypoxic areas28. In addition, the MS/MS spectrum for m/z 428 obtained from the tissue samples (Fig. 5), shows a fragment peak at 369, representative of the loss of a trimethylamine group from the carnitine. The MS/MS imaging shows that this fragment co-localises with the m/z 428 parent peak (Fig. 5, inset images).
Acyl-carnitines act as intermediates to shuttle fatty acids into the mitochondria. Long chain fatty acids cannot cross the mitochondrial membranes for reasons of permeability and must be converted to fatty acyl-CoAs, which are in turn converted to acylcarnitines. In the mitochondria, acylcarnitines are converted back to fatty acyl-CoAs to undergo β-oxidation and eventually produce energy. The import into the mitochondria is the rate limiting step of fatty acid oxidation and is carried out by various carnitine transferases.
Lin et al. reported that fatty acid oxidation is prevalent in cells derived from human gliomas and that it plays an important role in proliferation. They also found that inhibition of fatty acid oxidation leads to tumour growth and increased survival in a syngeneic mouse glioma model29. This finding concurs with the work of Huang et al., who demonstrated the importance of the hypoxia inducible factor-1α (HIF-1α) protein in inhibiting fatty acid oxidation at low oxygen tensions, resulting in cancer cell proliferation30. Interestingly, we report an increase in β-oxidation within a subset of tumour hypoxia (relative to the normoxic regions of the U87 xenograft model), eluding to complex mechanisms at play within specific oxygen tensions.
Zaugg et al. discussed the role of carnitine palmitoyltransferase 1C (CPT1C), a brain-specific enzyme responsible for transporting carnitines across the mitochondrial membrane. Their findings suggest that hypoxia can cause upregulation of CPT 1C in vivo31, in keeping with our evidence of an increase in palmitoylcarnitine in hypoxic regions of the U87 xenograft tumour model. These findings support the complex role of hypoxic regulation of carnitines in GBM, where their upregulation at specific oxygen tensions confers a metabolic advantage to hypoxic cells.
Conclusion
In conclusion, we have devised a method for 3D DESI-MS imaging of a glioma model and analysed its heterogeneity. Automation of experiment and of 3D visualisation facilitates its use as a high throughput imaging modality that is suitable for large cohorts of samples. Furthermore, the use of consecutive sections allows the acquisition of a considerable amount of information on the biology of the tumour analysed. Despite the limitations of our U87 xenograft model, distinct regions of heterogeneous lipid expression that correlate with the varying levels of local oxygen tension and palmitoyl- and stearoyl-carnitines, appear to be playing key roles to aid β-oxidation in hypoxic areas of GBMs. Mapping on tissue the metabolic shift to lipid metabolism in regions with low oxygen tension is relevant to better understanding tumour metabolic heterogeneity in GBM and the impact of hypoxia on the dynamic changes of tumour microenvironment and its consequences on resistance to radiation treatment and chemotherapy32,33,34,35.
Methods
Materials
Phosphate buffered saline (PBS) was made using tablets purchased from Oxoid, UK, and dissolved in dH2O.
1 ml Tween (T) was added to PBS to make PBS-T. BSA (Sigma-Aldrich, UK) was dissolved at 1 mg/ml PBST to make PBST-BSA solution.
All solvents were purchased from Sigma-Aldrich, UK.
In-vivo experiments
Animal experiments were conducted in accordance with the U.K. Animals (Scientific Procedures) Act 1986, and had institutional and regulatory ethical approval (University of Manchester Animal Welfare and Ethical Review- AWERB) under Home Office license PPL707760 (granted to KJW). 5 × 106 U87 cells were injected intradermally into the flank of a female cba nu/nu mice (age 10–14 weeks/weight 20–24 g). Mice were maintained under dawn-til-dusk 12 h light/dark cycles at an average ambient temperature of 21 °C (range 19–23 °C), relative humidity average 55% (range 45–65%) in pathogen free housing (Techniplast GM500 Mouse IVC Green Line) with sterilized bedding, environmental enrichment and access to food water ad libitum with water supplied by Hydropac watering system. Weights, body condition and behaviour were monitored routinely throughout the experiments as measures of animal well-being. After 4 weeks animals were culled and tumours flash frozen in isopentane.
Sample preparation
A xenograft glioblastoma tumour was sectioned using a cryostat (Leica 3050 s, UK), and tissue sections were thaw mounted onto glass slides. Fifteen sections were taken at 120 μm steps through the tumour, at a 10 μm thickness for DESI-MS imaging. Serial sections (8 μm) were taken for immunofluorescence.
DESI-MS 3D imaging
Experiments were carried out using a 2D DESI stage (Prosolia, Indianapolis,USA) attached to a Xevo-G2-XS quadrupole- time of flight (Q-TOF) (Waters, UK). A modified top plate of the DESI stage (Prior Scientific, Cambridge) was created to allow the automated PL200 microscope slide loader (Prior Scientific, Cambridge) to place slides onto the stage, and then move them back to the holding cassette. A snapshot of the slide and object definition algorithms were used to identify the location of the tissue(s) on the slide. These co-ordinates were then written to the experiment file and imaging commenced. This process continued until all 15 slides were analysed. Spatial resolution was set to 120 μm, in order to make even sized voxels. A mass range of m/z 50–1200 was used. Spray conditions used were 5 µl/min, 95% methanol 5% water, with a nebulising gas of nitrogen at 6 bar pressure. Typical positions of the sprayer were used (sprayer 1.5 mm above surface, 6 mm sprayer to capillary distance, 75° sprayer impact angle, 5° collection capillary angle). Imaging was carried out in both negative and positive ion mode, at 10 scans/sec. The capillary voltage was 2.3 kV in negative ion mode, and 3.42 kV in positive ion mode.
HDImaging v1.4 (Waters, Milford, USA) was used for visualisation of individual 2D images, where images were normalised to the total ion current (TIC). Recently developed software from Waters, UK allowed reconstruction of the sections into a 3D image of the molecularly distinct regions within the whole tissue volume. The reconstruction was carried out automatically, and involved peak picking, object alignment and rotation, as well as a non-negative matrix factorisation (NNMF).
DESI-MS/MS
Conditions were the same as DESI-MS imaging. MS/MS was carried out using 7 bar of argon gas. Lipidomics Gateway database https://www.lipidmaps.org27 was used for identification of species.
Immunofluorescence
Tissue sections were fixed in ice-cold acetone for 10 min and then left to dry. A DAKO pen (Agilent Technologies, UK) was used to draw around each of the sections, and 70 μl of blocking solution was pipetted onto each section (10% goat serum in PBST), and incubated at room temperature for 15 min. Slides were then washed twice for 3 min in PBST-BSA. Anti-CA-9 (mouse, Biosciences, Slovakia) at 1:1000 was made up in PBST-BSA, and 10% normal goat serum. ~ 70 μl of primary antibody solution was left on tissue sections at 4 °C overnight. The primary antibody solution was tapped off and the sections washed three times for four mins in PBS. A secondary antibody solution containing 1:100 Alexafluor 488 goat anti-mouse (Life technologies, UK) was prepared in PBST-BSA; and 70 μl was added to each section and left for 60 min in the dark at room temperature. Slides were washed again with PBS (3 × 4 min). Coverslips (VWR, UK) were then mounted using fluorescent mounting media (Abcam, UK). Images were acquired using a [20x/0.80 Plan Apo] objective using the 3D Histech Pannoramic 250 Flash II slide scanner, and processed using Panoramic viewer (3D HISTECH, Hungary). Images were also processed in Image J36, where images were converted to a 32-bit greyscale image, and a default threshold applied.
H&E
Sections adjacent to those used for DESI-MS were stained using haematoxylin and eosin (H&E). Following submersion in xylene (Sigma-Aldrich, UK) for 2 min, slides were immersed 4 times for 2 min in industrial methylated spirit (Sigma-Aldrich, UK). Following washing in tap water, sections were stained using Harris haematoxylin (3 min; Sigma-Aldrich, UK), rinsed in hot water, and dipped for one second in eosin (Leica, UK). Following further industrial methylated spirit (4 × 2 min) and xylene (4 × 2 min) washes, slides were left to dry. Samples were mounted with coverslips using aqueous mounting media (Abcam, UK) and left overnight to dry. Images were acquired using the 3D Histech Pannoramic 250 Flash II slide scanner with [20x/0.80 Plan Apo] objective and processed using Pannoramic viewer (3D HISTECH, Hungary).
Data availability
The datasets generated during and/or analysed during the current study are not publicly available as they are still being mined with the hope of a future publications, but are available from the corresponding author on reasonable request.
References
Louis, D. N. et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 131, 803–820 (2016).
Soeda, A. et al. The evidence of glioblastoma heterogeneity. Sci. Rep. 5, 1–7 (2015).
Parker, N. R. et al. Intratumoral heterogeneity identified at the epigenetic, genetic and transcriptional level in glioblastoma. Sci. Rep. 6, 22477 (2016).
Prados, M. D. et al. Toward precision medicine in glioblastoma: the promise and the challenges. Neuro-Oncol. 17, 1051–1063 (2015).
Srivastava, N. K., Pradhan, S., Gowda, G. A. & Kumar, R. In vitro, high-resolution 1H and 31P NMR based analysis of the lipid components in the tissue, serum, and CSF of the patients with primary brain tumors: one possible diagnostic view. NMR Biomed. 23, 113–122 (2010).
Tugnoli, V. et al. Characterization of lipids from human brain tissues by multinuclear magnetic resonance spectroscopy. Biopolymers 62, 297–306 (2001).
Zhang, Y. J., Wang, H., Zhang, J., Lv, J. W. & Huang, Y. R. Positive feedback loop and synergistic effects between hypoxia-inducible factor-2 and stearoylCoA desaturase-1 promote tumorigenesis in clear cell renal cell carcinoma. Cancer Sci. 104, 416–422 (2013).
Furuta, E. et al. Fatty acid synthase gene is up-regulated by hypoxia via activation of Akt and sterol regulatory element binding protein-1. Cancer Res. 68, 1003–1011 (2008).
Zhao, W. et al. Fatty acid synthase: a novel target for antiglioma therapy. Br. J. Cancer 95, 869–878 (2006).
Orita, H., Coulter, J., Tully, E., Kuhajda, F. P. & Gabrielson, E. Inhibiting fatty acid synthase for chemoprevention of chemically induced lung tumors. Clin. Cancer Res. 14, 2458–2464 (2008).
Rossi, S. et al. Fatty acid synthase expression defines distinct molecular signatures in prostate cancer. Mol. Cancer Res. 1, 707–715 (2003).
Tao, B. B. et al. Up-regulation of USP2a and FASN in gliomas correlates strongly with glioma grade. J. Clin. Neurosci. 20, 717–720 (2013).
Willaime, J. M. Y., Turkheimer, F. E., Kenny, L. M. & Aboagye, E. O. Quantification of intra-tumour cell proliferation heterogeneity using imaging descriptors of 18F fluorothymidine-positron emission tomography. Phys. Med. Biol. 58, 187–203 (2013).
Eary, J. F., O’Sullivan, F., O’Sullivan, J. & Conrad, E. U. Spatial heterogeneity in sarcoma 18F-FDG uptake as a predictor of patient outcome. J. Nucl. Med. 49, 1973–1979 (2008).
Asselin, M. C., O’Connor, J. P. B., Boellaard, R., Thacker, N. A. & Jackson, A. Quantifying heterogeneity in human tumours using MRI and PET. Eur. J. Cancer 48, 447–455 (2012).
Takats, Z., Wiseman, J. M., Gologan, B. & Cooks, R. G. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science 306, 471–473 (2004).
Abbassi-Ghadi, N. et al. Repeatability and reproducibility of desorption electrospray ionization-mass spectrometry (DESI-MS) for the imaging analysis of human cancer tissue: a gateway for clinical applications. Anal. Methods 7, 71–80 (2015).
Banerjee, S. et al. Diagnosis of prostate cancer by desorption electrospray ionization mass spectrometric imaging of small metabolites and lipids. Proc. Natl. Acad. Sci. USA 114, 3334–3339 (2017).
Chen, R. B., Hui, L. M., Sturm, R. M. & Li, L. J. Three dimensional mapping of neuropeptides and lipids in crustacean brain by mass spectral imaging. J. Am. Soc. Mass Spectrom. 20, 1068–1077 (2009).
Giordano, S. et al. 3D mass spectrometry imaging reveals a very heterogeneous drug distribution in tumors. Sci. Rep. 6, 1–8 (2016).
Eberlin, L. S., Ifa, D. R., Wu, C. & Cooks, R. G. Three-dimensional vizualization of mouse brain by lipid analysis using ambient ionization mass spectrometry. Angewandte Chemie-International Edition 49, 873–876 (2010).
Tata, A. et al. Rapid detection of necrosis in breast cancer with desorption electrospray ionization mass spectrometry. Sci. Rep. 6, 35374 (2016).
Jarmusch, A. K. et al. Lipid and metabolite profiles of human brain tumors by desorption electrospray ionization-MS. Proc. Natl. Acad. Sci. USA 113, 1486–1491 (2016).
Jayashree, B. et al. Monitoring of changes in lipid profiles during PLK1 knockdown in cancer cells using DESI MS. Anal. Bioanal. Chem. 408, 5623–5632 (2016).
Eberlin, L. S. et al. Classifying human brain tumors by lipid imaging with mass spectrometry. Cancer Res. 72, 645–654 (2012).
Eberlin, L. S. et al. Ambient mass spectrometry for the intraoperative molecular diagnosis of human brain tumors. Proc. Natl. Acad. Sci. USA 110, 1611–1616 (2013).
Sud, M. et al. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 35, D527–D532 (2007).
Chughtai, K., Jiang, L., Greenwood, T. R., Glunde, K. & Heeren, R. M. Mass spectrometry images acylcarnitines, phosphatidylcholines, and sphingomyelin in MDA-MB-231 breast tumor models. J. Lipid Res. 54, 333–344 (2013).
Lin, H. et al. Fatty acid oxidation is required for the respiration and proliferation of malignant glioma cells. Neuro Oncol. 19, 43–54 (2016).
Huang, D. et al. HIF-1-mediated suppression of Acyl-CoA dehydrogenases and fatty acid oxidation is critical for cancer progression. Cell Rep. 8, 1930–1942 (2014).
Zaugg, K. et al. Carnitine palmitoyltransferase 1C promotes cell survival and tumor growth under conditions of metabolic stress. Genes Dev. 25, 1041–1051 (2011).
Bronisz, A., Salinska, E., Chiocca, E. A. & Godlewski, J. Hypoxic roadmap of glioblastoma-learning about directions and distances in the brain tumor environment. Cancers 12, 1213 (2020).
Colwell, N. et al. Hypoxia in the glioblastoma microenvironment: shaping the phenotype of cancer stem-like cells. Neuro-Oncol. 19, 887–896 (2017).
Monteiro, A. R., Hill, R., Pilkington, G. J. & Madureira, P. A. The role of hypoxia in glioblastoma invasion. Cells 6, 45 (2017).
Vartanian, A. et al. GBM’s multifaceted landscape: highlighting regional and microenvironmental heterogeneity. Neuro-Oncol. 16, 1167–1175 (2014).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Acknowledgements
The University of Manchester was supported by the CRUK & EPSRC Cancer Imaging Centre in Cambridge & Manchester Grant (Cambridge C197/A16465, Manchester C8742/A18097). FH was funded through a BBSRC Ph.D. Studentship. The Bioimaging Facility microscopes used in this study were purchased with grants from BBSRC, Wellcome and the University of Manchester Strategic Fund. Special thanks go to Roger Meadows for his help with the microscopy and image analysis.
Author information
Authors and Affiliations
Contributions
F.H. carried out 2D DESI-MS image analysis, immunofluorescence, immunofluorescence image analysis, and H&E staining, with interpretation of the histopathology by F.R. E.J. performed all DESI-MS imaging and 3D DESI-MS image analysis. J.D. assisted in immunofluorescence H&E staining. R.C., E.H., and E.J. developed the automatic slide loader robot for 3D DESI-MS imaging. L.C. sectioned the glioblastoma. E.J. was supervised by EC who both facilitated the collaboration with Waters, UK. K.J.W. and A.M. supervised F.H. and provided advice and guidance throughout the research.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Henderson, F., Jones, E., Denbigh, J. et al. 3D DESI-MS lipid imaging in a xenograft model of glioblastoma: a proof of principle. Sci Rep 10, 16512 (2020). https://doi.org/10.1038/s41598-020-73518-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-73518-x
This article is cited by
-
Metabolic heterogeneity in cancer
Nature Metabolism (2024)
-
Mass spectrometry imaging discriminates glioblastoma tumor cell subpopulations and different microvascular formations based on their lipid profiles
Scientific Reports (2022)
-
Potential impact of tissue molecular heterogeneity on ambient mass spectrometry profiles: a note of caution in choosing the right disease model
Analytical and Bioanalytical Chemistry (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.