Abstract
In remote areas of malaria-endemic countries, rapid diagnostic tests (RDTs) have dramatically improved parasitological confirmation of suspected malaria cases, especially when skilled microscopists are not available. This study was designed to determine the frequency of Plasmodium falciparum isolates with histidine-rich protein 2 (pfhrp2) gene deletion as one of the possible factors contributing to the failure of PfHRP2-based RDTs in detecting malaria. A total of 300 blood samples were collected from several health centres in Nyala City, Western Sudan. The performance of PfHRP2-based RDTs in relation to microscopy was examined and the PCR-confirmed samples were investigated for the presence of pfhrp2 gene. A total of 113 out of 300 patients were P. falciparum positive by microscopy. Among them, 93.81% (106 out of 113) were positives by the PfHRP2 RDTs. Seven isolates were identified as false negative on the basis of the RDTs results. Only one isolate (0.9%; 1/113) potentially has pfhrp2 gene deletion. The sensitivity and specificity of PfHRP2-based RDTs were 93.81% and 100%, respectively. The results provide insights into the pfhrp2 gene deletion amongst P. falciparum population from Sudan. However, further studies with a large and systematic collection from different geographical settings across the country are needed.
Similar content being viewed by others
Introduction
Malaria remains a major public health problem particularly in sub Saharan Africa in which majority of the populations are at risk1. Late diagnosis and treatment of the disease can lead to life threatening conditions2. The World Health Organization (WHO) implements vector control, early diagnosis and prompt treatment strategies to eliminate malaria3. Accordingly, the early, rapid and accurate diagnosis is an essential component in malaria control and elimination programs. Microscopic detection of malaria parasites is still considered the gold standard for diagnosis of the disease; this method is relatively sensitive and allows parasitic quantitatification and species identification4,5. However, in remote areas, this procedure may not be easily available due to the requirement of high-quality equipment and the challenge in maintaining well-trained microscopists6. Alternatively, WHO recommends the use of rapid diagnostic tests (RDTs), which have later become an alternative way of establishing the rapid diagnosis of malaria infections7. Accordingly, RDTs were implemented as part of malaria case management along with microscopy for diagnosing malaria among suspected cases in Sudan8.
Most of the current malaria RDTs target the histidine-rich protein-2 (HRP2), which is a species-specific antigen of Plasmodium falciparum9. Recent reports indicate that PfHRP2-based RDTs may provide false negative results10. Though among the causes of false negative results are product quality, transportation, storage distribution, parasite density and due to the existence of P. falciparum isolates that lack the pfhrp2 gene. Such deletions have been reported in a broader range of malaria-endemic countries in Africa11,12,13, South America14,15 and Asia16.
Recent evidence suggested that P. falciparum isolates with pfhrp2 gene deletion also circulate in the neighbouring countries17,18 with more than 80% of microscopically confirmed isolates found to carry pfhrp2 gene deletion in Eritrea19. Recently, P. falciparum isolates with deleted pfhrp2 was also reported in UK travellers from Sudan and south Sudan11. This condition highlights the importance of molecular surveillance to detect these deletions because they could lead to false-negative diagnoses when using PfHRP2-based malaria RDTs20.
Despite the diagnostic threat posed by P. falciparum parasites lacking the pfhrp2 gene, sporadic data is available about the existence of P. falciparum parasites with pfhrp2 deletion in Sudan, which has high burden of malaria in the region. Screening of P. falciparum parasites with pfhrp2 deletions in Sudan is essential to asses RDTs performance and the impact on clinical case management and malaria elimination efforts. Therefore, the present study aimed to investigate the pfhrp2 gene deletion in P. falciparum isolates in Nayala City, Western Sudan.
Results
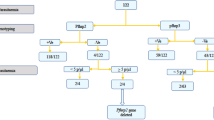
A total of 300 suspected patients with malaria were screened for malaria parasite by microscopy; 113 patients were positive for P. falciparum infection. Subsequently, the microscopically confirmed specimens were tested against HRP2-based malaria RDTs. Amongst the isolates, the PfHRP2 RDTs identified 106 (93.81%) positives. A total of 7 out of 113 of P. falciparum microscopy-confirmed cases were RDT negative. This finding resulted in overall 93.81% [95% confidence interval (CI) 87.65–97.47] and 100% (95% CI 98.05–100) sensitivities and specificities of the PfHRP2-based RDTs respectively (Table 1). The parasite density of the seven RDT-negative specimens was 96–5,120 asexual parasites/µl with a mean of 1,540 parasites/µl.
Confirmation of P. falciparum infection by PCR
All the microscopy-confirmed P. falciparum cases were P. falciparum infections (113 out of 113) because all of them generated PCR products for P. falciparum specific ssrRNA primers. Therefore, the 106 RDT-positive isolates were found not eligible for further pfhrp2 deletion analysis. The remaining seven isolates were further investigated to examine whether pfhrp2 gene was lacking given the negative RDT results of these specimens.
Detection of Pfhrp2 gene
Plasmodium falciparum ssrRNA gene was amplified in all seven RDT-negative isolates. The samples were subjected to another round of PCR to amplify the flanking regions of pfhrp2. The pfhrp2 gene was successfully amplified in most of the isolates (six out of seven, 85.7%), this exclude the deletion of the entire gene in all these samples. Amongst the seven samples, the entire pfhrp2 gene was deleted in one sample (14.3%). The pfcsp gene was successfully amplified to confirm this deletion. In addition the parasitic density of this sample was relatively high (Table 2).
Discussion
Despite the substantial drop in malaria cases and deaths over the past decade, efforts should be intensified to ensure prompt and sensitive diagnosis and effective treatment. PfHRP2-based RDTs are valuable tools for P. falciparum malaria diagnosis, especially in areas where routine microscopic diagnosis is inaccessible. However, studies across the globe have reported the reduced diagnostic performance of PfHRP2-based RDTs. Evidence from many African countries revealed that HRP2-based RDTs failed to detect P. falciparum parasites19. This observed failure is attributed to various factors, including the lack of the pfhrp2 gene, which is an important factor responsible for false negative results. In Sudan, there were a few reports of false negative RDT results associated with pfhrp2-negative, though the availability of data regarding the Pfhrp2 deletion in Sudanese population are limited21. Given that pfhrp2-deleted P. falciparum isolates have been reported in the country11,22, this study investigated the possible circulation of parasites with pfhrp2 deletions in western part of Sudan.
Our study reports, for the first time, the presence of P. falciparum parasites with pfhrp2 deletion in western Sudan. Of the 7 false negative RDT results, one isolate was smear-positive and pfhrp2 PCR-negative. The presence of parasite DNA and its quality was verified by two other independent genes, suggesting that the sample has pfhrp2 gene deletion. While the cause of false negative RDT for the 6 samples (5.3%) is unknown, 0.9% (1/113) of the false negative can be explained by pfhrp2 deletion. The six samples determined as positive by microscopy and negative using HRP2 RDT were confirmed positive by PCR as indicated by the amplified pfhrp2 gene. This finding could be attributed to the sequence variability among the pfhrp2 region or low parasitaemia as some of the samples had parasitaemia below the detection limit of RDT12,23. Among the false negative isolates by RDT, the number of parasites density varies from 96 to 5,120 asexual parasites/µl. A very low parasite densities or target antigen concentrations could also contributed to false negative results. Another possible explanation is that other factors, including the product quality, transportation, storage and distribution, possibly affected the performance of RDTs. This is very unlikely as SD Bioline Pf/Pv/Mixed Combo cassettes used in this study was prequalified by WHO, and the shipment and storage conditions in the field were optimal and followed WHO recommendations. Thus, they are less likely to be the reasons for false negative results in this study. The most likely explanation could be low parasitaemia and/or sequence variation of the repeat region of the pfhrp2, which is targeted by the HRP2-based RDT.
The only one pfhrp2 negative isolate may not reflect the actual number of the pfhrp2 negative P. falciparum among the study population. This is because the 106 RDT positive isolates were not screened for pfhrp2 gene deletion. The RDT positive signal could be generated from HRP3 due to the cross reactivity of HRP3 antigen with the HRP2. In this context, the actual number of pfhrp2 deletion could be higher than the estimated prevalence. Nevertheless, whether the deletion of pfhrp2 is compensated by the presence of HRP3 could not be determined and this is one of the limitations of the study.
Limited information is available regarding the deletions of pfhrp2 gene in samples obtained from Sudanese patients. A similar recent study with a small sample size conducted by our group in the centre of Sudan reported genetic variations in pfhrp2 and pfhrp2-negative samples but deletion was not confirmed21. Our current study, however, found one deletion that caused RDT negative result. Study area, sample size and collection period could explain the differences in findings. Other studies also reported genetic variation and suspected deletion of pfhrp2 gene22 as well as pfhrp2 deletions in malaria patients among UK travellers from Sudan11, though it is not clear which part of Sudan the UK patient travelled from.
To date, parasites lacking the pfhrp2 gene have been unequivocally identified in many malaria endemic regions16. Despite the low prevalence of pfhrp2 deletion in the current study, the rates of deletions vary considerably in Africa and globally. Remarkably high rates of pfhrp2 gene deletions were reported from neighbouring Eritrea17. Moreover, findings from several countries indicated that P. falciparum isolate with pfhrp2 gene deletion is circulating in east Africa, including Kenya18, Tanzania and Uganda24, South Sudan11 and Rwanda25. By contrast, reports of presence of pfhrp2 gene deletions in West Africa have been limited, including in Mali12 and Senegal13. Recent studies provide evidence of deletion of the pfhrp2 gene as reported from Equatorial Guinea26 and Nigeria27. However, in many highly malaria-endemic countries, data about pfhrp2 deletion are missing. This underscores the importance of conducting comprehensive molecular surveillance to investigate the pfhrp2 gene deletion in all endemic countries.
The study was not designed to systematically identify pfhrp2 deletion in western Sudan and the prevalence does not reflect the true prevalence in the whole region. However, the prevalence of pfhrp2 deletion is considerably lower than 5%, which is the minimum threshold needed to change the RDT types based on WHO guidelines. Evidently, malaria RDTs tests that rely on PfHRP2 for the detection of P. falciparum infections are still suitable for P. falciparum malaria diagnosis in Sudan.
Despite the importance of monitoring the deletion of pfhrp2 gene across the whole country, this study was limited by the collection of samples from a single location in the western part of Sudan. The research needs to be expanded to cover other geographic regions given that this finding may not be representative of the situation in the whole country. Moreover, screening of pfhrp2 gene as well as the pfhrp3 gene among RDTs positive isolates could help in determining the actual number of HRP gene deletion among the study population, which is one of the limitations of the current study.
In conclusion, this study revealed that P. falciparum isolates with pfhrp2 gene deletion are present in the parasite populations in western Sudan. The PfHRP2-based RDTs used in this study detected the majority of P. falciparum infections. Further studies, including a large and systematic collection from different geographical settings across the country, will be essential to estimate the pfhrp2 deletion prevalence and to better assess the performance of PfHRP2-based RDTs.
Methods
Study population and site
A total of 300 suspected patients with malaria from 4 health centres in Nyala City were enrolled in a cross-sectional study conducted between July 2018 and April 2019. Nyala is the capital of the South Darfur State, Western Sudan and covers an area of about 127,300 km2. The city lies in a savannah zone between 9° 30′ 13″ N latitude and 15° 27′ 28″ E longitude. Patients presented with symptoms consistent with uncomplicated malaria attending the selected health centres were enrolled and blood smears for Malaria were obtained from each patient.
Sample collection
A total of 300 venous blood samples (3 ml each) were collected from each participant for the preparation of thin and thick blood films and for rapid diagnostic tests (RDTs) and for DNA extraction. An aliquot of blood was saved on a Whatman 3MM filter paper (Whatman, Clifton, NJ, USA) for parasite specimen preservation.
Microscopy and RDT analyses
Thick and thin blood smears were prepared and stained with 10% Giemsa for 15 min. Subsequently, the stained slides were examined under a 100 × oil immersion microscope by two independent expert microscopists (conducted by a senior laboratory technician in malaria reference laboratory in the state ministry of health). Parasitaemia was calculated by counting the number of parasites observed per 200 leukocytes and assuming a total of 8,000 leukocytes/µl. Simultaneously, all blood samples were assayed with SD BIOLINE Malaria Ag P.f/P.v RDT, which is a qualitative and differential test for the immunological detection of HRP2 and parasite lactate dehydrogenase of P. falciparum and P. vivax, respectively. The RDT assays were performed, and the results were interpreted according to the instructions provided by the manufacturers.
DNA extraction
For molecular analysis, genomic DNA was extracted from blood samples by using a Geneius™ Micro gDNA Extraction kit (Geneaid Biotech Ltd., Taiwan) in accordance with the manufacturer’s instructions. DNA was resuspended in 100 µl elution buffer and stored at − 20 °C.
Confirmation of P. falciparum infection by polymerase chain reaction (PCR)
The presence of P. falciparum infection was further confirmed by species-specific nested PCR by using a small subunit ribosomal RNA (ssrRNA) gene via a previously described two-step procedure28. Plasmodium-specific primers were first used, and the PCR product of the positive samples was used as a template in a multiplex PCR system through species-specific primers.
Detection of pfhrp2 gene
Microscopic and PCR positive P. falciparum samples but RDT negative were subjected to pfhrp2 PCR amplification. Specific primers Pfhrp2-F (5′ ATTCCGCATTTAATAATAACTTG TGTAGC 3′) and Pfhrp2-R (5′ ATGGCGTAGGCAATGTGTGG 3′) targets exon 2 of pfhrp2 were used as described previously12.
The PCR mixture had a total volume of 20 µl containing 4 µl of 5 × HOT FIREPol Blend Master Mix, 0.75 µM each primer and 2 µl DNA. The thermoprofile process consisted of an initial denaturation step at 95 °C for 4 min, followed by 35 cycles at 95 °C for 30 s, 58 °C for 45 s, 72 °C for 1 min and a final extension step at 72 °C for 5 min. Furthermore, the samples in which no pfhrp2 was amplified were subjected to P. falciparum circumsporozoite gene (pfcsp) PCR amplification, with the assumption that successful ssrRNA and csp gene amplification indicates reasonable quantity and quality of DNA that would allow pfhrp2 amplification. Thus, all the samples that were not amplified by pfhrp2 PCR may have pfhrp2 gene deletion.
Ethical statement
This study protocol was reviewed and approved by the Institute of Molecular Biology, University of Nyala and the committee of the Research directorate, Federal Ministry of Health (fmoh/nhrc/rd/rec). Prior to blood samples collection, written informed consent was obtained from all subjects and also from a parent or guardian for children less than 18 years of age. All methods were performed in accordance with the relevant guidelines and regulations.
Data availability
Any further requested information regarding the experimental and data analysis during the current study is available from the corresponding author on reasonable request.
References
Adusei, K. A. & Owusu-Ofori, A. Prevalence of Plasmodium parasitaemia in blood donors and a survey of the knowledge, attitude and practices of transfusion malaria among health workers in a hospital in Kumasi, Ghana. PLoS ONE 13, e0206303 (2018).
Landier, J. et al. The role of early detection and treatment in malaria elimination. Malar. J. 15, 363 (2016).
WHO. Guidelines for the Treatment of Malaria 12th edn. (World Health Organization, Geneva, 2012).
Boncy, P. J. et al. Malaria elimination in Haiti by the year 2020: an achievable goal?. Malar. J. 14, 237 (2015).
Bailey, J. W., Williams, J., Bain, B. J., Parker-Williams, J. & Chiodini, P. L. Haematology GHTFotBCfSi. Guideline: the laboratory diagnosis of malaria. Br. J. Haematol. 163, 573–580 (2013).
Viana, G. M. R. et al. Histidine-rich protein 2 (pfhrp2) and pfhrp3 gene deletions in Plasmodium falciparum isolates from select sites in Brazil and Bolivia. PLoS ONE 12, e0171150 (2017).
Kiemde, F. et al. Accuracy of a Plasmodium falciparum specific histidine-rich protein 2 rapid diagnostic test in the context of the presence of non-malaria fevers, prior anti-malarial use and seasonal malaria transmission. Malar. J. 16, 294 (2017).
Federal Ministry of Health, Sudan. Sudan Malaria Diagnosis and Treatment Protocol 2017 https://reliefweb.int/report/sudan/sudan-malaria-diagnosis-and-treatment-protocol-2017 (2017).
Rogier, E. et al. Conventional and high-sensitivity malaria rapid diagnostic test performance in 2 transmission settings: Haiti 2017. J. Infect. Dis. 221, 786–795 (2020).
Cheng, Q. et al. Plasmodium falciparum parasites lacking histidine-rich protein 2 and 3: a review and recommendations for accurate reporting. Malar. J. 13, 283 (2014).
Grignard, L., Nolder, D., Sepúlveda, N., Berhane, A., Mihreteab, S., Kaaya, R., et al. A Novel Multiplex qPCR Assay for Detection of Plasmodium falciparum with Histidine-rich Protein 2 and 3 (pfhrp2 and pfhrp3) Deletions in Polyclonal Infections. bioRxiv. (2020).
Koita, O. A. et al. False-negative rapid diagnostic tests for malaria and deletion of the histidine-rich repeat region of the hrp2 gene. Am. J. Trop. Med. Hyg. 86, 194–198 (2012).
Wurtz, N. et al. Pfhrp2 and pfhrp3 polymorphisms in Plasmodium falciparum isolates from Dakar, Senegal: impact on rapid malaria diagnostic tests. Malar. J. 12, 34 (2013).
Gamboa, D. et al. A large proportion of P. falciparum isolates in the Amazon region of Peru lack pfhrp2 and pfhrp3: implications for malaria rapid diagnostic tests. PLoS ONE 5, e8091 (2010).
Houzé, S., Hubert, V., Le Pessec, G., Le Bras, J. & Clain, J. Combined deletions of pfhrp2 and pfhrp3 genes result in Plasmodium falciparum malaria false-negative rapid diagnostic test. J. Clin. Microbiol. 49, 2694–2696 (2011).
Kojom, L. P. & Singh, V. Prevalence of Plasmodium falciparum field isolates with deletions in histidine-rich protein 2 and 3 genes in context with sub-Saharan Africa and India: a systematic review and meta-analysis. Malar. J. 19, 46 (2020).
Berhane, A. et al. Major threat to malaria control programs by Plasmodium falciparum lacking histidine-rich protein 2, Eritrea. Emerg. Infect. Dis. 24, 462–470 (2018).
Beshir, K. B. et al. Plasmodium falciparum parasites with histidine-rich protein 2 (pfhrp2) and pfhrp3 gene deletions in two endemic regions of Kenya. Sci. Rep. 7, 1–10 (2017).
Berhane, A. et al. Rapid diagnostic tests failing to detect Plasmodium falciparum infections in Eritrea: an investigation of reported false negative RDT results. Malar. J. 16, 105 (2017).
Abdallah, J. F. et al. Prevalence of pfhrp2 and pfhrp3 gene deletions in Puerto Lempira, Honduras. Malar. J. 14, 19 (2015).
Mussa, A., Talib, M., Mohamed, Z. & Hajissa, K. Genetic diversity of Plasmodium falciparum histidine-rich protein 2 (PfHRP2) and its effect on the performance of PfHRP2-based rapid diagnostic tests. BMC Res. Notes 12, 334 (2019).
Hamid, M. A., Awad-Elgeid, M. & Nasr, A. Gene variation and suspected Plasmodium falciparum histidine-rich protein 2 gene deletion and its impact on sensitivity of malaria rapid diagnostic tests in Sudan. BMJ Glob. Health 2, A21-A (2017).
Deme, A. B. et al. Analysis of pfhrp2 genetic diversity in Senegal and implications for use of rapid diagnostic tests. Malar. J. 13, 34 (2014).
Thomson, R. et al. pfhrp2 and pfhrp3 gene deletions that affect malaria rapid diagnostic tests for Plasmodium falciparum: analysis of archived blood samples from 3 African countries. J. Infect. Dis. 220, 1444–1452 (2019).
Kozycki, C. T. et al. False-negative malaria rapid diagnostic tests in Rwanda: impact of Plasmodium falciparum isolates lacking hrp2 and declining malaria transmission. Malar. J. 16, 123 (2017).
Berzosa, P. et al. First evidence of the deletion in the pfhrp2 and pfhrp3 genes in Plasmodium falciparum from equatorial Guinea. Malar. J. 19, 1–9 (2020).
Funwei, R. et al. Molecular surveillance of pfhrp2 and pfhrp3 genes deletion in Plasmodium falciparum isolates and the implications for rapid diagnostic tests in Nigeria. Acta Trop. 196, 121–125 (2019).
Eshag, H. A. et al. Molecular epidemiology of malaria parasite amongst patients in a displaced people’s camp in Sudan. Trop. Med. Health 48, 1–7 (2020).
Acknowledgements
This work was partially supported by the Grants of the Commission of Scientific Research and Innovation, Ministry of Higher Education and Scientific Research, Sudan, Grant No. SRIC/2017/RP761.
Author information
Authors and Affiliations
Contributions
K.H., K.B.B., Z.M. conceived and designed the study; M.A.B., M.A.D., A.M., A.B.M. and M.T. conducted field and laboratory work; and MA carried out statistical analysis. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Boush, M.A., Djibrine, M.A., Mussa, A. et al. Plasmodium falciparum isolate with histidine-rich protein 2 gene deletion from Nyala City, Western Sudan. Sci Rep 10, 12822 (2020). https://doi.org/10.1038/s41598-020-69756-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-69756-8
This article is cited by
-
Spatiotemporal mapping of malaria incidence in Sudan using routine surveillance data
Scientific Reports (2022)
-
Plasmodium falciparum is evolving to escape malaria rapid diagnostic tests in Ethiopia
Nature Microbiology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.