Abstract
Kawasaki disease (KD) is an acute febrile systemic vasculitis of unknown etiology that affects infants and young children. Considerable evidence supports the hypothesis that there is a genetic basis for KD susceptibility. Genome-wide association studies (GWAS) have identified several genetic variants associated with KD. This study aims to replicate three novel KD-associated single nucleotide polymorphisms (SNPs), identified by GWAS in Japanese, in a Taiwanese population. Associations between these SNPs and development of coronary artery lesions (CALs) were also investigated. The rs2254546 A/G, rs2857151 A/G, and rs4813003 C/T SNPs were genotyped in 681 children with KD and 563 ethnically-matched healthy controls using TaqMan Assay or DNA sequencing. We found rs2254546 and rs4813003 SNPs were significantly associated with KD (G allele, odds ratio [OR] = 1.54, P = 1.0 × 10–5; C allele, OR = 1.32, P = 8.1 × 10–4). However, no evidence for associations with CAL development was observed. Our study successfully validates associations of the rs2254546 and rs4813003 SNPs with KD in a Taiwanese population. Further functional studies of the SNPs are important in understanding the pathogenesis of KD.
Similar content being viewed by others
Introduction
Kawasaki disease (KD; OMIM 611775) is an acute febrile vasculitis syndrome that occurs predominantly in children under 5 years of age1. The major clinical symptoms of KD are fever lasting for longer than 5 days, bilateral non-purulent conjunctivitis, cervical lymphadenopathy, erythema of the palms and soles, diffuse mucosal inflammation, and polymorphous skin rashes2,3. If left untreated, around 15–25% children with KD develop coronary artery lesions (CALs), making this disease the most important cause of acquired heart disease in children of developed countries4. Although its etiology remains unclear, higher incidence rates in east Asian populations and higher risk in siblings imply that genetic factors play important roles in the development of KD5,6.
Genome-wide association study (GWAS) is a systematic approach to analyze the correlation between genetic markers and human diseases. This method allows one to find out disease-associated variants across the genome in large population groups. GWAS has been successfully undertaken for different kinds of complex genetic diseases and discovers a large number of associated loci. As of 2018, the GWAS catalog contains over 5,600 studies and 71,673 variant-trait associations from 3,567 publications7. KD, a complex disorder, has also been studied by GWAS to seek potentially associated variants. Until now, a total of 7 KD GWAS have been conducted in different races and some novel associated KD loci are identified8,9,10,11,12,13,14.
Because of difference in genetic architectures based on ethnicity, replications of GWAS findings in other racial groups are needed to verify the susceptible genes for KD. Therefore, we aim to determine whether GWAS-identified single nucleotide polymorphisms (SNPs) can be replicated in a Taiwanese population by using a case–control study design with 681 children with KD and 563 healthy controls. In addition, we examined associations between the GWAS-significant SNPs and CAL development in children with KD.
Results
Association between SNPs and KD susceptibility
The genotyping of SNPs rs2254546, rs2857151, and rs4813003 was successful in 563 controls and 681 children with KD (Tables 1, 2, 3). The observed genotype frequencies for these polymorphisms were in agreement with Hardy–Weinberg equilibrium in the controls. Association tests revealed significant differences in the distribution of genotypes and alleles of SNPs rs2254546 A/G (P = 3.5 × 10–5 and 1.0 × 10–5) and rs4813003 C/T (P = 2.0 × 10–3 and 8.1 × 10–4) between controls and KD children, which remained significant after Bonferroni correction (Pc < 0.05) (Tables 1, 3). The frequencies of rs2254546 G/G genotype and G allele (OR 1.69, 95% CI 1.34–2.13; OR 1.54, 95% CI 1.27–1.86) and rs4813003 C/C genotype and C allele (OR 1.50, 95% CI 1.18–1.90; OR 1.32, 95% CI 1.12–1.54) significantly increased in children with KD than in the controls. The genotype and allele frequencies for SNP rs2857151 A/G did not differ significantly (P = 0.81 and 0.69) (Table 2).
Association between SNPs and CAL development
Based on the CAL data of children with KD, we further examined the association between investigated polymorphisms and CAL formation. No statistically significant differences were observed in genotype and allele frequencies of any of the 3 SNPs between children with CAL and those without CAL (P = 0.87 and 0.63 for rs2254546, P = 0.34 and 0.25 for rs2857151, P = 0.67 and 0.46 for rs4813003) (Tables 1, 2, 3).
Discussion
Replication of positive findings from genetic association studies in independent populations has become the gold standard to validate susceptibility genes. In this study, we attempted to replicate 3 GWAS-identified SNPs for Japanese children with KD in a Taiwanese population. We found that rs2254546 G/G genotype and G allele and rs4813003 C/C genotype and C allele were associated with increased risk of KD. These findings validated rs2254546 G allele and rs4813003 C allele confer risk to KD in our study population, which was consistent with the Japanese GWAS12. However, the association between rs2857151 G allele and KD cannot be replicated in our study. When the analysis was restricted to the CAL outcome, we found that none of these replicated SNPs was risk factor for CAL development in Taiwanese children with KD.
The advent of GWAS has made it a reality to study the genetic mechanisms underlie KD pathogenesis in an unbiased and efficient way. The first GWAS of KD performed in a Caucasian population found that rs17531088 (EXOC1) and rs7199343 (ZFHX3) were the most significantly associated SNPs8. A Korean GWAS reported by Kim et al. revealed that KD was associated with rs527409 (1p31)9. A GWAS conducted in Taiwanese population showed that 10 SNPs located in 3 novel loci (COPB2, ERAP1, and IGHV) were associated with KD10. Khor et al. demonstrated that significant associations with KD at the GWAS level were observed in rs1801274 (FCGR2A), rs2233152 (near MIA and RAB4B), and rs28493229 (ITPKC) in a combined European and Asian population11. Interestingly, a Japanese GWAS and a Taiwanese GWAS published concurrently in the same journal reported BLK, CD40, and HLA as susceptibility loci for KD12,13. The most recent GWAS in Korean population discovered NMNAT2, BLK, and HCP5 loci contributed to KD risk14.
SNP rs2254546 is located between FAM167A and BLK gene loci, a region known to have genetic associations with autoimmune diseases like systemic lupus erythematosus15,16,17, rheumatoid arthritis18,19, and systemic sclerosis20,21. BLK encodes B lymphoid kinase (Blk), a member of the Src family of kinases, which mediates downstream signaling of B cell receptors and affects the proliferation, differentiation and tolerance of B cells22. It has also been found that Blk is required to regulate T-cell mediated proinflammatory cytokine production23. FAM167A gene was recently reported to contribute to immune function including antibody isotype determination and immunoglobulin production24. In addition to our current study, genetic associations of rs2254546 with KD have been validated in Chinese population25,26. Together, these findings support both humoral and cellular immunity may be involved in KD pathogenesis.
rs4813003, located 4.9 kb downstream of CD40 gene, was also replicated successfully in our study population. CD40 is a cell surface receptor that belongs to the tumor necrosis factor receptor superfamily and has been found expressed on antigen-presenting cells such as B cells, macrophages, and dendritic cells27. The dyadic interaction of CD40 and CD40 ligand plays an essential role for proliferation, differentiation, and activation of B and T cells28,29. Besides, CD40 has been proposed as a contributing factor for immune-mediated inflammatory diseases such as systemic lupus erythematosus30, rheumatoid arthritis31, Crohn’s disease32, Graves’ disease33, and psoriasis34. Replication of the rs4813003 SNP in Chinese population also revealed it was nominally associated with KD susceptibility35. It is therefore conceivable to infer that CD40 is prominent in determining the risk for KD.
In summary, our study confirmed the associations of rs2254546 and rs4813003 polymorphisms with KD susceptibility in a Taiwanese population but these SNPs did not contribute to the development of clinically evident CAL. It will be necessary to validate or replicate these associations in other independent large-scale cohorts of different ethnicities.
Methods
Study population
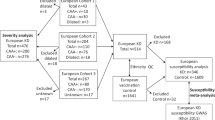
The case group contained 681 unrelated Taiwanese children with KD (423 boys, 258 girls; mean age 1.8 years, range 0.1–8.3 years), which were consecutively recruited from MacKay Memorial Hospital in Taiwan between 2005 and 2019. A portion of the KD cohort has ever been included in our previous studies36,37,38,39. Diagnosis of KD was made according to the diagnostic criteria of the American Heart Association2. Oral aspirin and intravenous gamma-globulin therapy were prescribed as soon as the diagnosis was made. All children were examined by 2-dimensional echocardiography during the febrile stage and after hospital discharge. CALs were defined by an internal arterial diameter was ≥ 3 mm in children < 5 years old, ≥ 4 mm in children ≥ 5 years old, or > 1.5 times that of an adjacent artery40. Of all patients studied, 157 developed CAL while the remaining revealed no evidence of CAL.
The control group consisted of 563 unrelated ethnic-matched healthy people (223 males, 340 females; mean age 35.8 years, range 10.1–66.4 years) without a history of KD, autoimmune, or allergic disease. The Institutional Review Board of MacKay Memorial Hospital approved this study and written informed consent was obtained from either the participants or their parents/guardians. All procedures of this study conformed to the principles of the Declaration of Helsinki. All methods were conducted in accordance with the relevant guidelines and regulations.
SNP selection and analysis
Inclusion criteria of the candidate SNPs was set at genome-wide significance of combined P value less than 5.0 × 10–8 and was not previously validated in our KD patients. Based on these conditions, a total of three polymorphisms identified in the Japanese GWAS12 were chosen for replication: rs2254546 at 8p22-23 (FAM167A-BLK), rs2857151 at 6p21.3 (HLA-DQB2–DOB), and rs4813003 at 20q13 (CD40). Genomic DNA was extracted from peripheral blood sample of each participant using the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany). The rs2254546 and rs4813003 SNPs were genotyped by the TaqMan Allelic Discrimination Assay (Applied Biosystems, Foster City, CA) as previously described41.
Genotyping for the rs2857151 polymorphism was analyzed by PCR amplification followed by DNA sequencing. The primer pairs used were 5′-CCT ATC ATT TTG TGG GAG GTA GT-3′ (forward) and 5′-CCC AGC TGT ACC TGC CTT AG-3′ (reverse). PCR was performed at a 50 μL reaction volume containing 100 ng of genomic DNA, 0.2 μM of each primer, and 25 μL of 2 × EmeraldAmp MAX PCR Master Mix (Takara Biotechnology, Dalian City, Liaoning, China). PCR cycling parameters were: one cycle of 98 °C for 1 min, 25 cycles of 98 °C for 10 s, 49 °C for 10 s and 72 °C for 30 s, followed by one cycle of 72 °C for 5 min. The PCR fragments were sequenced on an ABI PRISM 3700 Automated Sequencer (Applied Biosystems) using the PCR primers.
Statistical analysis
The Hardy–Weinberg equilibrium for genotypes in controls and genotype and allele frequencies associated with the KD susceptibility and CAL formation were assessed by the χ2 test. Odds ratios and 95% confidence intervals were also calculated. The Bonferroni correction was used to calculate corrected P (Pc) values. Two-tailed Pc values of less than 0.05 were considered to be statistically significant. Prior to the study, statistical power to detect effects of these SNPs on KD susceptibility was calculated using the Quanto Ver. 1.1 software (Department of Preventive Medicine, University of Southern California, CA, USA). The study was designed to have a power of over 98% at a 5% significance level to determine a relative risk of 1.5 conferred by the risk genotype of each SNP with the KD prevalence of 65.3 per 10,000 children42.
References
Kawasaki, T., Kosaki, F., Okawa, S., Shigematsu, I. & Yanagawa, H. A new infantile acute febrile mucocutaneous lymph node syndrome (MLNS) prevailing in Japan. Pediatrics 54, 271–276 (1974).
Newburger, J. W. et al. Diagnosis, treatment, and long-term management of Kawasaki disease: A statement for health professionals from the Committee on Rheumatic Fever, Endocarditis and Kawasaki Disease, Council on Cardiovascular Disease in the Young, American Heart Association. Circulation 110, 2747–2771 (2004).
Burns, J. C. & Glode, M. P. Kawasaki syndrome. Lancet 364, 533–544 (2004).
Kato, H. et al. Long-term consequences of Kawasaki disease. A 10- to 21-year follow-up study of 594 patients. Circulation 94, 1379–1385 (1996).
Uehara, R. & Belay, E. D. Epidemiology of Kawasaki disease in Asia, Europe, and the United States. J. Epidemiol. 22, 79–85 (2012).
Fujita, Y. et al. Kawasaki disease in families. Pediatrics 84, 666–669 (1989).
Buniello, A. et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 47, D1005–D1012 (2019).
Burgner, D. et al. A genome-wide association study identifies novel and functionally related susceptibility Loci for Kawasaki disease. PLoS Genet. 5, e1000319 (2009).
Kim, J. J. et al. A genome-wide association analysis reveals 1p31 and 2p13.3 as susceptibility loci for Kawasaki disease. Hum. Genet. 129, 487–495 (2011).
Tsai, F. J. et al. Identification of novel susceptibility Loci for kawasaki disease in a Han chinese population by a genome-wide association study. PLoS ONE 6, e16853 (2011).
Khor, C. C. et al. Genome-wide association study identifies FCGR2A as a susceptibility locus for Kawasaki disease. Nat. Genet. 43, 1241–1246 (2011).
Onouchi, Y. et al. A genome-wide association study identifies three new risk loci for Kawasaki disease. Nat. Genet. 44, 517–521 (2012).
Lee, Y. C. et al. Two new susceptibility loci for Kawasaki disease identified through genome-wide association analysis. Nat. Genet. 44, 522–525 (2012).
Kim, J. J. et al. A genome-wide association analysis identifies NMNAT2 and HCP5 as susceptibility loci for Kawasaki disease. J. Hum. Genet. 62, 1023–1029 (2017).
Hom, G. et al. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N. Engl. J. Med. 358, 900–909 (2008).
Ito, I. et al. Replication of the association between the C8orf13-BLK region and systemic lupus erythematosus in a Japanese population. Arthritis Rheum. 60, 553–558 (2009).
Yang, W. et al. Population differences in SLE susceptibility genes: STAT4 and BLK, but not PXK, are associated with systemic lupus erythematosus in Hong Kong Chinese. Genes Immun. 10, 219–226 (2009).
Gregersen, P. K. et al. REL, encoding a member of the NF-kappaB family of transcription factors, is a newly defined risk locus for rheumatoid arthritis. Nat. Genet. 41, 820–823 (2009).
Ito, I. et al. Replication of association between FAM167A(C8orf13)-BLK region and rheumatoid arthritis in a Japanese population. Ann. Rheum. Dis. 69, 936–937 (2010).
Gourh, P. et al. Association of the C8orf13-BLK region with systemic sclerosis in North-American and European populations. J. Autoimmun. 34, 155–162 (2010).
Ito, I. et al. Association of the FAM167A-BLK region with systemic sclerosis. Arthritis Rheum. 62, 890–895 (2010).
Reth, M. & Wienands, J. Initiation and processing of signals from the B cell antigen receptor. Annu. Rev. Immunol. 15, 453–479 (1997).
Samuelson, E. M. et al. Reduced B lymphoid kinase (Blk) expression enhances proinflammatory cytokine production and induces nephrosis in C57BL/6-lpr/lpr mice. PLoS ONE 9, e92054 (2014).
Mentlein, L. FAM167A-BLK is a Susceptibility Locus in Autoimmune Diseases: Characterization of the FAM167 Gene Family (Doctoral Dissertation) (Karolinska Institutet, Stockholm, 2018).
Yan, Y. et al. Combined analysis of genome-wide-linked susceptibility loci to Kawasaki disease in Han Chinese. Hum. Genet. 132, 669–680 (2013).
Wang, W. et al. 8p22-23-rs2254546 as a susceptibility locus for Kawasaki disease: A case-control study and a meta-analysis. Sci. Rep. 4, 4247 (2014).
van Kooten, C. & Banchereau, J. Functional role of CD40 and its ligand. Int. Arch. Allergy Immunol. 113, 393–399 (1997).
van Kooten, C. & Banchereau, J. Immune regulation by CD40–CD40-L interactions. Front. Biosci. 2, d1-11 (1997).
van Kooten, C. & Banchereau, J. CD40–CD40 ligand. J. Leukoc. Biol. 67, 2–17 (2000).
Pyrovolaki, K. et al. Increased expression of CD40 on bone marrow CD34+ hematopoietic progenitor cells in patients with systemic lupus erythematosus: Contribution to Fas-mediated apoptosis. Arthritis Rheum. 60, 543–552 (2009).
Gotoh, H. et al. Increased CD40 expression on articular chondrocytes from patients with rheumatoid arthritis: Contribution to production of cytokines and matrix metalloproteinases. J. Rheumatol. 31, 1506–1512 (2004).
Battaglia, E. et al. Expression of CD40 and its ligand, CD40L, in intestinal lesions of Crohn’s disease. Am. J. Gastroenterol. 94, 3279–3284 (1999).
Faure, G. C., Bensoussan-Lejzerowicz, D., Bene, M. C., Aubert, V. & Leclere, J. Coexpression of CD40 and class II antigen HLA-DR in Graves’ disease thyroid epithelial cells. Clin. Immunol. Immunopathol. 84, 212–215 (1997).
Ohta, Y. & Hamada, Y. In situ expression of CD40 and CD40 ligand in psoriasis. Dermatology 209, 21–28 (2004).
Lou, J. et al. Systematic confirmation study of GWAS-identified genetic variants for Kawasaki disease in a Chinese population. Sci. Rep. 5, 8194 (2015).
Huang, F. Y. et al. Genetic variations of HLA-DRB1 and susceptibility to Kawasaki disease in Taiwanese children. Hum. Immunol. 68, 69–74 (2007).
Huang, F. Y. et al. The -590 C/T and 8375 A/G interleukin-4 polymorphisms are not associated with Kawasaki disease in Taiwanese children. Hum. Immunol. 69, 52–57 (2008).
Huang, F. Y. et al. Lack of association of the vascular endothelial growth factor gene polymorphisms with Kawasaki disease in Taiwanese children. J. Clin. Immunol. 28, 322–328 (2008).
Huang, F. Y. et al. Genetic polymorphisms in the CD40 ligand gene and Kawasaki disease. J. Clin. Immunol. 28, 405–410 (2008).
Arjunan, K. et al. Coronary artery caliber in normal children and patients with Kawasaki disease but without aneurysms: An echocardiographic and angiographic study. J. Am. Coll. Cardiol. 8, 1119–1124 (1986).
Yang, Y. C. et al. Genetic polymorphisms in the ITPKC gene and cervical squamous cell carcinoma risk. Cancer Immunol. Immunother. 61, 2153–2159 (2012).
Chang, C. L., Wong, C. S., Yang, Y. C. & Chiu, N. C. Influence of latitude on the prevalence of Kawasaki disease: A retrospective cohort study from the Taiwan National Health Insurance database and review of the literature. Int. J. Environ. Res. Public Health 15, 845 (2018).
Acknowledgements
This research was funded by Ministry of Science and Technology (MOST 104-2314-B-195-020) and the MacKay Memorial Hospital of Taiwan (MMH E-107-07 and MMH E-108-07).
Author information
Authors and Affiliations
Contributions
M.R.C., T.Y.C., L.Y.C. and Y.J.L. conceived and designed the studies. M.R.C., N.C.C., H.C., K.D.Y., L.C., D.T.N.H., F,Y.H., Y.P.L., W.S.L., and C.L.L. collected samples and performed laboratory experiments. M.R.C. and T.Y.C. and Y.J.L. analyzed and interpreted the data. M.R.C. and T.Y.C. wrote the manuscript. L.Y.C.. and Y.J.L. edited the manuscript. L.Y.C. and Y.J.L. are guarantors for the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chen, MR., Chang, TY., Chiu, NC. et al. Validation of genome-wide associated variants for Kawasaki disease in a Taiwanese case–control sample. Sci Rep 10, 11756 (2020). https://doi.org/10.1038/s41598-020-68673-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-68673-0
This article is cited by
-
Whole-exome sequencing analysis identifies novel variants associated with Kawasaki disease susceptibility
Pediatric Rheumatology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.