Abstract
Background
Single-nucleotide polymorphisms (SNPs) of several genes are linked to the etiopathogenesis of Kawasaki disease (KD). Association of SNPs of inositol 1,4,5-triphosphate-3-kinase C (ITPKC) gene with susceptibility to KD and coronary artery lesions (CALs) has been observed in children of certain ethnicities, but not from others. The present study was planned to explore this genetic association in the North Indian cohort.
Methods
Fifty children with KD and 50 age- and sex-matched controls were studied for two SNPs (rs28493229 and rs2290692) of the ITPKC gene using polymerase chain reaction and restriction fragment length polymorphism. Findings were confirmed by Sanger sequencing. A meta-analysis was also carried out for GG and CC genotypes of the SNPs.
Results
There was significant association between KD susceptibility and CG + GG genotype of rs2290692 (p = 0.015, odds ratio = 4.1, 95% confidence interval = 1.38–13.83). None of the single alleles or genotypes of the SNPs of ITPKC were, however, significantly associated with KD susceptibility. A meta-analysis also did not show any significant association of these SNPs to KD susceptibility.
Conclusions
Our findings suggest that ITPKC gene SNPs (rs28493229 and rs2290692) did not have a significant association with susceptibility to KD in children from North India. Larger multicentric studies incorporating different ethnicities are required to understand the genetic basis of KD.
Impact
-
While SNP rs28493229 of the ITPKC gene is not found to be associated with susceptibility to KD, the combined genotype of SNP rs2290692 is shown to be associated.
-
Impact of ITPKC gene SNP on KD is different across different races and ethnicities. We could find an association of the combined genotype of rs2290692 with it in the Indian population.
-
This study highlights that phenotype and genotypic association of KD varies with ethnicities. Larger multicentric studies are required to reach a conclusion regarding the genetic association of KD.
Similar content being viewed by others
Introduction
Kawasaki disease (KD) is one of the common childhood vasculitides and is being increasingly recognized as an important cause of acquired heart disease in young children.1 The highest incidence of KD has been reported from Japan, Korea, and Taiwan.2,3,4 Reports from several centers in India over the past two decades suggest that the incidence of KD may be increasing.2,5,6 Two hospital-based studies from Chandigarh, India showed that KD is the most common cause of vasculitis in young children.7,8,9 One of the main concerns in KD is its propensity to affect the heart and cardiovascular system and this can result in the development of coronary artery lesions (CALs).10,11,12 Treatment with IVIG not only reduces the occurrence of CAL but also decreases myocardial inflammation.13,14
Although therapy of KD has evolved over the past three decades,1,15,16 the etiology of this disease remains a mystery. Many etiological associations have been proposed for KD, but the cause and effect relationship has not yet been established.17 Genetic association of KD has been suspected because of the high incidence of the disease in children in families of Japanese ancestry and siblings or offsprings of patients with KD.18,19 Inositol 1,4,5 triphosphate 3-kinase C (ITPKC) is a negative regulator of Ca2+/nuclear factor of activated T cell (NFAT) pathway in T cells.20 Onouchi et al. reported the association of polymorphism of rs28493229 of ITPKC with susceptibility to KD and CAL in Japanese and American KD cohorts.21 This finding was, however, not replicated in studies from other countries like China and Taiwan.22,23 Similarly, while Peng et al. reported an association of polymorphism of rs2290692 of ITPKC with susceptibility to KD in China,22 the finding was not replicated in studies from Taiwan and Korea.24,25 There is no study on SNPs of ITPKC in association with KD from India to date. As the phenotype of KD in India is different, it is important to study the genotype of KD in Indian children.26,27,28 In this study, we have studied the SNPs rs28493229 at intron 1 and rs2290692 at 3′-UTR (untranslated region) of the ITPKC gene in North Indian KD patients (with or without CAL) and compared the results with healthy controls.
Methods
Study population
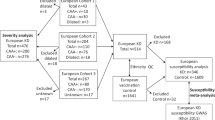
This study was carried out in Pediatric Allergy Immunology Unit, Advanced Pediatrics Centre, Postgraduate Institute of Medical Education and Research, Chandigarh, India. Our institute serves as a not-for-profit tertiary care referral center for North India. Patients and controls were of North Indian ethnicity. Fifty cases (25 KD patients without CAL and 25 KD patients with CALs) and 50 age- and sex-matched controls were selected for sampling over a period of 14 months (January 2018–April 2019). The mean (SD) age of children with KD was 3.9 (2.53) years, whereas the mean (SD) age of controls was 3.92 (2.46) years. There were 30 (60%) boys and 20 (40%) girls (M:F = 1.5:1) in both the KD group and control group.
Diagnosis of KD was based on American Heart Association (AHA) guidelines.1,29 A CAL was defined as any coronary artery abnormality and included ectasia, dilatation (with the coronary z-score value of ≥2 but <2.5), and aneurysms (with the coronary z-score value of ≥2.5).1 Written informed consent was taken from all the subjects for the purposes of this study. The study protocol was approved by Institute Ethics Committee and Institute Thesis Committee. The manuscript has been approved by Departmental Review Board. Two-dimensional echocardiographic coronary assessment of patients was carried out in the febrile phase and ~4–6 weeks later on follow-up.
Genetic study
Peripheral venous blood sample (2–3 ml) was drawn from cases and control subjects in ethylenediaminetetraacetic acid (EDTA) vacutainer.
Genomic DNA was extracted from peripheral blood using QIAmp DNA Blood Mini Kit (Qiagen, Hilden, Germany). DNA quality was determined using 1% agarose gel electrophoresis, followed by staining with ethidium bromide. The purity of DNA was determined by taking optical density (OD) of samples at 260 and 280 nm using TECAN infinite M200 Pro with Nanoquant plate (Tecan group (Life-sciences and Diagnostics) AG, Switzerland) and was stored at −80 °C till further analysis.
Genotyping for SNPs rs28493229 and rs2290692 was carried out using PCR and restriction fragment length polymorphism (RFLP). 3′-UTR and intron 1 of ITPKC gene were amplified using PCR at controlled conditions using specific oligonucleotide primers as previously used by Peng et al.22 PCR primers (Integrated DNA Technologies, Lowa), restriction enzymes (BanI and AvaI) (New England Biolabs Inc., Massachusetts), length of the PCR products, and the digested fragments are shown in Table 1A. Genotyping was performed by the PCR-RFLP method. Primer sequence, restriction enzymes, and band size details have been mentioned in Table 1A. PCR cycle conditions for amplification are available on request.
Representative gel pictures showing the genotypes of both the SNPs have been given in Fig. 1a, b. PCR products of samples with the presence of C allele and G allele were confirmed with bidirectional Sanger sequencing (Applied Biosystems, Foster, CA) (Fig. 1c, d). Results were reproducible with no discrepancy in genotyping.
a Gel electrophoresis for confirmation of CG genotypes with restriction fragments highlighted as bands of 36, 60, 141 and 177 bp in digested column and band of 237 bp in an undigested column of rs28493229 in intron 1 of ITPKC. b Gel electrophoresis picture showing bands of restriction fragments (digested and undigested columns) of rs2290692 in 3′-UTR of ITPKC. c Sanger sequencing of control and representative cases of different genotypes of rs28493229 at intron 1 of ITPKC. d Sanger sequencing results of control and representative cases of rs2290692 at 3′-UTR of ITPKC.
Meta-analysis
In addition, all previous studies on SNPs rs28493229 and rs2290692 of the ITPKC gene in association with susceptibility and coronary complication of KD were gathered. A meta-analysis was performed for genotypes of both SNPs and their association with susceptibility to KD.
Statistical analysis
Allele and genotype frequencies of SNPs were compared between patients with KD and control subjects. Similar comparisons were also carried out between patients of KD with and without CALs. The association of categorical variables with KD patients was analyzed using χ2 test/Fisher’s exact tests, whereas comparisons of quantitative variables between two study groups were carried out using an independent-sample t test (parametric) or Mann–Whitney U test (nonparametric). A p value of ≤0.05 was taken as significant. Data analysis was done using R software version 5.3.0 with RStudio IDE (The R Foundation) in Windows 10 platform. Deviation of SNPs from Hardy–Weinberg law was evaluated with Stata IC version 14 (StataCorp LLC, TX) using “genass” package. Meta-analyses were performed with R and Stata software.
Results
Allele and genotype frequencies
Genotype distribution of SNPs rs28493229 (Pearson’s χ2 0.378, p = 0.54) and rs2290692 (χ2 = 0.263, p = 0.76) were in Hardy–Weinberg equilibrium. We analyzed the association of these two SNPs with KD and control groups. G and C allele frequency of rs28493229 observed among cases was 89% and 11%, respectively. Among controls, 92% G alleles and 8% C alleles were found (Table 2A). GG and CG genotype of this SNP was found to be 39 (78%) and 11 (22%) among cases. We could not get any case or control with the CC genotype of rs28493229 in our study. Similarly, the allele frequency of rs2290692 at 3′-UTR among cases was 63% and 37% for the G and C allele, respectively. Nineteen GG (38%), 25 CG (50%), and 6 CC (12%) genotypes of SNP rs2290692 were found in cases, whereas among controls, the frequencies were 20 (40%), 16 (32%), and 14 (28%), respectively. No statistical differences were found in allele, carrier, and genotype frequencies of both SNPs between cases and controls (Table 2A).
Association between ITPKC polymorphism and occurrence of KD
On multivariate logistic regression, adjusted odds of KD occurrence were increased by nearly one and half times (odds ratio (OR) 1.53; 95% confidence interval (CI): 0.54–4.65; p = 0.43) in subjects with CG genotype on rs28493229. Similarly, adjusted odds of KD occurrence were increased by 1.8 times (OR 1.8; 95% CI: 0.72–4.56; p = 0.21) in subjects with CG genotype on rs2290692. However, these associations failed to reach statistical significance (Table 2A). We observed an over-representation of CG genotype in rs28493229 as well as rs2290692 at 3′-UTR in patients with KD. Adjusted odds of occurrence of KD using SNP predictors are also demonstrated in a forest plot (Supplemental Fig. 1A).
Effects of combined genotypes for the risk of KD were also analyzed. On multivariate logistic regression, adjusted odds of occurrence of KD were increased by more than 4 times in subjects with any genotype with G allele (i.e., CG + GG) of rs2290692 (OR 4.14; 95% CI: 1.38–13.83; p = 0.015) after adjusting the confounding effects of sex and another SNP (Table 2C).
Association between ITPKC polymorphism and occurrence of coronary abnormality in KD
Allele, genotype, and carrier frequencies were analyzed between the KD patients with and without CALs. Effect size in genetic characteristics of both SNPs failed to reach statistical significance (Table 2B). Adjusted odds of occurrence of coronary abnormality in KD were lowered by 50% in subjects with CC genotype on rs2290692 at 3′-UTR (OR 0.50; 95% CI: 0.06–3.35; p = 0.49) after adjusting the confounding effects of sex and other SNPs (Supplemental Fig. 1B).
Association of ITPKC polymorphism and size of aneurysms
The z-scores for the size of aneurysms were divided into tertiles. The highest tertile scores represented higher z-scores. No significant association of SNPs was observed with the size of aneurysms. These effect sizes were adjusted for the age at which the illness occurred. Predictive margins of two SNPs with a probability of largest aneurysms (z-score) are illustrated in Supplemental Fig. 1C, D.
Meta-analyses
We further performed a meta-analysis for a possible association of both SNPs and susceptibility and complications of KD by combining the results of our study with all previous studies on rs28493229 and rs2290692 of the ITPKC gene. In the meta-analysis for the association of CC genotype of rs28493229, combined data confers a 1.5 times higher risk for this genotype to develop KD (OR = 1.46, 95% CI: 0.96–2.23) (Table 3A). Latter just crossed the line of null hypothesis showing only the trend for significance (Fig. 2a).
a Forest plot for the association between CC genotype of rs28493229 and risk of having KD (estimates of odds ratio and its 95% CI are plotted with a box and horizontal line). b Forest plot for the association between GG genotype of rs2290692 and risk of having KD (estimates of odds AQ8ratio and its 95% CI are plotted with a box and horizontal line). c Forest plot for the association between CC genotype of rs2290692 and risk of having KD (estimates of odds ratio and its 95% CI are plotted with a box and horizontal line).
Similarly, a meta-analysis was performed individually for the association of CC and GG genotypes of rs2290692 with the risk of having KD. In the meta-analysis for the association of GG genotype of rs2290692, combined effect showed that odds of occurrence of GG in KD was 21% lower than in controls in this meta-analysis with an OR of 0.7881 and its 95% CI of 0.6085–1.0205 (Table 3B). The combined average only showed a trend for significance (Fig. 2b).
In the meta-analysis of CC genotype of our study and all previous studies of SNP rs2290692, combined OR did not confer any significant association with the susceptibility of KD (OR = 1.07, 95% CI: 0.663–1.731) (Table 3C and Fig. 2c).
Discussion
KD is a common medium vessel vasculitis of young children. It may lead to complications like myocarditis, KD shock syndrome, and the development of CALs.10,14 Etiology of KD still remains an enigma and its pathogenesis is poorly understood.3,17,30 A high incidence of KD in some Asian populations, particularly Japanese and in native Japanese population residing in Hawaii, points towards some genetic link of KD.3,19 A study by Onouchi et al. in 2008 revealed a significant association of polymorphisms of rs28493229 of ITPKC gene with susceptibility and severity of KD in Japanese and American patients.21 Functional SNPs of ITPKC have been shown to result in altered messenger RNA (mRNA) splicing and gene transcription in patients with KD. Polymorphisms of ITPKC are reported as an important predisposing factor for KD.21,31 We studied the association of two SNPs of the ITPKC gene in patients with KD and healthy controls of North Indian ethnicity.
Although certain polymorphisms of the ITPKC gene are shown to be associated with susceptibility to KD from some ethnicities, the results have not been uniformly replicated across ethnicities (Table 1B, C).22,23,24,25,26,27,28,29,30,31,32 Such associations between SNP and KD need to be studied extensively in different ethnicities before coming to a conclusion. Moreover, the phenotype of KD in India is different.26,27,28 Contrary to the pattern of KD in Eastern Asian and North American children, proportionately greater number of male patients, higher incidence of older children (almost half of them of age around 5 or more than that), early appearance of periungual peeling (prior to day 10 of fever), and early appearance of thrombocytosis were observed in Indian cohorts of KD. This suggests the possibility of different genotypic associations of these SNPs in patients with KD from India.
In the present study, we performed PCR-RFLP and representative bidirectional Sanger sequencing for two SNPs (rs28493229 at intron 1 and rs2290692 at 3′-UTR) of the ITPKC gene in North Indian children with KD. Diagnosis of KD was made on the basis of AHA guidelines.1,29 According to the 1000 Genome Project, the prevalence of C and G allele of rs28493229 in the South Asian population is 13% and 87%, respectively.33 In our study, C and G Allele frequency of rs28493229 in the KD group was 11% and 89%, respectively. Ancestral GG genotype of rs28493229 was found in 78% of patients with KD (Table 1B).
We failed to show an association of any allele or genotype of rs28493229 with susceptibility to disease. This result is in accordance with the findings of studies on Chinese, Taiwanese, and Filipino patients (Table 1B).22,23,32,34,35 Although two previous meta-analyses had shown significant association of this SNP with KD,36,37 our meta-analysis combining our study with all previous studies on rs2849229 did not show any association with KD (Table 3A and Fig. 2a). This suggests that lowering of splicing efficiency of mRNA level due to the presence of the C allele of rs2849229 may not be a universal phenomenon.21 This may also highlight the remarkable heterogeneity of functional SNP on KD across ethnicities.
Although the association of the C allele of rs2290692 of 3′-UTR and KD was reported from China,22 our study did not show any association of any single allele or genotype with KD. Similar findings were reported in children with KD from Taiwanese and Korean ethnicities (Table 1C).24,25 Meta-analyses combining our study with previous studies on rs2290692 also did not show any significant association with susceptibility to KD (Table 3B, C and Fig. 2b, c). This may simply reflect ethnic variation or the limited role of a single allele or genotype of the SNP on the pathogenesis of KD.
However, analyzing any genotype containing G allele (i.e., CG + GG) in rs2290692, we observed a statistically significant association of genotypes with G allele with KD occurrence in both univariate and multivariate analysis (p = 0.015; OR = 4.16, 95% CI: 1.38–13.83) (Table 2C). Such an association of rs2290692 is not described in previous studies. It is postulated that polymorphism in any allele may result in altered regulation efficiency of miRNAs. These miRNAs are involved in the regulation of gene expression. Our result showing an overrepresentation of the G allele in the KD group suggests that associated miRNAs may bind to the G allele altering the gene expression.38 In addition, there may be evolutionary and epigenetic effects on the genetic structure of individuals in different ethnicities.
It appears that no specific SNP of the ITPKC gene may have a uniform association with susceptibility to KD and CAL formation across ethnicities. KD is undoubtedly a multifactorial disease and these SNPs may only confer some predisposition to develop KD.39 Pathogenesis of KD may reflect the effect of several genes involved in calcium-NFAT signaling, which ultimately activates gene transcription for T cell activation and cytokine secretion.35 Larger multicentric studies incorporating different ethnicities are required to understand the genetic basis of KD.
The relevance, strength, and limitations of the study
This is the first study of ITPKC SNP association with KD and CAL from India. Results from RFLP were confirmed by Sanger sequencing. Age and sex matching eliminate possible differences due to exposure prevalence and the effect of variable exposure in individuals. More importantly, the likelihood of false negatives due to the population stratification effect is also nullified by matching age and sex. However, this study has some limitations. This was a single-center hospital-based study and the numbers are understandably small. We were unable to conduct the functional level of studied gene polymorphism. Further, the effect of other genes (e.g., Ca2+/NFAT or tumor growth factor-β pathway) in the studied cohort also cannot be excluded.
Conclusions
In conclusion, this is the first study in the Indian population that has explored the association of KD with two SNPs (rs28493229 and rs2290692) of the ITPKC gene. The frequency of the C allele of rs28493229 of ITPKC was found to be 11%, which is lesser than that in the Japanese population (22%), but comparable with KD cohorts from Taiwan and China. We documented a higher frequency of G allele (63%) of rs2290692 in patients with KD in our study. This study, and meta-analysis, did not provide any evidence to support the association of rs28493229 with KD susceptibility or CAL in children with KD of North Indian ethnicity. Combined genotypes containing the G allele of rs2290692 (CG + GG) were found to be significantly associated with the risk of KD but not with CAL. Further studies with a larger sample size are required to confirm this finding. In addition, studies on multiple SNPs of the ITPKC gene together with haplotype analysis are needed to explore the relevance of ITPKC gene polymorphisms in KD in the Indian population.
Data availability
All data generated or analysed during this study are included in this published article (and its supplementary information file).
References
McCrindle, B. W. et al. Diagnosis, treatment, and long-term management of Kawasaki disease: a scientific statement for health professionals from the American Heart Association. Circulation 135, e927–e999 (2017).
Singh, S., Vignesh, P. & Burgner, D. The epidemiology of Kawasaki disease: a global update. Arch. Dis. Child. 100, 1084–1088 (2015).
Rowley, A. H. & Shulman, S. T. The epidemiology and pathogenesis of Kawasaki disease. Front. Pediatr. 6, 374 (2018).
Kim, G. B. Reality of Kawasaki disease epidemiology. Korean J. Pediatr. 62, 292–296 (2019).
Jiao, F. et al. The emergence of Kawasaki disease in India and China. Glob. Cardiol. Sci. Pract. 2017, e201721 (2017).
Suresh, N., Varadarajan, V. V. & Ranjith, M. S. Kawasaki disease in south India: a prospective, case-control study. Ann. Trop. Paediatr. 27, 277–283 (2007).
Singh, S. & Aulakh, R. Kawasaki disease and Henoch Schonlein purpura: changing trends at a tertiary care hospital in north India (1993-2008). Rheumatol. Int. 30, 771–774 (2010).
Singh, S. et al. Is Kawasaki disease incidence rising in Chandigarh, North India? Arch. Dis. Child. 96, 137–140 (2011).
Singh, S. & Bhattad, S. Kawasaki disease incidence at Chandigarh, North India, during 2009-2014. Rheumatol. Int. 36, 1391–1397 (2016).
Dahdah, N. Not just coronary arteritis, Kawasaki disease is a myocarditis, too. J. Am. Coll. Cardiol. 55, 1507–1508 (2010).
Reddy, M. et al. Pro-brain natriuretic peptide (ProBNP) levels in North Indian children with Kawasaki disease. Rheumatol. Int. 36, 551–559 (2016).
Dionne, A. & Dahdah, N. Myocarditis and Kawasaki disease. Int. J. Rheum. Dis. 21, 45–49 (2018).
Eleftheriou, D. et al. Management of Kawasaki disease. Arch. Dis. Child. 99, 74–83 (2014).
Routhu, S. K. et al. Assessment of endothelial dysfunction in acute and convalescent phases of Kawasaki disease using automated edge detection software: a preliminary study from North India. J. Clin. Rheumatol. 27, 143–149 (2019).
Newburger, J. W. et al. The treatment of Kawasaki syndrome with intravenous gamma globulin. N. Engl. J. Med. 315, 341–347 (1986).
Mori, M. et al. Meta-analysis of the results of intravenous gamma globulin treatment of coronary artery lesions in Kawasaki disease. Mod. Rheumatol. 14, 361–366 (2004).
Burgner, D. & Harnden, A. Kawasaki disease: what is the epidemiology telling us about the etiology? Int. J. Infect. Dis. 9, 185–194 (2005).
Holman, R. C. et al. Racial/ethnic differences in the incidence of Kawasaki syndrome among children in Hawaii. Hawaii Med. J. 69, 194–197 (2010).
Kumrah, R., Vignesh, P., Rawat, A. & Singh, S. Immunogenetics of Kawasaki disease. Clin. Rev. Allergy Immunol. https://doi.org/10.1007/s12016-020-08783-9 (2020).
Berridge, M. J. Inositol trisphosphate and calcium signalling mechanisms. Biochim. Biophys. Acta 1793, 933–940 (2009).
Onouchi, Y. et al. ITPKC functional polymorphism associated with Kawasaki disease susceptibility and formation of coronary artery aneurysms. Nat. Genet. 40, 35–42 (2008).
Peng, Q. et al. Single-nucleotide polymorphism rs2290692 in the 3’UTR of ITPKC associated with susceptibility to Kawasaki disease in a Han Chinese population. Pediatr. Cardiol. 33, 1046–1053 (2012).
Chi, H. et al. ITPKC gene SNP rs28493229 and Kawasaki disease in Taiwanese children. Hum. Mol. Genet. 19, 1147–1151 (2010).
Kuo, H. C. et al. Single-nucleotide polymorphism rs7251246 in ITPKC is associated with susceptibility and coronary artery lesions in Kawasaki disease. PLoS ONE 9, e91118 (2014).
Kim, K. Y. et al. ITPKC and SLC11A1 gene polymorphisms and gene-gene interactions in Korean patients with Kawasaki disease. Yonsei Med. J. 59, 119–127 (2018).
Kushner, H. I., Macnee, R. & Burns, J. C. Impressions of Kawasaki syndrome in India. Indian Pediatr. 43, 939–942 (2006).
Singh, S., Gupta, M. K., Bansal, A., Kumar, R. M. & Mittal, B. R. A comparison of the clinical profile of Kawasaki disease in children from Northern India above and below 5 years of age. Clin. Exp. Rheumatol. 25, 654–657 (2007).
Singh, S., Bansal, A., Gupta, A., Kumar, R. M. & Mittal, B. R. Kawasaki disease: a decade of experience from North India. Int. Heart J. 46, 679–689 (2005).
Newburger, J. W. et al. Diagnosis, treatment, and long-term management of Kawasaki disease: a statement for health professionals from the Committee on Rheumatic Fever, Endocarditis, and Kawasaki Disease, Council on Cardiovascular Disease in the Young, American Heart Association. Pediatrics 114, 1708–1733 (2004).
Rodó, X. et al. Tropospheric winds from northeastern China carry the etiologic agent of Kawasaki disease from its source to Japan. Proc. Natl Acad. Sci. USA 111, 7952–7957 (2014).
Dietz, S. M. et al. Dissecting Kawasaki disease: a state-of-the-art review. Eur. J. Pediatr. 176, 995–1009 (2017).
Natividad, M. F., Torres-Villanueva, C. A. T. & Saloma, C. P. Superantigen involvement and susceptibility factors in Kawasaki disease: profiles of TCR Vβ2+ T cells and HLA-DRB1, TNF-α and ITPKC genes among Filipino patients. Int. J. Mol. Epidemiol. Genet. 4, 70–76 (2013).
The International Genome Sample Resource. 1000 Genomes: a deep catalog of human genetic variation. http://www.internationalgenome.org/ (2019).
Yan, Y. et al. Combined analysis of genome-wide-linked susceptibility loci to Kawasaki disease in Han Chinese. Hum. Genet. 132, 669–680 (2013).
Wang, W. et al. The roles of Ca2+/NFAT signaling genes in Kawasaki disease: single- and multiple-risk genetic variants. Sci. Rep. 4, 5208 (2014).
Khor, C. C. et al. Genome-wide association study identifies FCGR2A as a susceptibility locus for Kawasaki disease. Nat. Genet. 43, 1241–1246 (2011).
Lou, J. et al. A functional polymorphism, rs28493229, in ITPKC and risk of Kawasaki disease: an integrated meta-analysis. Mol. Biol. Rep. 39, 11137–11144 (2012).
Sethupathy, P. & Collins, F. S. MicroRNA target site polymorphisms and human disease. Trends Genet. 24, 489–497 (2008).
Yeter, D. & Deth, R. ITPKC susceptibility in Kawasaki syndrome as a sensitizing factor for autoimmunity and coronary arterial wall relaxation induced by thimerosal’s effects on calcium signaling via IP3. Autoimmun. Rev. 11, 903–908 (2012).
Lin, M. T. et al. Clinical implication of the C allele of the ITPKC gene SNP rs28493229 in Kawasaki disease: association with disease susceptibility and BCG acar reactivation. Pediatr. Infect. Dis. J. 30, 148–152 (2011).
Kuo, H. C. et al. ITPKC single nucleotide polymorphism associated with the Kawasaki disease in a Taiwanese population. PLoS ONE 6, e17370 (2011).
Onouchi, Y. et al. ITPKC and CASP3 polymorphisms and risks for IVIG unresponsiveness and coronary artery lesion formation in Kawasaki disease. Pharmacogenomics J. 13, 52–59 (2013).
Kuo, H. C. et al. A replication study for association of ITPKC and CASP3 two-locus analysis in IVIG unresponsiveness and coronary artery lesion in Kawasaki disease. PLoS ONE 8, e69685 (2013).
Acknowledgements
We wish to thank Prof. Bhavneet Bharti for their help in statistical analysis.
Funding
The authors thankfully acknowledge the Indian Council of Medical Research, New Delhi, India for financial assistance—Thesis Grant No. 3/2/June-2017/PG-Thesis-HRD (50) dated 13 March 2018. The funding agency had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
D.B.: conception and design, acquisition of data, data analysis, data interpretation, drafting manuscript, editing, and critical revision. R.K.: acquisition of data and data analysis. A.K.: acquisition of data and data analysis. An.K.: concept and design of the study, data interpretation, editing of the draft, and critical revision. P.S.: data interpretation, editing draft, and critical revision. A.R.: design of the study, acquisition of data, and data analysis. S.S.: concept and design of the study, clinical data, editing, critical revision, and approval of the final version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Consent to participate
Informed written consent is taken from parents of all subjects and controls. An assent was obtained from all children above 7 years of age. This study was approved by Institute Ethics Committee and Institute Thesis Committee and Departmental Review Board.
Consent for publication
Yes.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Bhattarai, D., Kumrah, R., Kaur, A. et al. Association of ITPKC gene polymorphisms rs28493229 and rs2290692 in North Indian children with Kawasaki disease. Pediatr Res 92, 1090–1098 (2022). https://doi.org/10.1038/s41390-021-01830-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41390-021-01830-x