Abstract
The biodiversity and evolution of fungal communities were monitored over a period of 3 vintages in a new winery. Samples were collected before grape receipt and 3 months after fermentation from 3 different wine related environments (WRE): floor, walls and equipment and analyzed using Illumina Mi-Seq. Genera of mold and filamentous fungi (294), non-enological (10) and wine-associated yeasts (25) were detected on all WREs before the arrival of the first harvest. Among them, genera like Alternaria and Aureobasidium persisted during two vintages. Therefore, these genera are not specific to winery environment and appear to be adapted to natural or anthropic environments due to their ubiquitous character. Some genera like Candida were also detected before the first harvest but only on one WREs, whereas, on the other WREs they were found after the harvest. The ubiquitous character and phenotypic traits of these fungal genera can explain their dynamics. After the first harvest and during 3 vintages the initial consortium was enriched by oenological genera like Starmerella introduced either by harvest or by potential transfers between the different WREs. However, these establishing genera, including Saccharomyces, do not appear to persist due to their low adaptation to the stressful conditions of winery environment.
Similar content being viewed by others
Introduction
Over the years, extensive research has been conducted on microbial biodiversity during the winemaking process. Microorganisms including filamentous fungi, bacteria and yeasts have been identified throughout the whole process using conventional culture-dependent techniques or molecular methods resulting in the description of this process as a true microbial ecosystem1,2. To understand the different roles of these microorganisms, especially yeasts, in wine fermentation and their impact on wine quality, in-depth knowledge of their population dynamics is needed. The development of recent technologies such as Next-Generation Sequencing (NGS) provided more complete understanding of the complexities of microbial communities3,4. These methods have uncovered that the microbial diversity is higher than expected5,6 and new microbial species (Cytospora, Gigaspora, Naganishia, Rhodosporidiobolus, Vuilleminia) have also been described7,8,9. Moreover, these methods made it possible to study diversity in more varied ecosystems like Wine-Related Environments (WREs: floor, walls and equipment)10. All these works described the presence of the same genera or species in different WREs and during the winemaking process, suggesting the persistence of these microorganisms during several vintages and their transfer between different WREs or between WREs and must or wine.
Intraspecific fingerprinting techniques were used to confirm the persistence and the transfer of microorganisms like Saccharomyces cerevisiae. Thus, techniques like mtDNA-RFLPs and interdelta PCR, were used to demonstrate the implantation and/or persistence of commercial strains of S. cerevisiae in must/wine and in WREs11,12,13,14. Indeed, a previous study showed that, although no longer used, commercial strains of S. cerevisiae can survive in the winery ecosystem for one year12. Concerning autochthonous S. cerevisiae, despite its low presence in the vineyard15, studies have shown that certain strains implicated in spontaneous fermentations originated from the winery environment13,16,17, or in some cases from the vineyard17. Moreover, implantation and persistence on the WREs and during the alcoholic fermentation (AF) were also highlighted for several non-Saccharomyces species18,19. Using FT-IR spectroscopy, the persistence of the species Hanseniaspora guilliermondii and Hanseniaspora uvarum during two vintages in the winery environment and their implantation in grape must have been previously demonstrated19.
All these findings highlighted: (i) the important role of WREs as an ecological niche for winery flora, and (ii) the implication of winery resident flora in the winemaking process.
From the beginning of the 21st century, the number of new wine holdings has been increasing worldwide20,21. To our knowledge, only 3 studies were focused on yeast biodiversity in newly established wineries during several vintages22,23,24. But, none of them assessed the total fungal population (yeasts, mold and filamentous fungi) present in WREs and especially before the arrival of the first harvest. Moreover, the authors focused only on the diversity of the yeast population using a culture-dependent technique (mtDNA-RFLP) and on the effect of using commercial strains on the diversity of yeast populations found during the AF. Indeed, the results showed the capacity of starter strains to persist and become resident in the environment of new wineries22,24; however, no real evidence of their presence was proven in WREs.
Considering the growing interest in vinification with indigenous flora, the study of the evolution of microbial populations on the WREs of a new winery without using commercial strains is of high interest.
In this direction, in the present study we collected several samples from three different winery surfaces (floor, walls and equipment), for three consecutive vintages (2016, 2017 and 2018) and at separate time points (before grape harvest and 3 months after fermentation) and used NGS technologies to study the fungal biodiversity populations in the WREs of a new winery. We were able to study the microbial dynamics over time focusing on the contribution of fungal populations present on grapes and in must to WREs populations. We gained new insights on the capacities and mechanisms developed in the establishment or colonization of a fungal consortium in the winery environment.
Results and Discussion
Fungal status of the new winery before the arrival of the first harvest
The fungal diversity on the WREs of a new winery operating exclusively in spontaneous fermentation was studied to determine the fungal status of the winery before the arrival of the first harvest (T0 2016). Samples were collected from the floor, walls, new (NE) and used (UE) equipment. After Illumina sequencing, fungal OTUs were classified into 3 categories: (i) the mold and filamentous fungi genera, (ii) the yeast genera never described before in the winemaking process (named non-enological yeasts), and (iii) the yeast and yeast-like genera already described in the winemaking process (named wine-associated yeasts) (Fig. 1). Wine-associated yeasts include all the genera described in the literature in the vineyard, vine, grapes, must, wine and winery surfaces25,26,27,28,29.
Mold and filamentous fungi dominated the fungal genera identified on all the surfaces before the first harvest, with 294 different genera (Fig. 1 and Table 1). Yeast genera (non-enological and wine-associated) were identified before the arrival of any harvest in any of the WREs (Fig. 1 and Table 1) and a total of 35 different genera were found (Table 1). Wine-associated yeasts were identified on all the studied surfaces of the new winery and dominated the yeast population detected (Fig. 1 and Table 1). The identification of non-enological yeast genera may have resulted from the use of high-performance Illumina sequencing and the constant enrichment of the databases used7,30. The new winery surfaces (floor, walls and new equipment) showed a higher percentage (average 13%) of unidentified genera compared to the used equipment (1%) (Fig. 1).
Diversity and dynamics of mold and filamentous fungi genera
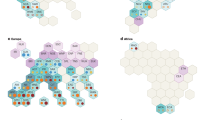
The different genera of mold and filamentous fungi identified at T0 2016 are represented using their relative abundances. Only the genera that represented >1% of relative abundance were selected (Fig. 2). Thus, a total of 32 mold and filamentous fungi genera among the 294 were identified on all the winery surfaces. At T0 2016, the taxonomic profiles observed on the floor, walls and the new equipment were quite close (with high representativeness of the genus Alternaria) compared to the used equipment which was dominated by the genus Cladosporium with the highest relative abundance (79%). These two genera had opposing relative abundances (a high abundance of Alternaria occurs with a low abundance of Cladosporium and vice-versa), suggesting potential interaction.
After two years of operations, a considerable modification of taxonomic profiles was observed, especially for the winery floor and equipment. These modifications resulted in a significant decrease in the total number of different identified genera (32 genera at T0 2016 and only 16 genera at T0 2018), as well as evolutions in the dominant genera (Fig. 2). So, the dominant genera Alternaria and Cladosporium described at T0 2016 were detected to a lesser extent at T0 2018, but persisted after 2 vintages. Their persistence throughout the whole study can be due to the presence of spores that might have been deposited from the surrounding atmosphere of the winery. Indeed, spores of Alternaria and Cladosporium are known to be widespread airborne spores31. Opposite abundances between the genera Alternaria and Cladosporium, observed at T0 2016, were also observed at T0 2018, suggesting again potential interaction (Fig. 2). After two years of operations, some genera became more widespread, like the genera Penicillium on the floor (57%) and Didymella on the equipment (68%). On the walls, Alternaria remained the most detected genus (35%), but new genera such as Cladosporium, Cyphellophora, Fusicolla, Knufia and Penicillium were also detected. The genus Acremonium was newly detected on all the WREs and the genus Aspergillus on the floor and equipment (Fig. 2).
Based on these results, implantation, colonization and persistence of some genera of mold and filamentous fungi were observed. For example: the genera Alternaria and Didymella present on all WREs between the vintages 2016 and 2018 can be considered as resident genera capable of persistence. After 2 years, genera Cladosporium and Penicillium are able to establish in all environments and persist. All these genera can therefore be qualified as ubiquitous genera. As for the genera Acremonium and Aspergillus, their implantation on environments might be linked to human activity and/or ventilation, which is an important factor of transfer of microorganisms between winery surfaces32. In addition, humidity and temperature described as two parameters influencing the growth of mold and filamentous fungi on surfaces like building material33,34,35 may also be factors influencing the establishment and persistence of the genera present on WREs.
Diversity and dynamics of non-enological yeast genera
Ten different genera of non-enological yeasts were detected before the arrival of the first harvest on all WREs. The genera Exophiala and Hannaella were the only genera present on all the winery surfaces with the highest percentage on winery floor (25% and 10%, respectively) (Table 2). After the 2 genera mentioned above, the genus Leucosporidium is the one with the highest percentages on winery floor (9%) and on walls (5%). These three genera are usually found in natural ecosystems like plants, soil, water and decaying wood material36,37,38,39. Therefore, it was not surprising to find these 3 genera on the surfaces of this new winery. These genera can therefore be qualified as ubiquitous genera.
Between T0 2016 and T0 2018, a significant increase in the biodiversity and quantity of non-enological yeasts were observed. Biodiversity-wise, 21 genera were detected at T0 2018 while only 8 genera were found at T0 2016. Quantity-wise, a 7-fold increase in the percentage of non-enological yeast was observed between these two time points on the walls (Table 2). Transfers of yeasts between different surfaces can be carried out by air flow and bioaerosol activity40 and over a period of one year27. Thus, these different transfers may explain the increase and dynamics of non-enological yeasts between T0 2016 and T0 2018. The two most abundant genera Exophiala and Hannaella, found initially at T0 2016, persisted in the winery environment throughout all the vintages and on all the winery surfaces. These two genera presented very high percentages, especially at T0 2017 on winery floor (73%) and T0 2018 on the winery walls (59%), however they did not exceed 5% on the winery equipment (Table 2). So, these results suggest that the conditions of every WRE plays an important role in retaining and shaping the fungal consortium of the winery. Indeed, the proliferation of fungi detected on different building material or in a particular area depends on the growth material and the conditions (e.g. humidity and temperature) under which they are found40,41. Compared to Exophiala and Hannaella, the genus Leucosporidium also persisted and colonized all WREs but with low percentages. Other genera previously detected at T0 2016, like, Buckleyzyma, Cutaneotrichosporon and Yamadazyma were detected frequently on all WREs but at very low levels (highest percentage: 2%) over the three vintages. Eleven new genera (e.g. Cystofilobasidium, Kondoa, Kregervanrija, Kwoniella, Lodderomyces, Malassezia, Oberwinklerozyma, Occultifur, Ogataea, Papilioterma, Piskurozyma, Pseudomicrostroma, Saitozyma, Schwanniomyces, Trichosporon and Udeniomyces) were detected punctually (1 or 2 times between T0 2016 and T0 2018) and the majority of them were detected after the fermentation activity (Table 2).
As for mold and filamentous fungi, different behaviors of non-enological yeasts were observed: some genera that can be considered as resident (environmental flora) and capable of persisting (Exophiala and Hannaella), genera capable of establishing and persisting (Buckleyzyma, Cutaneotrichosporon, Leucosporidium and Yamadazyma) and finally genera brought by the fermentation activity (e.g. Kregervanrija and Ogataea).
Diversity and dynamics of wine-associated yeast genera
At T0 2016, the taxonomic profiles for the floor, walls and the new equipment are quite close compared to the used equipment (Fig. 3), as described previously for mold and filamentous fungi. Relative abundance profiles show that the winery floor, walls and new equipment are primarily dominated by the yeast-like genus Aureobasidium (30%, 55% and 70%, respectively) and the genus Filobasidium (22%, 29% and 15%, respectively); however the used equipment is dominated by the genera Meyerozyma (45%), Wickerhamomyces (30%) and Aureobasidium (14%) (Fig. 3). Genus Aureobasidium was a dominant and common genus to all WREs, probably due to its capacity to persist in stress conditions linked to the sporulation ability of this genus42.
Before the arrival of the first harvest, yeast genera of enological interest like Candida, Hanseniaspora, Pichia and Saccharomyces were detected. Their presence on the used equipment is not surprising and is linked to the presence of must residues and/or wine, particularly in areas that are difficult to clean. The detection of the genera Candida, Pichia and Saccharomyces on the winery floor could be explained by the potential transfer between this environment and the used equipment due to human activities. Before the arrival of the first harvest, none of the yeast genera of enological interest were identified on the walls (Fig. 3).
Dynamics of wine-associated yeasts through three vintages on floor
On the winery floor at T0 2016, the 5 genera Aureobasidium, Candida, Cystobasidium, Filobasidium and Wickerhamomyces represented more than 65% of the total abundance (Fig. 4a). These genera were detected over time with the highest abundance: at T3 2016 (95%), at T0 2017 (92%), at T3 2017 (79%) and at T0 2018 (70%), but also with changes in their percentage of abundance. Indeed, a 75-fold increase of the genus Candida was observed at T3 2016, probably due to the fermentation activity (transfer from must and wine). The genera Bullera, Vishniacozyma and Metschnikowia detected at T0 2016 were also detected over time. The genus Saccharomyces detected at T0 2016 (<1%) was detected only at T3 2016 till T3 2017 but not at T0 2018. The genera Hanseniaspora, Priceomyces and Starmerella described on grapes and must19,42 were newly detected probably brought in during the harvest with the grapes and harvest equipment.
During the 3 vintages, the presence of spoilage microorganisms like the genera Dekkera (as from T3 2017) and Zygosaccharomyces (as from T3 2016) was observed on the winery floor (Fig. 4a). The presence of these yeasts 3 months after the AF could be strongly linked to the fermentation activity (transfer from must and wine). However, their implantation and persistence took place on the winery floor only after the second harvest.
Therefore, we showed that the communities found at T0 2016 conditioned the future resident flora (T0 2018) on the winery floor by the presence over time of the same 16 genera, particularly the genera Aureobasidium, Candida, Cystobasidium, Filobasidium, Metschnikowia and Wickerhamomyces, while others (Solicoccozyma and Torula) showed punctual presence. We also showed that the AF might underlie the enrichment (Candida) or introduction of genera of interest (Hanseniaspora and Starmerella) and/or spoilage (Dekkera and Zygosaccharomyces) yeasts on the winery floor.
Dynamics of wine-associated yeasts through three vintages on walls
Some dominant genera on the walls were in common with those found on the floor, like Aureobasidium, Bullera, Filobasidium, Vishniacozyma and Wickerhamomyces. Their evolutions were similar as they were detected over the three vintages but changes in their percentage of abundance are observed. New genera like Candida, Cryptococcus, Cystobasidium, Hanseniaspora, Pichia, Saccharomyces and Starmerella were detected after the first or the second harvest (Fig. 4b). Their presence on the walls may have been due to transfer via grape berries and/or must because they are also commonly described on grape berries and must26,43 or to the result of transfers between WREs by air flow or bioaerosol activity. In addition, the increase in the abundance of some genera already present on the walls at T0 2016, such as Debaryomyces, may also have been due to fermentation activity and/or to the transfer of these genera between WREs. The spoilage genera Dekkera and Zygosaccharomyces were detected only at T3 2016 and at T0 2018 (Fig. 4b), and did not show real persistence.
Thus, after 2 years, the resident flora of winery walls included 6 wine-associated yeast genera (Aureobasidium, Bullera, Debaryomyces, Filobasidium, Vishniacozyma and Wickerhamomyces) found between T0 2016 and T0 2018, as previously described for winery floor. The AF activity also impacted the yeast flora of the walls (particularly the first fermentation) by the enrichment of certain genera like Debaryomyces and Wickerhamomyces and by the occurrence of new genera like Candida, Cryptococcus, Dekkera, Metschnikowia, Pichia, Saccharomyces, Starmerella and Zygosaccharomyces. The genus Saccharomyces was only detected during the 2017 vintage (T0 and T3), and did not persist in 2018. However, certain genera persisted until 2018 (e.g. Candida, Hanseniaspora). In addition, the flora of the walls showed radical changes at different time points. Thus, the resident flora of the walls appeared to have a higher state of instability than that of the winery floor. This instability may also be due to the fact that several genera, mainly of enological interest, were not in an environment favorable for their installation. Their presence appeared to be the result of transfers from other surfaces.
Dynamics of wine-associated yeasts through three vintages on equipment
At T0 2016 an average taxonomic profile was established by the association of the 2 profiles obtained from the used and new winery equipment (Fig. 4c). The genera Aureobasidium and Filobasidium, which were more abundant for the 2016 vintage (as for floor and walls), presented very low abundances afterwards (10% at T0 2017, 2% at T3 2017 and 1% at T0 2018). Throughout the vintages, the taxonomic profiles became enriched by other genera, most importantly, Candida, Debaryomyces, Hanseniaspora, Metschnikowia, Saccharomyces, Sporobolomyces and Wickerhamomyces which became more widespread (Fig. 4c). The presence of these genera, and more particularly for the genus Saccharomyces, probably resulted from must handling (beginning of AF in a fermentation tank) before the sampling of the winery equipment at T0 2017. After this time point, the genus Saccharomyces was not detected, so does not seem to persist. The higher presence of non-Saccharomyces yeasts compared to Saccharomyces on winery equipment have been previously reported44.
The spoilage genera Dekkera and Zygosaccharomyces were both probably provided by must and/or wine, particularly for the 2017 vintage. The highest abundance of the genus Dekkera was observed at T3 2017 and persisted to T0 2018 (Fig. 4c).
Thus, the changes of wine-associated yeasts on winery equipment showed specific dynamics compared to the floor and walls. Shifts in the taxonomic profiles were observed between the 2016 vintage and the next vintages. These observations and interpretations must take into account that winery equipment is regularly cleaned and is in direct contact with must and wine. Also, the results show that the evolution of wine-associated yeasts on equipment is not conditioned by the initial yeast population (T0 2016) as found for the winery floor and walls. Despite this, some wine-associated yeast genera appear to be ubiquitous and adapted to winery conditions. Indeed, the genera Aureobasidium, Bullera, Filobasidium and Vishniacozyma were detected throughout the whole study no matter which WRE it is.
Establishment of a winery consortium
The presence of mold, filamentous fungi and yeasts on the WREs of a new winery before the arrival of the first harvest was described. Considering the different abundances of all the genera (not only those which represent >1%), a total of 329 different fungal genera were identified at T0 2016 (294 mold and filamentous fungi and 35 yeasts) on all the WREs, which have never been described before. After 2 years, 172 fungal genera (132 mold and filamentous fungi and 40 yeasts) were identified and 17 fungal genera were newly detected (5 mold and filamentous fungi and 12 yeasts).
Among the total fungal genera and in terms of abundance, a total of 40 genera (mold, filamentous fungi and yeasts) presented different evolutions over time (Fig. 5). On the one hand, 10/40 genera (Alternaria, Aureobasidium, Bullera, Didymella, Exophiala, Filobasidium, Hannaella, Holtermanniella, Sporobolomyces and Vishniacozyma) were detected before the first harvest (T0 2016) and throughout the whole study on all the WREs until T0 2018 (Fig. 5). The majority of these 10 genera are commonly described on the berries and/or in must and have been considered, so far, as related to vineyard and wine-growing environments. However, according to the literature, these genera are described as ubiquitous genera widely distributed in natural ecosystems (soil, air, water and plants) and can colonize buildings45,46,47. In fact, the genus Alternaria is able to survive in cold, hot and dry weather and all year round in debris and seeds48,49,50. In addition, the sporulation capacity described for mold and fungi genera could contribute toward enhancing their persistence in adverse conditions like the winery environment34,51. Certain phenotypic traits described for the persistent genus Aureobasidium like the production of extracellular polysaccharides52 and film forming capacity53 may explain its persistence throughout the study. The persistence of Bullera, Filobasidium and Vishniacozyma genera may be related to their belonging to the Basidiomycetous yeast division and more particularly to the oxidative group of Basidiomycetous. Indeed, this group is described as being usually found in higher dominance in stressful environments and persisting well in adverse environments54,55. Several studies demonstrated the tolerance of the genus Exophiala to different stresses56 and the Hannaella yeast genus has been described in the phyllosphere of various plant species including Vitis vinifera and in soil35,57 which may explain their persistence over time. Thus, according to the results obtained in this study and the specific capacities of the genera described previously, we confirm that these 10 genera are not specific to a given environment like that of wineries. These genera are resident environmental flora that can persist in stressful environments including a winery (Fig. 5).
In addition, 20/40 of the fungal genera were detected at T0 2016 and showed persistence after 2 years on 1 or 2 WREs (Fig. 5). As previously described, ubiquitous character of these fungal genera linked to some phenotypic traits can explain their persistence37,39,54,55,58,59,60,61,62,63,64,65,66,67,68,69,70. These genera are considered as persistent and/or establishing winery flora. Among them, some genera became established via the harvest or transfers between WREs, especially on the walls and equipment (Fig. 5). After their implantation, these genera were able to persist until 2018, except the genus Saccharomyces, which persisted only on the floor 2 years after the arrival of the first harvest (Fig. 5). After the implantation linked to harvest, the persistence of the genus Candida may have been due to phenotypic traits like invasive growth71 and film formation53. Regarding the genus Pichia, the capacity of some species, isolated from grapes, to form ascospores72,73 and to produce killer toxins (proteins secreted by a fungi inhibiting the growth of other fungi)74 potentially contribute to the implantation and persistence of this genus in winery environment. Establishment and persistence of the genus Metschnikowia may be linked to its ability to outgrow other microorganisms due to the production of reddish pulcherrimin involved in iron chelation75,76. Furthermore, among the genus Wickerhamomyces, the species W. anomalus (previously known as Pichia anomala)77 is known for its resistance to osmotic pressure and the production of killer toxins78,79,80.
Finally, 10/40 of the fungal genera (Acremonium, Aspergillus, Barnettozyma, Cryptococcus, Dekkera, Hanseniaspora, Priceomyces, Saccharomycopsis, Starmerella and Zygosaccharomyces) were not detected at T0 2016 but exhibited implantation after the 2016 and/or 2017 harvests and certain of them persisted over time depending on the WREs (Fig. 5). The capacity of the genus Aspergillus to produce conidia81 and the antagonism activity of the genus Acremonium82 can explain their establishment in winery environment. Regarding the genus Hanseniaspora, the implantation and the persistence of 2 species of Hanseniaspora during 2 vintages has been previously demonstrated19. The genus Priceomyces has never been described in a winery environment before, only on grape berries43. Nevertheless, this genus was detected over one year in brewery air83. Thus, all yeast genera showed potential for implantation in all WREs and to become part of the established winery flora as well as the spoilage genus Dekkera previously described as a persistent genus in several steps of the winemaking process and in WREs84,85. So, the implantation of all 10 genera can be explained by human activities (fermentation and/or transfer) in the winery and are considered as establishing winery flora (Fig. 5).
In conclusion, this work provided new insights into the establishment of the fungal consortium in a new winery. This fungal consortium consisted of persistent environmental flora that is not specific to the winery environment. These environmental flora included mold, filamentous fungi (e.g. Alternaria, Didymella) and yeasts (e.g. Aureobasidium, Filobasidium). Certain genera (e.g. Candida) were found before the arrival of the first harvest on one WREs, mainly on the winery floor, and persisted over time. Meanwhile, on the other WREs, the same genera were detected following the start of the harvest. Therefore, this finding suggests that these genera are well adapted to one specific environment of the winery and could be classified as persistent flora and/or establishing flora. However, during 2 vintages, the environmental flora was enriched by other fungal genera (e.g. Acremonium, Hanseniaspora). These genera were considered as establishing winery flora and are less adapted to the winery environment. Moreover, the majority of establishing flora are genera that present a fermentative metabolism that could also to be a prerequisite state preventing their adaptation. Future works could be carried out to understand the mechanisms implicated in the establishment and persistence of this flora on the WRE of this new winery and more particularly, flora of enological interest.
Material and Methods
Facility description
All the samples were collected from a newly established winery located in Nuits-Saint-Georges (Bourgogne, France). The new winery was built in 2016 and started operating in September 2016. The winery covers a surface area of 1000 m2 and is situated in an industrial zone with no surrounding vineyards. All the red wines from this winery were obtained exclusively from Pinot Noir grapes.
Sampling of wine-related environments (WREs)
Samples were taken during three consecutive vintages (2016, 2017 and 2018). For the first vintage, 2016, the samples were collected at two separate time points: before the receipt of grapes (T0), three months after the end of the fermentations (T3), and from three different winery surfaces: floor (F), walls (W), equipment (E). At T0 time point, the majority of the equipment in the new winery was new, except for certain vats (36/108) that were formerly used in another winery. Therefore, the samples collected from the winery equipment (E) at T0 2016 were separated between used equipment (UE) and new equipment (NE). The equipment sampled included lift conveyor belts, destemmers, presses, stainless steel vats, wooden vats, pumps, and transfer pipes.
For the samples collected after T0 2016, no separation between UE and NE was realized since all the equipment was in contact with the must and/or wine and therefore considered as UE.
For the 2017 vintage, the same sampling methodology was followed; however, to obtain more representative samples of the winery, the number was doubled for the samples collected from the floor and walls (Table 3). In the same way, the last samples were taken only at T0 of the 2018 harvest, which means that the sampling was carried out for two full years. All the samples were collected by streaking specific areas of each winery surface using sterile cotton swabs, which were then placed in an equivalent volume (40 mL for 40 samples) of physiological water solution (NaCl at 0.9% w/v).
A total of 280 samples (2016 vintage), 400 samples (2017 vintage) and 200 samples (T0 2018) were collected on all the winery surfaces (Table 3). Given this significant number, the samples were pooled to obtain average samples per environment and for each sampling time point. For example, for the 2016 vintage, the 40 samples from the floor at T0 were pooled to form an average sample. Thus, a total of 7 average samples (2016 vintage), 6 average samples (2017 vintage) and 3 average samples (T0 2018) were analyzed afterwards.
DNA extraction
10 mL of each average sample was centrifuged at 4000 rpm for 4 min at 4 °C. The supernatant was discarded and the cell pellet was suspended in 200 µL of DNA-Yeast extraction buffer (2% Triton X-100 (v/v), 1% SDS (w/v), 100 mM NaCl, 10 mM Tris and 1 mM EDTA at pH 8). Then, 60 µL of phenol/chloroform/isoamyl alcohol (25:24:1) and 0.3 g of glass beads (0.5 mm in diameter; Scientific Industries) were added. The cells were lysed by Precellys 24-Dual (Bertin Technologies) for 3 ×45 s and placed on ice for 2 min. Afterwards, 200 µL of TE Buffer was added (10 mM Tris and 1 mM EDTA pH 8) and the mixture was centrifuged at 13 700 rpm for 10 min at 4 °C. The supernatant was collected and the DNA was precipitated with 1 mL of 100% (v/v) ethanol solution and centrifuged at 13 700 rpm for 10 min at 20 °C. The DNA pellet was washed with 70% (v/v) ethanol solution and centrifuged at 13 700 rpm for 5 min at 20 °C. Finally, the DNA pellet was dried at 95 °C for 5 min to remove the excess ethanol and re-suspended in 40 µL of Milli-Q water and stored at −20 °C.
ITS amplicon library preparation
The nuclear ribosomal internal transcribed spacer 2 (ITS2) region of the fungal DNA was amplified for this analysis using a mono index approach. The fusion primers selected for this study were 5.8S-Fun (5′-CAAGCAGAAGACGGCATACGAGAT-NNNNNNNNNNNN-AGTCAGTCAG-GG-AACTTTYRRCAAYGGATCWCT-3′) and ITS4-Fun (5′-AATGATACGGCGACCACCGAGATCTACAC-TATGGTAATT-AA-AGCCTCCGCTTATTGATATGCTTAART-3′). The fusion primers contained the 24- to 29-bp Illumina sequencing adaptor, the 12-bp Golay barcode (5.8S-Fun exclusively), a 10-bp primer pad, a 2-bp linker, and the 21- to 27-bp core primer (see Supplementary Table S1)86.
PCR amplification was carried out in a final volume of 50 μL, including 2 μL DNA template, 0.3 µL of Platinum Taq DNA Polymerase 2.5 U (Invitrogen™, Waltham, MA, USA), 1 × Taq buffer, 20 μM dNTP and 1 μL of each primer 10 µM. The PCR program consisted in denaturing for 5 min at 94 °C, followed by 30 cycles of 30 s at 94 °C, 1 min at 60 °C, 1 min at 72 °C and final extension at 72 °C for 5 min. Finally, the pooled PCR products were size-selected with an Invitrogen® 2% E-Gel, purified using the QIAquick purification kit (Qiagen, Germantown, MD). Their concentration was determined with the Qubit® dsDNA HS Assay Kit (Thermo Fisher Scientifics, USA). The pooled PCR products were sequenced with an Illumina® MiSeq sequencer (2 × 300 cycles) (Illumina, San Diego, CA, USA). 20–25% PhiX control DNA was spiked in the run to add base diversity.
Sequence analysis
Data from Mi-Seq sequencing were analyzed with PIPITS, an automated pipeline for the detection and differentiation of fungal ITS87 that produces Operational Taxonomic Unit (OTUs) abundance tables for each metagenomic sample.
Data availability
All relevant data are available upon request.
References
Amerine, M. A. & Kunkee, R. E. Microbiology of Winemaking. Annual Review of Microbiology 22, 323–358 (1968).
Ivey, M. L. & Phister, T. G. Detection and identification of microorganisms in wine: a review of molecular techniques. J. Ind. Microbiol. Biotechnol. 38, 1619–1634 (2011).
Belda, I., Zarraonaindia, I., Perisin, M., Palacios, A. & Acedo, A. From Vineyard Soil to Wine Fermentation: Microbiome Approximations to Explain the “terroir” Concept. Front. Microbiol. 8, 821, https://doi.org/10.3389/fmicb.2017.00821 (2017).
Morgan, H. H., du Toit, M. & Setati, M. E. The Grapevine and Wine Microbiome: Insights from High-Throughput Amplicon Sequencing. Front. Microbiol. 8, 820, https://doi.org/10.3389/fmicb.2017.00820 (2017).
David, V. et al. High-throughput sequencing of amplicons for monitoring yeast biodiversity in must and during alcoholic fermentation. J. Ind. Microbiol. Biotechnol. 41, 811–821 (2014).
Wang, C., García-Fernández, D., Mas, A. & Esteve-Zarzoso, B. Fungal diversity in grape must and wine fermentation assessed by massive sequencing, quantitative PCR and DGGE. Front. Microbiol. 6, 1156, https://doi.org/10.3389/fmicb.2015.01156 (2015).
Bokulich, N. A., Ohta, M., Richardson, P. M. & Mills, D. A. Monitoring Seasonal Changes in Winery-Resident Microbiota. PLoS One 8, e66437, https://doi.org/10.1371/journal.pone.0066437 (2013).
Kecskeméti, E., Berkelmann-Löhnertz, B. & Reineke, A. Are Epiphytic Microbial Communities in the Carposphere of Ripening Grape Clusters (Vitis vinifera L.) Different between Conventional, Organic, and Biodynamic Grapes? PLoS One 11, e0160852, https://doi.org/10.1371/journal.pone.0160852 (2016).
Mezzasalma, V. et al. Grape microbiome as a reliable and persistent signature of field origin and environmental conditions in Cannonau wine production. PLoS One 12, e0184615, https://doi.org/10.1371/journal.pone.0184615 (2017).
Bokulich, N. A. et al. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods. 10, 57–59 (2013).
Vezinhet, F., Hallet, J.-N. & Valade, M. & Poulard, A. Ecological Survey of Wine Yeast Strains by Molecular Methods of Identification. Am. J. Enol. Vitic. 43, 83–86 (1992).
Santamaría, P., Garijo, P., López, R., Tenorio, C. & Rosa Gutiérrez, A. Analysis of yeast population during spontaneous alcoholic fermentation: Effect of the age of the cellar and the practice of inoculation. Int. J. Food Microbiol. 103, 49–56 (2005).
Mercado, L., Dalcero, A., Masuelli, R. & Combina, M. Diversity of Saccharomyces strains on grapes and winery surfaces: analysis of their contribution to fermentative flora of Malbec wine from Mendoza (Argentina) during two consecutive years. Food Microbiol. 24, 403–412 (2007).
Blanco, P., Orriols, I. & Losada, A. Survival of commercial yeasts in the winery environment and their prevalence during spontaneous fermentations. J. Ind. Microbiol. Biotechnol. 38, 235–239 (2011).
Valero, E., Cambon, B., Schuller, D., Casal, M. & Dequin, S. Biodiversity of Saccharomyces yeast strains from grape berries of wine-producing areas using starter commercial yeasts. FEMS Yeast Res. 7, 317–329 (2007).
Ciani, M., Mannazzu, I., Marinangeli, P., Clementi, F. & Martini, A. Contribution of winery-resident Saccharomyces cerevisiae strains to spontaneous grape must fermentation. Antonie Van Leeuwenhoek 85, 159–164 (2004).
Grangeteau, C. Biodiversité fongique du raisin au vin: Impact de l’activité anthropique. PhD thesis, Université de Bourgogne Franche Comté, (2016).
Grangeteau, C. et al. Diversity of yeast strains of the genus Hanseniaspora in the winery environment: What is their involvement in grape must fermentation? Food Microbiol. 50, 70–77 (2015).
Grangeteau, C. et al. Persistence of Two Non-Saccharomyces Yeasts (Hanseniaspora and Starmerella) in the Cellar. Front. Microbiol. 7, 268, https://doi.org/10.3389/fmicb.2016.00268 (2016).
Bruyas, P. Area under vines in the EU down by 15.7% between 1989 and 1999. www.publications.europa.eu/en/publication-detail/-/publication/971742b5-1a0f-4772-ba5a-345e41844619. (2003).
Wines Vines Analytics. U.S. Wineries Statistics. www.winesvinesanalytics.com/statistics/winery/. (2018)
Constantí, M., Poblet, M., Arola, L., Mas, A. & Guillamón, J. M. Analysis of Yeast Populations During Alcoholic Fermentation in a Newly Established Winery. Am. J. Enol. Vitic. 48, 339–344 (1997).
Beltran, G. et al. Analysis of yeast populations during alcoholic fermentation: A six year follow-up study. Syst. Appl. Microbiol. 25, 287–293 (2002).
Clavijo, A., Calderón, I. L. & Paneque, P. Effect of the use of commercial Saccharomyces strains in a newly established winery in Ronda (Málaga, Spain). Antonie Van Leeuwenhoek 99, 727–731 (2011).
Pretorius, I. S., Westhuizen, T. Jvander & Augustyn, O. P. H. Yeast Biodiversity in Vineyards and Wineries and Its Importance to the South African Wine Industry. A Review. S. Afr. J. Enol. Vitic. 20, 61–70 (1999).
Bisson, L. F. & Joseph, C. M. L. Yeasts. in Biology of Microorganisms on Grapes, in Must and in Wine (eds König, H., Unden, G. & Fröhlich, J.) 47–60 (Springer Berlin Heidelberg, https://doi.org/10.1007/978-3-540-85463-0-3 2009).
Díaz, C., Molina, A. M., Nähring, J. & Fischer, R. Characterization and Dynamic Behavior of Wild Yeast during Spontaneous Wine Fermentation in Steel Tanks and Amphorae. BioMed Res. Int. https://doi.org/10.1155/2013/540465 (2013).
Hendgen, M. et al. Effects of different management regimes on microbial biodiversity in vineyard soils. Scientific Reports 8, 9393, https://doi.org/10.1038/s41598-018-27743-0 (2018).
Drumonde-Neves, J., Franco-Duarte, R., Lima, T., Schuller, D. & Pais, C. Yeast Biodiversity in Vineyard Environments Is Increased by Human Intervention. PLoS One 11, e0160579, https://doi.org/10.1371/journal.pone.0160579 (2016).
Heather, J. M. & Chain, B. The sequence of sequencers: The history of sequencing DNA. Genomics 107, 1–8 (2016).
Grinn-Gofroń, A. & Strzelczak, A. Changes in concentration of Alternaria and Cladosporium spores during summer storms. Int. J. Biometeorol. 57, 759, https://doi.org/10.1007/s00484-012-0604-0 (2013).
Ocón, E. et al. Factors of influence in the distribution of mold in the air in a wine cellar. J. Food Sci. 76, M169–174, https://doi.org/10.1111/j.1750-3841.2011.02097.x. (2011).
Coppock, J. B. M. & Cookson, E. D. The effect of humidity on mould growth on constructional materials. Journal of the Science of Food and Agriculture 2, 534–537 (1951).
Pasanen, A.-L., Pasanen, P., Jantunen, M. J. & Kalliokoski, P. Significance of air humidity and air velocity for fungal spore release into the air. Atmospheric Environment. Part A. General Topics 25, 459–462 (1991).
Johansson, P., Ekstrand-Tobin, A., Svensson, T. & Bok, G. Laboratory study to determine the critical moisture level for mould growth on building materials. International Biodeterioration & Biodegradation 73, 23–32 (2012).
Landell, M. F. et al. Hannaella pagnoccae sp. nov., a tremellaceous yeast species isolated from plants and soil. Int. J. Syst. Evol. Microbiol. 64, 1970–1977 (2014).
Summerbell, R. C. The heterobasidiomycetous yeast genus Leucosporidium in an area of temperate climate. Can. J. Bot. 61, 1402–1410 (1983).
Vadkertiová, R., Dudášová, H. & Balaščáková, M. Yeasts in Agricultural and Managed Soils. in Yeasts in Natural Ecosystems: Diversity (eds Buzzini, P., Lachance, M. A. & Yurkov, A.) 117–144 (Springer International Publishing, 2017).
Yurkov, A. M., Schäfer, A. M. & Begerow, D. Leucosporidium drummii sp. nov., a member of the Microbotryomycetes isolated from soil. Int. J. Syst. Evol. Microbiol. 62, 728–734 (2012).
Theisinger, S. M. & Smidt, O. de. S. Bioaerosols in the Food and Beverage Industry. Ideas and Applications Toward Sample Preparation for Food and Beverage Analysis https://doi.org/10.5772/intechopen.69978 (2017).
Mensah-Attipoe, J. & Toyinbo, O. Fungal Growth and Aerosolization from Various Conditions and Materials. in Fungal Infection (ed. Silva de Loreto, E.) (IntechOpen, 2019).
Ramos, S. & García Acha, I. A vegetative cycle of Pullularia pullulans. Transactions of the British Mycological Society 64, 129–135 (1975).
Cordero-Bueso, G., Esteve-Zarzoso, B., Cabellos, J. M., Gil-Díaz, M. & Arroyo, T. Biotechnological potential of non-Saccharomyces yeasts isolated during spontaneous fermentations of Malvar (Vitis vinifera cv. L.). Eur. Food Res. Technol. 236, 193–207 (2013).
Ocón, E., Gutiérrez, A. R., Garijo, P., López, R. & Santamaría, P. Presence of non-Saccharomyces yeasts in cellar equipment and grape juice during harvest time. Food Microbiol. 27, 1023–1027 (2010).
Flannigan, B. Microorganisms in indoor air. in Microorganisms in Home and Work Environments: Diversity, Health Impacts, Investigation and Control (eds Samson, R. A., Miller, J. D. & Flannigan, B.) 27–47 (Taylor &Francis, 2001).
Nagahama, T. Yeast Biodiversity in Freshwater, Marine and Deep-Sea Environments. in Biodiversity and Ecophysiology of Yeasts (eds Péter, G. & Rosa, C.) 241–262 (Springer Berlin Heidelberg, 2006).
Egbuta, M. A., Mwanza, M. & Babalola, O. O. A Review of the Ubiquity of Ascomycetes Filamentous Fungi in Relation to Their Economic and Medical Importance. Advances in Microbiology 6, 1140–1158 (2016).
Thomma, B. P. H. J. Alternaria spp.: from general saprophyte to specific parasite. Mol. Plant Pathol. 4, 225–236 (2003).
Logrieco, A., Moretti, A. & Solfrizzo, M. Alternaria toxins and plant diseases: an overview of origin, occurrence and risks. World Mycotoxin Journal 2, 129–140 (2009).
Rotem, J. The genus Alternaria: biology, epidemiology, and pathogenicity. (American Phytopathological Society, 1994).
Musetti, R. et al. Inhibition of Sporulation and Ultrastructural Alterations of Grapevine Downy Mildew by the Endophytic Fungus Alternaria alternata. Phytopathology 96, 689–698 (2006).
Singh, R., Gaur, R., Tiwari, S. & Gaur, M. K. Production of pullulan by a thermotolerant Aureobasidium pullulans strain in non-stirred fed batch fermentation process. Braz. J. Microbiol. 43, 1042–1050 (2012).
Malfeito-Ferreira, M. Spoilage Yeasts in Red Wines. in Red Wine Technology (ed Morata, A.) 219–235 (Academic Press, 2019).
Fleet, G. H., Prakitchaiwattana, C., Beh, A. L. & Heard, G. M. The yeast ecology of wine grapes. in Biodiversity and Biotechnology of Wine Yeasts (ed Ciani, M.) 1–17 (Research Signpost, 2002).
Sabate, J., Cano, J., Esteve-Zarzoso, B. & Guillamón, J. M. Isolation and identification of yeasts associated with vineyard and winery by RFLP analysis of ribosomal genes and mitochondrial DNA. Microbiol. Res. 157, 267–274 (2002).
Gostinčar, C., Grube, M. & Gunde-Cimerman, N. Evolution of fungal pathogens in domestic environments? Fungal Biol. 115, 1008–1018 (2011).
Kaewwichian, R., Limtong, S., Am-In, S., Jindamorakot, S. & Sipiczki, M. Hannaella siamensis sp. nov. and Hannaella phetchabunensis sp. nov., two new anamorphic basidiomycetous yeast species isolated from plants. Int. J. Syst. Evol. Microbiol. 65, 1297–1303 (2015).
Flannigan, B. Air sampling for fungi in indoor environments. Journal of Aerosol Science 28, 381–392 (1997).
Bensch, K., Braun, U., Groenewald, J. Z. & Crous, P. W. The genus Cladosporium. Stud Mycol 72, 1–401 (2012).
Baxter, D. M., Perkins, J. L., McGhee, C. R. & Seltzer, J. M. A Regional Comparison of Mold Spore Concentrations Outdoors and Inside “Clean” and “Mold Contaminated” Southern California Buildings. Journal of Occupational and Environmental Hygiene 2, 8–18 (2005).
Basilico, M. et al. Influence of environmental factors on airborne fungi in houses of Santa Fe City, Argentina. Science of The Total Environment 376, 143–150 (2007).
Hawker, L. E. & Hendy, R. J. An Electron-Microscope Study of Germination of Conidia of Botrytis cinerea. Journal of General Microbiology 33, 43–46 (1963).
Kim, W. K. et al. Six Species of Penicillium Associated with Blue Mold of Grape. Mycobiology 35, 180–185 (2007).
Sempere, F. & Santamarina, M. P. The conidia formation of several Fusarium species. Annals of Microbiology 59, 663–674 (2009).
Mašínová, T. et al. Drivers of yeast community composition in the litter and soil of a temperate forest. FEMS Microbiol. Ecol. 93, fiw223, https://doi.org/10.1093/femsec/fiw223 (2017).
Atkin, C. L., Neilands, J. B. & Phaff, H. J. Rhodotorulic acid from species of Leucosporidium, Rhodosporidium, Rhodotorula, Sporidiobolus, and Sporobolomyces, and a new alanine-containing ferrichrome from Cryptococcus melibiosum. J. Bacteriol. 103, 722–733 (1970).
Skinner, F. A., Passmore, S. M. & Davenport, R. R. Biology and activities of yeasts. (London Academic Press, 1980).
ŌNishi, H. Osmophilic Yeasts. In Advances in Food Research (eds Chichester, C. O., Mrak, E. M. & Stewart, G. F.) 53–94 (Academic Press, 1963).
Buzzini, P., Branda, E., Goretti, M. & Turchetti, B. Psychrophilic yeasts from worldwide glacial habitats: diversity, adaptation strategies and biotechnological potential. FEMS Microbiol. Ecol. 82, 217–241 (2012).
Zaky, A. S., Tucker, G. A., Daw, Z. Y. & Du, C. Marine yeast isolation and industrial application. FEMS Yeast Res. 14, 813–825 (2014).
Loureiro, V. & Malfeito-Ferreira, M. Spoilage yeasts in the wine industry. Int. J. Food Microbiol. 86, 23–50 (2003).
Sibirnyĭ, A. A., Zharova, V. P., Kshanovskaia, B. V. & Shavlovskiĭ, G. M. Selection of a genetic strain of Pichia guilliermondii yeasts capable of forming a significant quantity of spores. Tsitol. Genet. 11, 330–333 (1977).
Vaughan-Martini, A., Kurtzman, C. P., Meyer, S. A. & O’Neill, E. B. Two new species in the Pichia guilliermondii clade: Pichia caribbica sp. nov., the ascosporic state of Candida fermentati, and Candida carpophila comb. nov. FEMS Yeast Res. 5, 463–469 (2005).
Urszula, B., Pawel, S. & Pawel, S. The influence of Pichia killer toxins on the wine spoilage yeasts. Potravinárstvo (2015).
Sipiczki, M. Metschnikowia Strains Isolated from Botrytized Grapes Antagonize Fungal and Bacterial Growth by Iron Depletion. Appl. Environ. Microbiol. 72, 6716–6724 (2006).
Türkel, S. & Ener, B. Isolation and characterization of new Metschnikowia pulcherrima strains as producers of the antimicrobial pigment pulcherrimin. Z. Naturforsch. C. J. Biosci. 64, 405–410 (2009).
Kurtzman, C. P. Phylogeny of the ascomycetous yeasts and the renaming of Pichia anomala to Wickerhamomyces anomalus. Antonie Van Leeuwenhoek 99, 13–23 (2011).
Comitini, F. et al. Pichia anomala and Kluyveromyces wickerhamii killer toxins as new tools against Dekkera/Brettanomyces spoilage yeasts. FEMS Microbiol. Lett. 238, 235–240 (2004).
Passoth, V., Fredlund, E., Druvefors, U. A. & Schnürer, J. Biotechnology, physiology and genetics of the yeast Pichia anomala. FEMS Yeast Res. 6, 3–13 (2006).
Fernández de Ullivarri, M., Mendoza, L. M. & Raya, R. R. Characterization of the killer toxin KTCf20 from Wickerhamomyces anomalus, a potential biocontrol agent against wine spoilage yeasts. Biological Control 121, 223–228 (2018).
Atoui, A., Mitchell, D., Mathieu, F., Magan, N. & Lebrihi, A. Partitioning of ochratoxin A in mycelium and conidia of Aspergillus carbonarius and the impact on toxin contamination of grapes and wine. J. Appl. Microbiol. 103, 961–968 (2007).
Lo Piccolo, S. et al. Identification of Acremonium isolates from grapevines and evaluation of their antagonism towards Plasmopara viticola. Ann. Microbiol. 65, 2393–2403 (2015).
Spitaels, F. et al. The Microbial Diversity of Traditional Spontaneously Fermented Lambic Beer. PLoS One 9, e95384, https://doi.org/10.1371/journal.pone.0095384 (2014).
Renouf, V., Miot-Sertier, C., Perello, M.-C., Revel, G. de & Lonvaud-Funel, A. Evidence for differences between B. bruxellensis strains originating from an enological environment. International Journal of Wine Research https://doi.org/10.2147/IJWR.S4612 (2009).
Suárez, R., Suárez-Lepe, J. A., Morata, A. & Calderón, F. The production of ethylphenols in wine by yeasts of the genera Brettanomyces and Dekkera: A review. Food Chemistry 102, 10–21 (2007).
Taylor, D. L. et al. Accurate Estimation of Fungal Diversity and Abundance through Improved Lineage-Specific Primers Optimized for Illumina Amplicon Sequencing. Appl. Environ. Microbiol. 82, 7217–7226 (2016).
Gweon, H. S. et al. PIPITS: an automated pipeline for analyses of fungal internal transcribed spacer sequences from the Illumina sequencing platform. Methods in Ecology and Evolution 6, 973–980 (2015).
Acknowledgements
This work was supported by the Regional Council of Bourgogne- Franche-Comté, the “Fonds Européen de Développement Régional (FEDER)” and the “Bureau Interprofessionnel des Vins de Bourgogne (BIVB)”. The authors would like to thank the wine company who provided the facility in which this study was realized.
Author information
Authors and Affiliations
Contributions
H.Ab. performed the experiments, conducted filed work, analyzed the data, constructed the figures and wrote the manuscript; C.R.C. performed the experiments; M.V. contributed to manuscript revision; P.D. analyzed the sequencing data; H.Al. contributed to manuscript revision; M.G.B. conducted filed work and contributed to manuscript revision; S.R. designed the experiment, conducted filed work and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Abdo, H., Catacchio, C.R., Ventura, M. et al. The establishment of a fungal consortium in a new winery. Sci Rep 10, 7962 (2020). https://doi.org/10.1038/s41598-020-64819-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-64819-2
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.