Abstract
Largely removed from anthropogenic delivery of nitrogen (N), Antarctica has notably low levels of nitrogen. Though our understanding of biological sources of ammonia have been elucidated, the microbial drivers of nitrate (NO3−) cycling in coastal Antarctica remains poorly understood. Here, we explore microbial N cycling in coastal Antarctica, unraveling the biological origin of NO3− via oxygen isotopes in soil and lake sediment, and through the reconstruction of 1968 metagenome-assembled genomes from 29 microbial phyla. Our analysis reveals the metabolic potential for microbial N2 fixation, nitrification, and denitrification, but not for anaerobic ammonium oxidation, signifying a unique microbial N-cycling dynamic. We identify the predominance of complete ammonia oxidizing (comammox) Nitrospira, capable of performing the entire nitrification process. Their adaptive strategies to the Antarctic environment likely include synthesis of trehalose for cold stress, high substrate affinity for resource utilization, and alternate metabolic pathways for nutrient-scarce conditions. We confirm the significant role of comammox Nitrospira in the autotrophic, nitrification process via 13C-DNA-based stable isotope probing. This research highlights the crucial contribution of nitrification to the N budget in coastal Antarctica, identifying comammox Nitrospira clade B as a nitrification driver.
Similar content being viewed by others
Introduction
Antarctica, a region largely insulated from anthropogenic nitrogen (N) deposition, exhibits notably low N concentrations1. These diminished N levels play an integral role in sustaining ecological equilibrium in the continent’s barren, ice-free terrains2,3. Consequently, elucidating the intricacies of microbial N-cycling pathways in Antarctic ecosystems—including diazotrophy (N2 fixation), nitrification, anaerobic ammonium oxidation (anammox), and denitrification—is paramount4. Diazotrophic Cyanobacteria are the primary producers of biologically accessible ammonium (NH4+) in these environments5. Alongside NH4+, nitrate (NO3−) constitutes a significant inorganic N pool in polar biomes6,7. The synthesis of NO3− in Antarctic ecosystems can be attributed to both abiotic atmospheric deposition, particularly within mineral-rich Antarctic soils8, and biotic nitrification processes. Still, the discrete contributions of these sources to the coastal Antarctic NO3− reservoir remain inadequately characterized, necessitating further investigation.
Nitrification plays a pivotal role in the global biogeochemical N cycle9 and contributes significantly to the emissions of nitrous oxide (N2O)10, a potent greenhouse gas. The process of nitrification encompasses the oxidation of ammonia (NH3) to NO3− via the intermediate nitrite (NO2−). Traditionally, these two metabolic steps have been attributed to distinct microbial guilds: ammonia-oxidizing bacteria (AOB)11 and archaea (AOA)12, responsible for converting NH3 to NO2−, and nitrite-oxidizing bacteria (NOB)13, which facilitate the subsequent oxidation of NO2− to NO3−. Beyond these canonical nitrifiers, the recently identified complete ammonia oxidizers (comammox) within the genus Nitrospira, specifically lineage II, perform full nitrification on its own14,15. These organisms are increasingly recognized for their significant role in nitrification across diverse ecosystems, including both engineered systems16 and natural environments17. The contribution of comammox bacteria to nitrification and subsequent NO3− production in Antarctic ecosystems, however, remains an enigma.
Phylogenetically, comammox Nitrospira cluster into clades A and B14, distinguished by their respective ammonia monooxygenase genes (amoA). Physiological assessments of clade A comammox Nitrospira have revealed an exceptionally high NH3 affinity, suggestive of an oligotrophic niche adaptation18,19. In stark contrast, clade B comammox Nitrospira, despite their widespread distribution in various habitats, including those characterized by low temperatures such as the Tibetan Plateau20,21 and Arctic permafrost22, have yet to be cultured for physiological scrutiny.
This study aims to unravel the complexities of the microbial N cycle in the ice-free regions of East Antarctica, identifying the primary sources of NO3− and examining the potential roles of microorganisms in the nitrification process. Our research is strategically centered on the Larsemann Hills (LH)23, a vast ice-free rocky landscape in East Antarctica, dotted with over a hundred oligotrophic lakes shrouded in ice. Through a comprehensive approach, we discover that (i) the origin of NO3− is primarily the biological nitrification process; (ii) the microbial N cycle is distinct, encompassing most microbial N-cycling processes except for the anammox pathway; and (iii) the comammox Nitrospira clade B serves as an abundant and active driver of nitrification, possessing various survival strategies against the cold and oligotrophic conditions of the coastal Antarctic environment.
Results and discussion
Isotopic signatures indicate the biological origin of nitrate
Depending on how NO3− is produced, the composition of its oxygen isotopes differs, allowing differentiation between abiotic and biotic sources24. For biologically produced NO3−, one oxygen atom (O) is expected from atmospheric oxygen (O2) and two from the surrounding water (H2O)24. Biologically produced NO3− would have a δ18O of ~0.6‰, since δ18O of atmospheric O2 is 23.9‰25 and the measured δ18O of Larsemann Hills (LH) lake water is –12.7 ± 1.5‰ (Supplementary Table 1). In addition, while biologically produced NO3− has a Δ17O of 0‰, which is identical to that of O2 and H2O, atmospheric NO3− is usually characterized by high oxygen isotopic ratios26,27 and can reach Δ17O ≥ 35‰ in the atmosphere in Antarctica26. Atmospheric NO3− in Antarctica is mainly produced by the reaction of nitrogen oxides (NOx), ozone (O3), and hydroxyl radicals (OH·)27.
The mean δ18O of NO3− in sediments from LH lake is 4.6‰. The annual mean δ18O of NO3− in snow and the atmosphere at the coastal Zhongshan station, situated in LH, is approximately 92‰28. From these, the contributions of atmospheric deposition and biological production to sedimentary NO3− pools can be quantified by isotope mass balancing, indicating that the nitrification process constitutes 95% of the NO3− in sediments. In addition, limited data of sediment NO3− Δ17O produced a mean of ~1.3‰, which strengthens the observed >90% contribution of nitrification considering the very high Δ17O of atmospheric NO3− at Zhongshan station (~32‰)28. Similarly, the isotope mass balance of δ18O and Δ17O suggested ~96% of soil NO3− in the two study areas is from nitrification. Thus, we demonstrated here that sediment and soil NO3− predominantly originated from biological production in and nearby the investigated lakes.
Unique microbial nitrogen cycle in coastal Antarctica
The primary production of NO3− is largely attributed to the microbial nitrification process, rather than atmospheric precipitation. Consequently, the origin of NH4+, the substrate for nitrification, can be traced back to biological N2 fixation, as well as mineralization. The qPCR-based quantification of functional N-cycling genes (Fig. 1) revealed a comparable amount of N2 fixation (nifH, encoding nitrogenase) to nitrification genes (amoA and nxrB, encoding ammonia monooxygenase subunit A and nitrite oxidoreductase subunit B, respectively), with quantities ranging from 102 to 104 copies ng−1 DNA. The relatively low quantity of the gene encoding hydroxylamine dehydrogenase (hao) (<10 copies ng−1 DNA) can be attributed to the low coverage of the applied primers (targeting AOB rather than comammox Nitrospira, Supplementary Table 2) and the absence of the bacterial hao gene in the genomes of AOA29, which are of notable abundance in the studied samples.
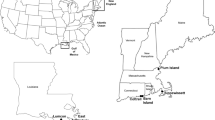
a A photograph illustrating the location of the Larsemann Hills (LH) and South Victoria Land (SVL) (left) in Antarctica, locations of studied surface soil and lake sediments in LH (center), and locations of surface soil samples in SVL (right). Red and green circles represent soil and sediment samples, respectively. Solid circles denote samples that have undergone metagenomic analysis, while empty circles denote samples that have been subjected solely to quantitative PCR analysis. The asterisk indicates samples that were used exclusively for metagenomic analysis and did not undergo comprehensive N-cycling functional gene screening. b–d Abundance of functional microbial N-cycling genes in the LH soils (b), LH lake sediments (c), and SVL soils (d). The studied functional N-cycling genes encompass N2 fixation (nifH), nitrification (amoA, hao, and nxrB), denitrification (napA, narG, nirS/K, norB, and nosZ); dissimilatory nitrate reduction to ammonium (DNRA; nrfA), assimilatory nitrite reduction (ANR; nasA and nirA), and anaerobic ammonium oxidation (anammox; hzo). The relative abundance of the associated functional genes is indicated by the line thickness. Source data are provided as a Source Data file.
In addition to N2 fixation and nitrification processes, we also observed the significant metabolic potential of denitrifiers, as indicated by the high abundances (>103 copies ng−1 DNA) of genes encoding the sequential reduction of NO3− (nar, nap), NO2− (nir), nitric oxide (NO, nor), and N2O (nos) (Fig. 1). The relatively higher abundances (>104 copies ng−1 DNA) of nirS and nirK genes (Fig. 1) can likely be attributed to their presence not only in denitrifiers but also in nitrifiers. Interestingly, we found substantial quantities (102 to 104 copies ng−1 DNA) of the functional gene for dissimilatory nitrate reduction to ammonium (DNRA, nrfA) and assimilatory nitrite reduction (ANR, nasA) (Fig. 1). The potential for DNRA and ANR is widespread among phylogenetically diverse microorganisms and these processes ensure the retention of bioavailable inorganic N in an ecosystem.
Strikingly, the hzo gene (encoding hydrazine oxidoreductase), a biomarker for the anammox process, was not detected in any of the tested sediment and soil samples. Anammox bacteria catalyze the anaerobic oxidation of NH4+ using NO2− as an electron acceptor, producing N2 as the final product. These bacteria were first identified in wastewater treatment systems30 and were subsequently discovered in various environments, including marine31, coastal32, terrestrial33, and engineering systems34. The absence of anammox functional markers in coastal Antarctica suggests unique microbial N-cycling properties in this remote region.
Diverse microbiomes and the microbial N-cycling processes
From an extensive dataset exceeding 450 gigabases of sequencing data, we managed to reconstruct a dereplicated collection of 724 high-quality and 1244 medium-quality35 metagenome-assembled genomes (MAGs) (Supplementary Data 1). The recovered genomes encompass 29 distinct phyla (Supplementary Data 1 and 2), marking, to the best of our knowledge, the most comprehensive inventory of Antarctica coastal soil and sediment genomes to date. On average, we obtained around 60 MAGs per sample site studied (Fig. 2a). The most abundant phyla included Actinobacteriota, Pseudomonadota, Bacteroidota, Chloroflexota, Verrucomicrobiota, Acidobacteriota, Patescibacteria, Planctomycetota, and Gemmatimonadota (Fig. 2a and Supplementary Data 1), aligning with findings from other Antarctic surveys5,36,37. Cyanobacteria also featured among the top ten most abundant MAGs (Supplementary Data 1), a deviation from the Mackay Glacier Region, where they were largely absent in most soil samples37. We identified only five archaeal MAGs within the Thermoproteota phylum, all of which were further classified as ammonia-oxidizing archaea (AOA) within Group I.1b.
a Overview of the microbial community composition derived from metagenome-assembled genomes (MAGs) extracted from six soil and six sediment samples in the Larsemann Hills (LH), as well as two soil samples from South Victoria Land (SVL). The statistical box in the top right corner presents the numerical distribution of the MAGs retrieved from the studied samples (Co-assemblies are not included in this statistic). b Distribution of selected key functional genes in the obtained MAGs, including those involved in respiration, N-cycling, carbon fixation, and trace gas metabolism. The adjacent heatmap displays the distribution of these genes across a set of 1968 MAGs, comprising 29 phyla. Summary and statistics of the presence/absence of key metabolic marker genes in the obtained MAGs are provided in Supplementary Data 1 and 2. Source data are provided as a Source Data file.
To comprehend the metabolic strategies that sustain the abundant bacterial life in these extremely nutrient-poor sediments and soils, we examined the distribution and affiliation of 52 marker genes conserved across different energy conservation and carbon acquisition pathways37 in the MAGs we retrieved. As predicted, genes for aerobic respiration were encoded by nearly all community members (Fig. 2b and Supplementary Fig. 1). Consistent with observations from the Mackay Glacier Region37, a significant number of the MAGs appear to fix carbon via the Calvin-Benson-Bassham (CBB) cycle (212 MAGs) or the 3-hydroxypropionate cycle (345 MAGs) (Fig. 2b). These mechanisms facilitate the generation of biomass through means that are either separate from or complementary to photoautotrophy, a role predominantly carried out by Cyanobacteria (Supplementary Fig. 1). Genomic analysis revealed that the most abundant and widespread community members encoded trace gas oxidation genes. In a pattern similar to previous discoveries37,38, carbon monoxide (CO) dehydrogenases (CoxL) were exclusive to Actinobacteriota and Chloroflexota (Fig. 2b). Interestingly, uptake [NiFe]hydrogenases were encoded by MAGs from Acidobacteriota, Chloroflexota, and Verrucomicrobiota (Fig. 2b), which aligns with observations from temperate soil where Acidobacteriota are known to be active atmospheric H2 consumers39, albeit with slight differences.
In the realm of N metabolism, a comprehensive array of functional genes was identified within the MAGs we analyzed (Fig. 2b). N2 fixation appears to be predominantly carried out by taxa within Acidobacteriota, Cyanobacteria, Desulfobacterota, Myxococcota, and Verrucomicrobiota (Fig. 2b). Consistent with the quantitative analyses, the hzs gene, indicative of the anammox process, was not present in the retrieved MAGs. While a total of 96 Planctomycetota MAGs were obtained, subsequent verification through taxonomic classification confirmed that none of these MAGs belong to the anammox family Brocadiaceae (Fig. 2b and Supplementary Data 2). The absence of anammox process may be attributed to the optimal temperature range of 12–17 °C, which is characteristic of anammox process found in similarly cold Arctic fjord sediments40, suggesting a limited tolerance to low temperatures. This assertion is further reinforced by the absence of anammox bacteria in the McMurdo Dry Valleys (an Antarctic desert)41 and in Arctic regions42 as well. The pervasive distribution of denitrification genes across nearly all phyla underscores the robust and widespread microbial denitrification activity in Antarctica, corroborating previous research that highlights the ubiquity of cold-adapted denitrifiers across diverse Antarctic ecosystems43. Regarding the nitrification process, the marker genes-amoA (associated with ammonia oxidation) and nxrB (linked to nitrite oxidation) were found exclusively in the Thermoproteota and Nitrospirota phyla (Fig. 2b). Although Antarctic soil nitrification was previously reported back in 199744, attributed to AOB genera Nitrosospira and Nitrosomonas, as well as AOA within group I.1b45,46,47,48, no investigation has yet been conducted into the presence and function of the recently discovered comammox bacteria within the Nitrospira genus14,15. Here, we present data showing the prevalence of the comammox Nitrospira amoA gene in Antarctica lake sediments and soils, strongly suggesting the activity of comammox Nitrospira in this region.
Abundant nitrification drivers in coastal Antarctica
The quantitative evaluation of nitrification-related functional genes, including amoA and nxrB (Supplementary Fig. 2), established that AOA and comammox bacteria were the predominant nitrifiers in the analyzed soils and sediments (Supplementary Fig. 3). Our metagenomic analysis corroborates these findings, having identified only AOA and Nitrospira MAGs among the various nitrifying groups (Fig. 2b), suggesting they outnumber AOB. Furthermore, a more detailed analysis of the abundance and community structure of all identified nitrifying groups was conducted using amplicon amoA and nxrB sequencing. Subsequent phylogenetic assessment revealed an unexpected dominance of clade B comammox Nitrospira in this environment (Supplementary Table 5 and Supplementary Fig. 8). This was surprising given the successful extraction of diversity information from AOA (Supplementary Fig. 6) and AOB (Supplementary Fig. 7) in the samples investigated (Supplementary Results and Discussion).
Among the dereplicated MAGs, five were classified as Group I.1b-AOA within the genera Nitrosocosmicus and Nitrospharea of the Thermoproteota phylum. This categorization stemmed from GTDB taxonomic classification, phylogenomic assessments, and average nucleotide identity (ANI) analyses (Supplementary Data 2 and Supplementary Fig. 4). These findings are in agreement with the data obtained from amoA amplicon sequencing, as discussed in the Supplementary Results and Discussion section. Together with their previous detection in Arctic soils49,50, this suggests a capacity of Nitrosocosmicus-like AOA to thrive in cold and nutrient-deficient conditions.
Of the seven Nitrospirota MAGs analyzed, four were verified as comammox Nitrospira, and the remaining three belonged to the exclusively nitrite-oxidizing lineages II and IV of Nitrospira. This classification was substantiated by phylogenomic and average nucleotide identity (ANI) analyses (Fig. 3a, b, Supplementary Table 4, and Supplementary Fig. 5), and these results concur with the phylogenetic assessment based on the amoA and nxrB gene of Nitrospira (Supplementary Figs. 8,9). The notably high abundance of these Nitrospira MAGs in the soil and sediment metagenomes studied (reaching up to 0.173%, Supplementary Table 3) further emphasizes their significant roles in nitrification within these ecosystems.
a Maximum-likelihood phylogenomic tree of the genus Nitrospira, constructed using a concatenated alignment of 91 single-copy core genes. b Average nucleotide identity (ANI) analysis of selected clade B comammox Nitrospira genomes. c Comparative cellular metabolic diagram for MAGs Nitrospira sp. La1-X1 and Nitrospira sp. La3-X1. Selected features are depicted, including core pathways of chemolithoautotrophic ammonia and nitrite oxidation, alternative energy metabolism, and potential detoxification pathways. Colors denote the presence of specific genes in La1-X1 (orange), La3-X1 (blue), or both (gray). Asterisks represent incomplete features. Key enzymes and pathways are abbreviated as follows: AMO ammonia monooxygenase, CAT catalase, CLD chlorite dismutase, FDH formate dehydrogenase, HAO hydroxylamine dehydrogenase, HYD 3b [NiFe] hydrogenase, MCP methyl-accepting protein, MSP methionine salvage pathway, NHase nitrile hydratase, NirK Cu-dependent nitrite reductase, NQR Na+-translocating NADH: ubiquinone oxidoreductase, NXR nitrite oxidoreductase, SOD superoxide dismutase. Enzyme complexes of the respiratory chains are labeled using Roman numerals. d Distribution of key metabolic features involved in N and alternative energy metabolism, the methionine salvage pathway, superoxide dismutase, and trehalose synthesis. In total, 19 Nitrospira genomes, including the two high-quality clade B comammox genomes (La1-X1 and La3-X1) obtained in this study, were analyzed. Features shown in white were not detected. Source data are provided as a Source Data file.
Two high-quality comammox MAGs, Nitrospira sp. La1-X1 and Nitrospira sp. La3-X1, have genome sizes of 4.26 Mb and 3.78 Mb, respectively (Supplementary Table 3). The amoA gene sequences from La1-X1 and La3-X1 align closely with the dominant amoA OTUs and with previously published sequences from clade B comammox (Fig. 4c and Supplementary Fig. 8). Consistent with the amoA-based phylogeny, phylogenomic analysis also demonstrates a distinct clustering of these comammox MAGs within clade B (Fig. 3a). For La1-X1, the ANI was highest with MAG Nitrospira sp. palsa1310, which was identified from Arctic permafrost soil22. The ANI between these genomes is 96.67% (Fig. 3b), surpassing the species threshold of 95%51, suggesting that these genomes represent different strains of the same Nitrospira species, potentially adapted to cold environments. In contrast, La3-X1 is grouped within a clade B subset that includes MAGs from drinking water treatment systems and glacier surface soil52, but it did not share high ANI values with any other clade B genomes (≤85%, Fig. 3b). This suggests that La3-X1 represents a unique so far undetected group within clade B.
a The concentration of produced NO3− (at 4 °C and 10 °C) and N2O (at 4 °C) after DNA-SIP incubations, using ammonium as substrate and chlorate as a specific inhibitor for comammox Nitrospira (t-test, adjusted P < 0.05, n = 3 biological replicates). The incubation period for 10 and 4 °C incubations is 8 weeks (56 days) and 12 weeks (84 days), respectively. “CK” denotes samples that have not been amended with any substrate or inhibitor. b The quantitative distribution and relative abundance of comammox Nitrospira amoA genes derived from DNA-SIP in 13CO2 and 12CO2-treated microcosms at 4 and 10 °C, respectively. Error bars represent standard errors (n = 3). c Maximum-likelihood phylogenetic tree of retrieved comammox Nitrospira amoA gene sequences from the original sample DNA and 13C-DNA from 4 and 10 °C DNA-SIP incubation samples. The heatmap on the right displays the relative abundance of specific operational taxonomic units (OTUs) in different DNA samples. The additional DNA-SIP data pertaining to the 10 °C incubations of LA2 and LS4, as well as the nxrB gene labeling data for LA1, are presented in the Supplementary Figs. 11–13.
Metabolic potential and surviving strategy
The clade B comammox MAGs Nitrospira sp. La1-X1 and Nitrospira La3-X1 harbor the complete genetic machinery for NH3 and NO2− oxidation, the respiratory chain, and the reduced tricarboxylic acid (rTCA) cycle, which is the conserved CO2 fixation pathway in Nitrospira (Fig. 3c, d). These core metabolic features are highly conserved in comammox Nitrospira, as reported in previous studies53,54. Similar to other Nitrospira genomes52, La1-X1 and La3-X1 do not encode nitric oxide reductase (NOR), which is crucial for enzymatic N2O production55. They do carry genes for urea transport and hydrolysis by the urease, indicating the use of urea as an alternative ammonia source, as shown for other comammox Nitrospira14,15,56. In addition, like some Nitrospira that have the confirmed ability to use hydrogen and formate as alternative energy sources57,58, the comammox MAGs La1-X1 and La3-X1 contain genes for formate and hydrogen oxidation. The formate dehydrogenases of comammox are similar to their nitrite-oxidizing counterparts. In contrast, comammox bacteria possess a 3b-type [NiFe] hydrogenase, which is rarely identified in canonical Nitrospira and its physiological role remains unclear54,59. While the capacity for formate oxidation is widely distributed in clade B, the 3b-type [NiFe], hydrogenase has been mainly identified in clade A comammox species54 (Fig. 3d). However, the presence of this hydrogenase type in a few clade B genomes52,54 including La3-X1 challenges the clade specificity of this feature.
In addition to the canonical F1F0 H+-driven ATPase, La1-X1 and La3-X1 encode a potentially Na+-pumping F1F0 ATPase, previously detected in the haloalkalitolerant nitrite-oxidizing Ca. Nitrospira alkalitolerans60, marine nitrite-oxidizing Ca. Nitronereus thalassa61 and clade A comammox Ca. Nitrospira kreftii19. Similar to these genomes, La3-X1 also possesses a Na+-translocating NADH:ubiquinone oxidoreductase (NQR), a feature missing in La1-X1 (Fig. 3c). These Na+-pumping enzyme complexes might represent an adaption of La3-X1 to saline or haloalkine conditions similar to other Nitrospira.
The disaccharide trehalose is one of several solutes known to protect bacteria against cold stress, and may also protect against other harmful environmental conditions, such as osmotic stress62. La1-X1 possesses a trehalose-6-phosphate synthase (OtsA) and the corresponding phosphatase (OtsB) to potentially produce the non-reducing disaccharide in two steps from UDP-glucose, as shown for E. coli63. In contrast, La3-X1 encodes several other trehalose synthesis enzymes in one gene cluster, indicating that this species might convert NDP-glucose (trehalose synthase) and maltose (trehalose synthase/amylase) to trehalose. However, all these trehalose synthesis pathways have also been identified in Nitrospira from mesophilic environments and might thus not be a distinguishing feature for cold adaptation. In addition to trehalose synthesis, La1-X1 and La3-X1 possess other features for coping with different stresses. Whereas a gene cluster encoding a nitrile hydratase has been detected in La1-X1 and La3-X1, the genomes lack an amidase to degrade the produced amides further to the corresponding carboxylic acid and ammonia, which could serve as an energy source. An example of a potential amidase is a putative formamidase identified in a clade B MAG obtained from the Rifle aquifer52. Other detoxification mechanisms include catalases and Fe or Mn superoxide dismutases (SOD) for reactive oxygen defense identified in both clade B genomes, and a periplasmic Cu-Zn SOD, which is present in La3-X1 and many other clade B genomes, but absent in La1-X1 (Fig. 3c, d).
An adaptation to oligotrophic conditions (Supplementary Table 1) might be the methionine salvage pathway (MSP) of La1-X1 (Fig. 3c). This pathway recycles the sulfur-containing intermediate 5’-methyl-thioadenosine back to methionine, thus allowing the use of reduced sulfur compounds under sulfur limitation. However, similar to the high-quality genome of the nitrite-oxidizing Nitrospira lenta57 and other clade B genomes, the mtnE gene encoding the last step in this pathway is missing in La1-X1. Whether other enzymes might complete the MSP in these Nitrospira species remains to be determined.
Nitrification and N2O production activity
We have demonstrated that comammox Nitrospira potentially serves as crucial drivers of nitrification in coastal East Antarctica, and further investigated their survival strategies from a genomic perspective. Additionally, we confirmed the nitrification activity of comammox Nitrospira using DNA-SIP on two lake sediment samples (LA1 and LA2) and one soil sample (LS4). DNA-SIP incubations were conducted at 4 °C (for LA1) and 10 °C (for LA1, LA2, and LS4), and the production of NO3− served as an indicator of nitrification activity (Fig. 4a and Supplementary Fig. 11). During the incubations, comammox Nitrospira were actively growing, as evidenced by the peak shifts of their DNA in the 13C-treatment (Fig. 4b and Supplementary Fig. 4). Subsequent sequencing of the amoA genes from the labeled DNA revealed clade B amoA OTUs, including those of comammox MAGs La1-X1 and La3-X1, in both the 4 °C and 10 °C incubations (Fig. 4c). Despite the temperature used in the DNA-SIP incubations not being identical to the in situ condition, our data strongly suggest an active role for clade B comammox Nitrospira in nitrification.
The addition of NH4+ resulted in an increase of NO3− production, however, did not stimulate higher N2O production compared to incubations without NH4+ addition (Fig. 4a), indicating the low N2O production potential of comammox Nitrospira55. The addition of chlorate, a potential comammox-specific inhibitor64, slightly reduced the production of NO3− and N2O (Fig. 4a). However, since chlorate can also influence other nitrite-oxidizing Nitrospira species as well as denitrifiers, the observed reductions in NO3− and N2O levels cannot be solely attributed to changes in comammox activity. Notably, prior research has shown the resilience of clade B comammox Nitrospira to freeze-thaw cycles, which underscores the endurance of this particular nitrifier group in environments with low and even subfreezing temperatures65. Raising the temperature (10 °C) stimulated the activity of AOA (in LA2) and AOB (in LA2 and LS4) (Supplementary Fig. 4, Supplementary Results and Discussion), indicating that these nitrifiers are more competitive under elevated temperature conditions, an important finding to predict ecosystem changes in response to global warming.
Implications and outlook
In this study, we elucidated the unique microbial N cycle in pristine and oligotrophic coastal Antarctic soil and lake sediments, identifying the microbial nitrification process as the primary pathway for NO3− production. Our metagenomic and quantitative functional gene-targeted analyses both revealed clade B comammox Nitrospira to be a key nitrifier in these environments. This finding not only expands our understanding of microbial diversity but also underscores the pivotal role of specific microbial groups in biogeochemical cycling in extreme environments. Our results also revealed fascinating patterns of niche differentiation between clade A and B comammox Nitrospira and canonical nitrifiers. Clade B species might have superior affinity for their substrate ammonia, and are potentially best adapted to survive and thrive in cold and oligotrophic environments. This niche differentiation could have significant implications for N-cycling in cold environments and it will be important to determine how the nitrifier and overall microbial communities will react to global warming.
In addition, we have successfully obtained in total of 1968 MAGs, significantly expanding the existing microbial genomic data for Antarctica. These MAGs provide invaluable insights into the microbial diversity and their metabolic capabilities in the extreme and unique environment of coastal Antarctica. The availability of these MAGs will undoubtedly facilitate further research into the intricate relationships between microbial communities, biogeochemical cycles, and climate change in polar regions and beyond.
This study underscores the importance of understanding the unique microbial N cycle in coastal Antarctica. We have uncovered clade B comammox Nitrospira to be a nitrification driver in low-temperature environments, and investigated their survival strategy and potential impact on climate change through the production of greenhouse gas N2O55,66. It is crucial to further investigate how comammox Nitrospira evolve and survive in Antarctic ecosystems, meeting one of the six priorities for Antarctic science67. Recognizing the climatological significance of N-cycling and biogeochemical processes, future research should continue to monitor these microbes, as they could hold the key to understanding the broader implications of microbial activity on our planet’s climate.
Methods
Sample collection and treatment
Samples were collected in Larsemann Hills (LH), the second-largest ice-free land in East Antarctica with an area of ~50 km2. LH has a cold and dry continental climate, with an annual mean temperature of ~−10 oC and the temperatures occasionally above 0 oC in summer68 lead to this region typically free of snow cover in summer. As a result of seasonal snow cover and the glacier melting, a number of landlocked lakes are developed in this region. Surface sediment (the upper ~5 cm) and surface water samples were collected from six lakes (LA1-LA6) in LHs in February 2020 (Fig. 1a). The surface sediment samples were collected using a stainless-steel spade on the shore of lakes, with typical water depths of ~150–200 cm. Approximately 1 L of surface water near the lake shore was also sampled with clean polyethylene (PE) bottles, a portion of which was used to measure temperature, pH, salinity, and conductivity utilizing a multi-probe water quality meter (YSI Professional Plus series; Supplementary Table 1). The remainder of the water samples was filtered using 0.22-μm polytetrafluoroethene (PTFE) filters for chemical analyses. Additionally, a surface soil sample (the top ~5 cm) was collected near each lake, ~100 m from the lake shore, using a clean stainless spatula. At each sampling site, larger gravels were removed firstly, and five soil sub-samples (four corners and the center of a square) were collected at a distance of 5–10 m and then mixed to obtain a representative sample (LS1-LS6). For a comprehensive understanding of N-cycling processes in the ice-free areas in coastal Antarctica, surface soil samples (SVL1–SVL10) were also collected in February 2022 at Inexpressible Island, South Victoria Land where the climate is comparable to that of LH, following the same sampling protocols. All of the sediment and soil samples were stored in sealed PE bags, and all samples were transported to the laboratory at temperatures of ~−20 °C for subsequent analysis.
Physiochemical analysis
In the laboratory, ~50 g of the sediment and soil samples were freeze-dried in 50-mL clean centrifuge tubes (ALPHA 1-4/LD, Martin Christ Inc.). After drying, the samples were homogenized using an agate mortar and pestle, and subsequently passed through a 1 mm sieve for further chemical analysis. For determining total organic carbon (TOC) content, approximately 5 g of samples were digested with 10% HCl (v/v) to remove carbonate. Then, TOC was measured with an automatic element analyser (Elementar, VARIO EL III), with acetanilide used as the external standard. The detection limit (DL) of the TOC was estimated to be ~0.005%. All samples were measured in triplicate, yielding a relative standard deviation (1σ) of <10% for each sample. For chemical ion analysis, ~5 g of freeze-dried samples were placed in sterile 50-mL centrifuge tubes and suspended in 25 mL Milli-Q water (18.2 MΩ). The solution was then ultrasonicated for 40 min. The supernatant was first centrifuged for 15 min at 3000×g, and then filtered through 0.22-μm PTFE filters for nutrient determination. Nutrient concentrations (NH4+, NO3−, and PO43−) in the sediment, soil extracts, and lake water samples were determined using an Aquion RFIC ion chromatograph (IC, Thermo Scientific, USA), equipped with the analytical columns CS12A (2 × 250 mm), AS11-HC (2 × 250 mm), methanesulfonic acid (MSA), and potassium hydroxide (KOH) as eluents for cations and anions, respectively. In addition, the concentrations of SiO32- were determined using an automated QuAAtro™ nutrient analyser (Seal Analytical Ltd., UK). During sample analysis, replicate determinations (n = 5) were performed, and 1σ for all species was <5%.
Isotopic analysis of nitrate
The isotopic composition of NO3− was determined using the bacterial denitrifier method at the Environmental Stable Isotope Laboratory of East China Normal University (ECNU-ESIL). Briefly, the denitrifying bacterium Pseudomonas aureofaciens, which lacks the N2O reductase enzyme, quantitatively transforms NO3− into gaseous N2O69,70. The δ15N and δ18O of the generated N2O were measured in duplicates using isotope-ratio mass spectrometry (IRMS, Thermo Scientific Delta V). The Δ17O of NO3− was separately analyzed through the thermal decomposition of N2O into N2 and O271, followed by measurements at m/z 32, 33, and 34 on the IRMS. The pooled standard deviation (1σp) was employed to ascertain the measurement precision of the overall denitrifier method72,73. The 1σp of all duplicate samples executed in at least two different batches was 0.6‰ for δ15N (n = 10), 0.3‰ for δ18O (n = 10), and 0.8‰ for Δ17O (n = 8). However, due to the limited amounts of NO3− in the samples, only three sediment and five surface soil samples were analyzed for Δ17O of NO3−.
In addition, lake water stable isotopes (δ18O and δ2H) were analyzed using laser absorption spectrometry (TIWA-45EP, Los Gatos Research, Inc.). To ensure quality control, replicate analyses (n = 5) were performed, yielding relative standard deviations of 0.05‰ and 0.2‰ for δ18O and δ2H, respectively.
DNA extraction, quantification, and sequencing analysis
Total DNA was extracted from 0.5 g sediment/soil samples using the Fast DNA SPIN kit (MP Biomedicals, Santa Ana, CA) according to the manufacturer’s protocols. The final DNA quality and quantity were determined using Quant-iT PicoGreen dsDNA Assay Kit (Thermo Fisher Scientific, China). A detailed description of the quantitative PCR (qPCR) analysis for functional N-cycling genes is provided in the Supplementary Methods section. It also elaborates on the procedures for PCR and high-throughput amplicon sequencing, as well as the subsequent phylogenetic analysis of nitrification genes.
Metagenomic sequencing
The total DNA from the original sediment and soil samples was sequenced on the Illumina HiSeq X ten platform using a 150-bp paired-end library at Beijing Novogene Biotech Co., Ltd. (Beijing, China). The NEXTFLEX Rapid DNA-Seq Library Prep Kit 2.0 (Bioo Scientific, Austin, TX, USA) was used for DNA library preparation with an insert size of ~300 bp according to the manufacturer’s recommendations. DNA was sheared using a Covaris S220 Focused Ultrasonicator to create 150 bp fragments. Subsequently, raw metagenomic sequencing data in FastQ format was generated using the RTA (Real-Time Analysis) v3.4.4 and bcl2fastq (btq) v2.16. Each individual sample yielded a range of 20–50 gigabases of sequencing data.
Assembly and binning of metagenomes
Raw reads were processed using fastp v0.19.774 for adapter trimming, quality filtering, and per-read quality trimming. The fourteen quality-controlled metagenomes were individually assembled and co-assembled using MEGAHIT v1.1.375 with default parameters (k-mers: 21, 29, 39, 59, 79, 99, 119, 141). Each of the assembly was initially binned using the binning module (–metabat2 –maxbin2 –concoct; –metabat2 for co-assembly) in the metaWRAP pipeline76 v1.3.2, and were consolidated using DAS Tool v1.1.277 with default parameters. After a dereplication check using dRep v3.0.078 (-comp 50 -con 10) and completeness and contamination evaluation using CheckM v1.1.379, 1968 MAGs were obtained, and the taxonomy of each MAG was assigned using GTDB-Tk v1.5.080 with the Genome Taxonomy Database (Release 06-RS202). In the process of annotating metabolic functions, the genomes that were extracted were examined using DIAMOND81 v0.9.14 against 52 tailor-made protein databases, which consisted of marker genes representing common metabolic traits for energy conservation as well as C and N fixation37. To confirm the existence of crucial metabolic genes in the MAGs, maximum-likelihood phylogenetic trees were generated to confirm their phylogenetic affiliation The relative abundances (in percentage) for all dereplicated MAGs were determined by aligning each sample’s clean paired-end reads to the MAGs utilizing CoverM v0.6.1 (https://github.com/wwood/CoverM) in genome mode, applying the default configurations. Moreover, the trimmed reads were incorporated into CLC Genomics Workbench version 20.0 (CLCBio, Qiagen, Germany), and the de novo assembly algorithm of CLC was employed to search for amoA and nxrB sequences. Contigs that contained either amoA or nxrB genes were selected for phylogenetic analysis, in conjunction with amplicon sequences.
Phylogenomic analysis and genome annotation
Genomes and MAGs classified by the GTDB-Tk database R20282 as Nitrospiracea, with estimated genome completeness ≥70% and contamination ≤10%, were downloaded from NCBI. Dereplication was performed using the drep v2.4.278 dereplicate workflow with cut-offs for estimated genome completion ≥70% and contamination ≤10%, but otherwise default settings. In addition to these 95 Nitrospiracea genomes, seven Nitrospira lineage IV and two clade B genomes were included in the phylogenetic analysis using the UBCG pipeline for the extraction and concatenated alignment of 91 single-copy core genes83. In addition, two Leptospirillum genomes (GCF_000284315.1, GCF_000299235.1) were included as outgroup. A maximum-likelihood phylogenetic tree was constructed using IQ-TREE v1.6.1284 with 1000 ultrafast bootstrap replications and the GTR + F + I + G4 model identified by the implemented Modelfinder85. ANI analysis of all clade B genomes was performed using the OrthoANI86. Gene calling and automatic genome annotation were performed using the MicroScope platform87, and annotations of selected features were manually checked and refined. For analyzing the distribution patterns of selected key features, 17 comammox clade B genomes were annotated using prokka v.1.14.688 with the “--gcode 11” and “--metagenome” options to obtain all protein sequences for generating a BLAST database. The distribution of selected key proteins within clade B was analyzed by conducting a BLASTp search against this database with default settings, except for an e-value cutoff of 1e-6. Only hits with an identity ≥35% (pident) and a query coverage ≥80% (qcovs) were reported as present. Phylogenomic and ANI analyses were carried out on the retrieved AOA MAGs, using representative genomes (which include Group I.1a and Group I.1b-AOA) and Group I.1b Nitrosocosmicus AOA genomes as references, respectively.
DNA-stable isotope probing (SIP) incubation and analysis
Lake sediments (LA1 and LA2) and the soil sample (LS4) were selected for DNA-SIP microcosm incubation experiments (Supplementary Fig. 10) to mimic a low-temperature oligotrophic environment. (Note that these three sampling locations are distributed almost uniformly throughout LH.) Microcosms were constructed in 120 mL serum bottles containing 10 g sediments or soil at 60% of maximum water-holding capacity and were incubated for 56 days at 10 °C in the dark. For each sample, two different treatments were established in triplicate microcosms. The 13CO2 microcosms were amended with 5% (v/v) 13CO2 (99 atom%; Sigma-Aldrich Co., St. Louis, MO, USA) plus approximately 5 µg 15N-NH4Cl-N g−1 dry weight soil/sediment (d.w.s.), while the 12CO2 control treatments received 5% (v/v) 12CO2 plus ~5 µg 14N-NH4Cl-N g−1 d.w.s. The water content was restored weekly over an 8-week incubation period by opening the bottles. Additional labeled or unlabeled NH4Cl was supplied (approximately once every 2–3 weeks) to maintain ~5 µg NH4Cl-N g−1 d.w.s. during the incubations. The sediment/soil samples were destructively sampled after 56 days of incubation and transferred immediately to –80 °C for subsequent molecular analysis. The remaining ~2 g of sediments were used for end-point quantification of NH4+, NO2−, and NO3− concentrations. The LA1 sediment sample was additionally chosen for incubation at 4 °C, adhering to the same procedure previously outlined for a period of 84 days (12 weeks). In addition to the ammonium substrate, 50 μM chlorate, a specific comammox inhibitor64, was introduced to study the activity of comammox Nitrospira. Moreover, the production of N2O64 was actively monitored during these 4 °C DNA-SIP incubations. The fractionation of DNA post-DNA-SIP incubations, along with the subsequent quantification analysis of functional nitrification groups, is described in the Supplementary Methods.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All AOA-, AOB-, and comammox Nitrospira-amoA and Nitrospira-nxrB OTU sequences obtained in this study were deposited in GenBank, with accession numbers MZ956347-MZ956585. Retrieved metagenome-assembled genomes (MAGs) have been uploaded to Figshare (https://doi.org/10.6084/m9.figshare.25435465). All raw sequencing reads of amplicon sequencing and metagenomes have been submitted to the National Center for Biotechnology Information (NCBI) under BioProject accession No. PRJNA855145. The database used in this study includes the GTDB database R06-RS202 (https://data.gtdb.ecogenomic.org/releases/release202/). The nitrifiers reference genomes used for comparative genomic analysis were downloaded from the NCBI Refseq database. Source data are provided with this paper.
References
Shi, G., Teng, J., Ma, H., Wang, D. & Li, Y. Metals in topsoil in Larsemann Hills, an ice-free area in East Antarctica: lithological and anthropogenic inputs. Catena 160, 41–49 (2018).
Ortiz, M. et al. Microbial nitrogen cycling in Antarctic soils. Microorganisms 8, 1442 (2020).
Mergelov, N. et al. Hypolithic communities shape soils and organic matter reservoirs in the ice-free landscapes of East Antarctica. Sci. Rep. 10, 10277 (2020).
Okabe, S., Satoh, H. & Watanabe, Y. In situ analysis of nitrifying biofilms as determined by in situ hybridization and the use of microelectrodes. Appl. Environ. Microbiol. 65, 3182–3191 (1999).
Cary, S. C., McDonald, I. R., Barrett, J. E. & Cowan, D. A. On the rocks: the microbiology of Antarctic Dry Valley soils. Nat. Rev. Microbiol. 8, 129–138 (2010).
Shiozaki, T. et al. Biological nitrogen fixation detected under Antarctic sea ice. Nat. Geosci. 13, 729–732 (2020).
Liu, X. et al. Nitrate is an important nitrogen source for Arctic tundra plants. Proc. Natl Acad. Sci. USA 115, 3398–3403 (2018).
Hopkins, D. W. et al. Isotopic evidence for the provenance and turnover of organic carbon by soil microorganisms in the Antarctic dry valleys. Environ. Microbiol. 11, 597–608 (2009).
Kuypers, M. M. M., Marchant, H. K. & Kartal, B. The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 16, 263–276 (2018).
Prosser, J. I., Hink, L., Gubry-Rangin, C. & Nicol, G. W. Nitrous oxide production by ammonia oxidizers: Physiological diversity, niche differentiation and potential mitigation strategies. Glob. Chang. Biol. 26, 103–118 (2020).
Arp, D. J. & Stein, L. Y. Metabolism of inorganic N compounds by ammonia-oxidizing bacteria. Crit. Rev. Biochem. Mol. Biol. 38, 471–495 (2003).
Konneke, M. et al. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437, 543–546 (2005).
Daims, H., Lucker, S. & Wagner, M. A new perspective on microbes formerly known as nitrite-oxidizing bacteria. Trends Microbiol. 24, 699–712 (2016).
Daims, H. et al. Complete nitrification by Nitrospira bacteria. Nature 528, 504–509 (2015).
van Kessel, M. A. et al. Complete nitrification by a single microorganism. Nature 528, 555–559 (2015).
Pinto, A. J. et al. Metagenomic evidence for the presence of comammox Nitrospira-like bacteria in a drinking water system. mSphere 1, e00054–15 (2016).
Sun, D. et al. Distribution and diversity of comammox Nitrospira in coastal wetlands of China. Front. Microbiol. 11, 589268 (2020).
Kits, K. D. et al. Kinetic analysis of a complete nitrifier reveals an oligotrophic lifestyle. Nature 549, 269–272 (2017).
Sakoula, D. et al. Enrichment and physiological characterization of a novel comammox Nitrospira indicates ammonium inhibition of complete nitrification. ISME J. 15, 1010–1024 (2021).
Zhang, S. et al. Ammonia oxidizers in high-elevation rivers of the Qinghai-Tibet Plateau display distinctive distribution patterns. Appl. Environ. Microbiol. 85, e01701–e01719 (2019).
Zhang, S. et al. Ammonia oxidizers in river sediments of the Qinghai-Tibet Plateau and their adaptations to high-elevation conditions. Water Res. 173, 115589 (2020).
Woodcroft, B. J. et al. Genome-centric view of carbon processing in thawing permafrost. Nature 560, 49–54 (2018).
Krishnan, K. P., Sinha, R. K., Krishna, K., Nair, S. & Singh, S. M. Microbially mediated redox transformations of manganese (II) along with some other trace elements: a study from Antarctic lakes. Polar Biol. 32, 1765–1778 (2009).
Granger, J. & Wankel, S. D. Isotopic overprinting of nitrification on denitrification as a ubiquitous and unifying feature of environmental nitrogen cycling. Proc. Natl Acad. Sci. USA 113, E6391–E6400 (2016).
Barkan, E. & Luz, B. High precision measurements of 17O/16O and 18O/16O ratios in H2O. Rapid Commun. Mass Spectrom. 19, 3737–3742 (2005).
Savarino, J., Kaiser, J., Morin, S., Sigman, D. M. & Thiemens, M. H. Nitrogen and oxygen isotopic constraints on the origin of atmospheric nitrate in coastal Antarctica. Atmos. Chem. Phys. 7, 1925–1945 (2007).
Shi, G. et al. Distinguishing summertime atmospheric production of nitrate across the East Antarctic ice sheet. Geochim. Cosmochim. Acta 231, 1–14 (2018).
Shi, G. et al. Isotopic constraints on sources, production, and phase partitioning for nitrate in the atmosphere and snowfall in coastal East Antarctica. Earth Planet. Sci. Lett. 578, 117300 (2022).
Stahl, D. A. & de la Torre, J. R. Physiology and diversity of ammonia-oxidizing archaea. Annu. Rev. Microbiol. 66, 83–101 (2012).
Strous, M. et al. Missing lithotroph identified as new planctomycete. Nature 400, 446–449 (1999).
Kuypers, M. M. et al. Anaerobic ammonium oxidation by anammox bacteria in the Black Sea. Nature 422, 608–611 (2003).
Dalsgaard, T., Canfield, D. E., Petersen, J., Thamdrup, B. & Acuna-Gonzalez, J. N2 production by the anammox reaction in the anoxic water column of Golfo Dulce, Costa Rica. Nature 422, 606–608 (2003).
Yao, Y. et al. Global variations and controlling factors of anammox rates. Glob. Chang. Biol. 29, 3622–3633 (2023).
Kartal, B., Kuenen, J. G. & van Loosdrecht, M. C. Engineering. Sewage treatment with anammox. Science 328, 702–703 (2010).
Bowers, R. M. et al. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat. Biotechnol. 35, 725–731 (2017).
Leung, P. M. et al. Energetic basis of microbial growth and persistence in fesert ecosystems. mSystems 5, e00495–19 (2020).
Ortiz, M. et al. Multiple energy sources and metabolic strategies sustain microbial diversity in Antarctic desert soils. Proc. Natl Acad. Sci. USA 118, e2025322118 (2021).
Ji, M. et al. Atmospheric trace gases support primary production in Antarctic desert surface soil. Nature 552, 400–403 (2017).
Giguere, A. T. et al. Acidobacteria are active and abundant members of diverse atmospheric H2-oxidizing communities detected in temperate soils. ISME J. 15, 363–376 (2021).
Canion, A. et al. Temperature response of denitrification and anaerobic ammonium oxidation rates and microbial community structure in Arctic fjord sediments. Environ. Microbiol. 16, 3331–3344 (2014).
Pascoal, F. et al. The spatial distribution and biogeochemical drivers of nitrogen cycle genes in an Antarctic desert. Front. Microbiol. 13, 927129 (2022).
Wright, K. E., Williamson, C., Grasby, S. E., Spear, J. R. & Templeton, A. S. Metagenomic evidence for sulfur lithotrophy by Epsilonproteobacteria as the major energy source for primary productivity in a sub-aerial arctic glacial deposit, Borup Fiord Pass. Front. Microbiol. 4, 63 (2013).
Cabezas, A. et al. Ubiquity and diversity of cold adapted denitrifying bacteria isolated from diverse Antarctic ecosystems. Front. Microbiol. 13, 827228 (2022).
Wilson, K., Sprent, J. I. & Hopkins, D. W. Nitrification in Antarctic soils. Nature 385, 404–404 (1997).
Hayashi, K. et al. Ammonia oxidation potentials and ammonia oxidizers of Lichen-Moss vegetatedsoils at two ice-free areas in East Antarctica. Microbes Environ. 35, ME19126 (2020).
Magalhaes, C. M., Machado, A., Frank-Fahle, B., Lee, C. K. & Cary, S. C. The ecological dichotomy of ammonia-oxidizing archaea and bacteria in the hyper-arid soils of the Antarctic Dry Valleys. Front. Microbiol. 5, 515 (2014).
Barnard, S. et al. Increased temperatures alter viable microbial biomass, ammonia oxidizing bacteria and extracellular enzymatic activities in Antarctic soils. FEMS Microbiol. Ecol. 96, fiaa065 (2020).
Leininger, S. et al. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442, 806–809 (2006).
Alves, R. J. E. et al. Ammonia oxidation by the Arctic terrestrial Thaumarchaeote Candidatus Nitrosocosmicus arcticus Is stimulated by increasing temperatures. Front. Microbiol. 10, 1571 (2019).
Alves, R. J. et al. Nitrification rates in Arctic soils are associated with functionally distinct populations of ammonia-oxidizing archaea. ISME J. 7, 1620–1631 (2013).
Richter, M. & Rossello-Mora, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl Acad. Sci. USA 106, 19126–19131 (2009).
Poghosyan, L. et al. Metagenomic recovery of two distinct comammox Nitrospira from the terrestrial subsurface. Environ. Microbiol. 21, 3627–3637 (2019).
Koch, H., van Kessel, M. & Lucker, S. Complete nitrification: insights into the ecophysiology of comammox Nitrospira. Appl. Microbiol. Biotechnol. 103, 177–189 (2019).
Palomo, A. et al. Comparative genomics sheds light on niche differentiation and the evolutionary history of comammox Nitrospira. ISME J. 12, 1779–1793 (2018).
Kits, K. D. et al. Low yield and abiotic origin of N2O formed by the complete nitrifier Nitrospira inopinata. Nat. Commun. 10, 1836 (2019).
Tang, X., Li, Y., Liu, M., Hou, L. & Han, P. Abundance, diversity and physiological preferences of comammox Nitrospira in urban groundwater. Sci. Total. Environ. 904, 167333 (2023).
Koch, H. et al. Expanded metabolic versatility of ubiquitous nitrite-oxidizing bacteria from the genus Nitrospira. Proc. Natl Acad. Sci. USA 112, 11371–11376 (2015).
Koch, H. et al. Growth of nitrite-oxidizing bacteria by aerobic hydrogen oxidation. Science 345, 1052–1054 (2014).
Bayer, B. et al. Metabolic versatility of the nitrite-oxidizing bacterium Nitrospira marina and its proteomic response to oxygen-limited conditions. ISME J. 15, 1025–1039 (2021).
Daebeler, A. et al. Exploring the upper pH limits of nitrite oxidation: diversity, ecophysiology, and adaptive traits of haloalkalitolerant Nitrospira. ISME J. 14, 2967–2979 (2020).
Mueller, A. J., Daebeler, A., Herbold, C. W., Kirkegaard, R. H. & Daims, H. Cultivation and genomic characterization of novel and ubiquitous marine nitrite-oxidizing bacteria from the Nitrospirales. ISME J. 17, 2123–2133 (2023).
Elbein, A. D., Pan, Y. T., Pastuszak, I. & Carroll, D. New insights on trehalose: a multifunctional molecule. Glycobiology 13, 17R–27R (2003).
Kandror, O., DeLeon, A. & Goldberg, A. L. Trehalose synthesis is induced upon exposure of Escherichia coli to cold and is essential for viability at low temperatures. Proc. Natl Acad. Sci. USA 99, 9727–9732 (2002).
Sun, D. et al. Chlorate as a comammox specific inhibitor reveals nitrification and N2O production activity in coastal wetland. Soil Biol. Biochem. 173, 108782 (2022).
Tang, X. et al. Ammonia-oxidizing archaea and comammox Nitrospira clade B as freeze-thaw resistant nitrifiers in wetland soils. Int. Biodeter. Biodegr. 178, 105570 (2023).
Han, P. et al. N2O and NOy production by the comammox bacterium Nitrospira inopinata in comparison with canonical ammonia oxidizers. Water Res. 190, 116728 (2021).
Kennicutt, M. C. I. I. et al. Polar research: six priorities for Antarctic science. Nature 512, 23–25 (2014).
Ma, Y., Bian, L., Xiao, C., Allison, I. & Zhou, X. Near surface climate of the traverse route from Zhongshan Station to Dome A, East Antarctica. Antarct. Sci. 22, 443–459 (2010).
Casciotti, K. L., Sigman, D. M., Hastings, M. G., Bohlke, J. K. & Hilkert, A. Measurement of the oxygen isotopic composition of nitrate in seawater and freshwater using the denitrifier method. Anal. Chem. 74, 4905–4912 (2002).
Sigman, D. M. et al. A bacterial method for the nitrogen isotopic analysis of nitrate in seawater and freshwater. Anal. Chem. 73, 4145–4153 (2001).
Kaiser, J., Hastings, M. G., Houlton, B. Z., Rockmann, T. & Sigman, D. M. Triple oxygen isotope analysis of nitrate using the denitrifier method and thermal decomposition of N2O. Anal. Chem. 79, 599–607 (2007).
Buffen, A. M., Hastings, M. G., Thompson, L. G. & Mosley-Thompson, E. Investigating the preservation of nitrate isotopic composition in a tropical ice core from the Quelccaya Ice Cap, Peru. J. Geophys. Res. Atmos. 119, 2674–2697 (2014).
Shi, G. et al. Investigation of post-depositional processing of nitrate in East Antarctic snow: isotopic constraints on photolytic loss, re-oxidation, and source inputs. Atmos. Chem. Phys. 15, 9435–9453 (2015).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Li, D. et al. MEGAHIT v1.0: a fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods 102, 3–11 (2016).
Uritskiy, G. V., DiRuggiero, J. & Taylor, J. MetaWRAP-a flexible pipeline for genome-resolved metagenomic data analysis. Microbiome 6, 158 (2018).
Sieber, C. M. K. et al. Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat. Microbiol. 3, 836–843 (2018).
Olm, M. R., Brown, C. T., Brooks, B. & Banfield, J. F. dRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J. 11, 2864–2868 (2017).
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P. & Tyson, G. W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055 (2015).
Chaumeil, P. A., Mussig, A. J., Hugenholtz, P. & Parks, D. H. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 36, 1925–1927 (2019).
Buchfink, B., Xie, C. & Huson, D. H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2015).
Parks, D. H. et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 36, 996–1004 (2018).
Na, S. I. et al. UBCG: up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J. Microbiol. 56, 280–285 (2018).
Nguyen, L. T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A. & Jermiin, L. S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589 (2017).
Lee, I., Ouk Kim, Y., Park, S. C. & Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 66, 1100–1103 (2016).
Vallenet, D. et al. MicroScope: an integrated platform for the annotation and exploration of microbial gene functions through genomic, pangenomic and metabolic comparative analysis. Nucleic Acids Res. 48, D579–D589 (2020).
Seemann, T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (NSFC) (Nos. 42371064 to P.H., 42276243 to G.S., 42030411 to L.H., and 42230505 to M.L.), and the Program of Shanghai Academic Research Leader (Grant No. 20XD1421600 to G.S.), and by the Dutch Research Council (NWO; Talent Program grants VI.Veni.192.086 and 016.Vidi.189.050 to S.L.). The LABGeM (CEA/Genoscope & CNRS UMR8030), the France Génomique, and the French Bioinformatics Institute national infrastructures (funded as part of Investissement d’Avenir program managed by Agence Nationale pour la Recherche, contracts ANR-10-INBS-09 and ANR-11-INBS-0013) are acknowledged for support within the MicroScope annotation platform.
Author information
Authors and Affiliations
Contributions
P.H., G.S. and M.L. conceived this study. P.H. and X.T. conducted the experimental work. P.H. and H.K. performed the comparative genomic analyses. X.T. analyzed the amplicon sequencing data. X.D. and P.H. analyzed the metagenomic data. D.W. and Z.L. collected the samples. Q.Z. did the physiochemical analyses. L.H. and S.L. contributed to the data interpretation. P.H. and G.S. wrote the manuscript, with contributions from all co-authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Maximiliano Ortiz and Cheng Han for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Han, P., Tang, X., Koch, H. et al. Unveiling unique microbial nitrogen cycling and nitrification driver in coastal Antarctica. Nat Commun 15, 3143 (2024). https://doi.org/10.1038/s41467-024-47392-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-47392-4
This article is cited by
-
All-inclusive nitrifiers in Antarctic soils
Nature Communications (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.