Abstract
“Antibody-breeding” has provided therapeutic/diagnostic antibody mutants with greater performance than native antibodies. Typically, random point mutations are introduced into the VH and VL domains of parent antibodies to generate diverse libraries of single-chain Fv fragments (scFvs), from which evolved mutants are selected. We produced an scFv against estradiol-17β with 11 amino acid substitutions and a >100-fold improved affinity constant (Ka = 1.19 × 1010 M−1) over the parent scFv, enabling immunoassays with >30-fold higher sensitivity. We systematically analyzed contributions of these substitutions to the affinity enhancement. Comparing various partial scFv revertants based on their Kas indicated that a revertant with four substitutions (VH-L100gQ, VL-I29V, -L36M, -S77G) exhibited somewhat higher affinity (Ka = 1.46 × 1010 M−1). Finally, the VH-L100gQ substitution, occurring in VH complementarity-determining region (CDR) 3, was found to be the highest-priority for improving the affinity, and VL-I29V and/or VL-L36M cooperated significantly. These findings encouraged us to reconsider the potential of VH-CDR3-targeting mutagenesis, which has been frequently attempted. The substitution(s) wherein might enable a “high rate of return” in terms of selecting mutants with dramatically enhanced affinities. The “high risk” of generating a tremendous excess of “junk mutants” can be overcome with the efficient selection systems that we developed.
Similar content being viewed by others
Introduction
As exemplified by the fact that pioneers in the field received the Nobel Prize in Chemistry in 20181, the “antibody-breeding” approach (i.e., in vitro molecular evolution of antibody molecules) revolutionized the generation of therapeutic and diagnostic antibody agents2,3,4,5. Standard strategies2,3,4,5 rely upon the introduction of random point mutations or site-directed mutations into the heavy and light chain variable (VH and VL) domains of a parent antibody to generate diverse libraries of mutated antibody fragments, e.g., single-chain Fv fragments (scFvs)6,7 or Fab fragments. Mutated fragments with improved binding characteristics contained therein are then selected and isolated with the aid of genotype-phenotype linking technology2, e.g., phage display8,9, ribosomal display10, or yeast cell-surface display11.
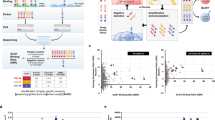
To date, we have produced affinity-matured scFv mutants binding to small biomarkers [e.g., estradiol-17β (E2)12,13,14, cotinine15, cortisol16, and Δ9-tetrahydrocannabinol (THC)17] to establish more sensitive immunochemical assay systems. Actually, these mutants enabled 3–100-fold higher sensitivity in competitive enzyme-linked immunosorbent assays (ELISAs), compared with the corresponding parental scFvs (Fig. 1). Because these scFvs were generated via mutagenesis based on error-prone polymerase chain reaction (PCR) experiments12,13,14,15,16,17,18, the missense mutations (causing amino acid substitutions) were introduced randomly. Consequently, some of them might function as “key mutations” for increasing in the affinity, whereas other mutations might be “junk mutations” that contribute little, nothing, or even decrease the affinity. Our previous results (Fig. 1) indicated the number of substitutions correlated with the extent of affinity improvement; thus, the anti-THC scFv with a single substitution gained 10-fold higher affinity (based on the calculated equilibrium affinity constant Ka), whereas the anti-E2 scFv with 11 substitutions exhibited a Ka that was increased (improved) by >100-fold. The scFv mutants against cortisol and cotinine showed intermediate improvement, exhibiting >30-fold and >40-fold higher Kas as the results of three and five substitutions, respectively. However, a reasonable explanation for such correlations requires evidence that most or many (if not all) of these multiple substitutions participated in increasing the affinity to at least some extent.
Summary of our previous “antibody-breeding” experiments with scFvs against (A) estradiol-17β12–14, (B) cotinine15, (C) cortisol16, and (D) Δ9-tetrahydrocannabinol17. Typical dose–response curves in competitive ELISAs using the wild-type scFv (scFv-wt; blue), wild-type Fab [Fab-wt; blue, shown only in (A)] and affinity-matured scFv mutants (scFv-m; magenta) are shown together with the respective Ka values. These scFv-ms were named in the original articles as (A) scFv#m3-a1814, (B) scFv#m1-5415, (C) scFv#m1-L1016, and (D) scFv#m1-3617. The vertical bars indicate the SDs for intra-assay variances (n = 4). The magnitude of improvements in the assay sensitivities (calculated based on the ratios of the midpoint values) are also shown. The primary structure of the scFv-ms, all assembled in the orientation of VH–linker–VL–FLAG tag, are illustrated. VH-CDR1, 2, 3, VL-CDR1, 2, and 3 are abbreviated as H1, H2, H3, L1, L2, and L3, respectively. The amino acid substitutions introduced are denoted with dark blue stars and one-letter codes. We should note that, in the original article where the anti-E2 scFv#m3-a18 (A) was generated14, we estimated the amino acid at the VH-100g position as glutamic acid, based on the behaviors of scFvs in ELISAs. Recently, however, we chemically assigned this residue as glutamine (Q) by LC/MS/MS49, as shown in this figure and discussed in this article.
Regarding the positions of the substitution(s), a remarkable difference was found between the anti-cortisol scFvs (only in the VL) and the anti-THC (only in the VH) (Fig. 1). In contrast, with the anti-E2 and anti-cotinine scFvs that showed more significant improvements, the substitutions were spread over both the VH and VL domains. The most successful example of in vitro affinity maturation for an antibody against a small compound was reported for an scFv against fluorescein derivative (Ka > 1 × 1012 M−1), which represented an affinity increase of>2,000-fold19. This “super” mutant, which is almost beyond native antibodies, was the product of 14 different substitutions, 12 of which were located in the VH domain (Supplementary Fig. S1A).
Recently, some approaches have been developed for improving antibody functions via computational analysis, in order to minimize the trial and error that is an inevitable aspect of the conventional strategies20,21,22,23. However, using an empirical approach toward more efficient mutagenesis strategies, e.g., targeting more limited regions (hotspots) with more controlled amino acid-substitution patterns is still important. To collect useful information, we sought the highest-priority substitutions for successful affinity maturation, among numerous randomly introduced multiple substitutions. We simultaneously pursued the greatest enhancement in affinity that was achievable with the fewest substitutions.
We selected our affinity-matured anti-E2 scFv with a 1010-order Ka and 11 substitutions14 as the subject of this study. Our novel approach employed here, i.e., systematic analysis using partial scFv revertants (scFvs with some substituted amino acids restored to the original sequence), revealed that the most critical substitution among the 11 was the one from leucine (L) to glutamine (Q) at the VH100g position, which occurred in complementarity-determining region (CDR) 3 of the VH domain (VH-CDR3). This mutation alone resulted in 17-fold enhanced Ka compared with the wild-type scFv (i.e., the parent scFv without any artificial mutations).
Results
Origin, and structural and binding characteristics of target anti-E2 scFv (scFv#M3rd)
The anti-E2 scFv focused on in this study, named scFv#M3rd(amb) here [this was originally reported as “scFv#m3-a18” (see Fig. 1A)], is an affinity-matured mutant that showed a 1010-range Ka value against free (i.e., not immobilized) E2 molecules. This scFv was our “third-generation mutant” that was generated previously in our laboratory after three iterative mutagenesis and selection steps performed on the wild-type scFv (scFv#WT)12,13,14 (Fig. 2A). scFv#WT was constructed by linking the VH and VL domains of a mouse anti-E2 antibody (Ab#E4-4)12 via a common linker sequence composed of glycine (G) and serine (S) in the sequence of (GGGGS)36,7 and attaching a FLAG tag24 at the C-terminus. The VH and VL domains contained 124 and 107 amino acids (Fig. 2A)12, which belonged to subgroups IIID and V25, respectively. In this study, we used the numbering and classifications defined by Kabat et al.25 The VH domain contained 15-residue CDR3, which is substantially longer than the average length of 8.7 residues for VH-CDR3 for mouse antibodies against any antigens26 or 8.50 residues for mouse antibodies against haptens27. Seven amino acids following the residue at position 100 (underlined in Fig. 2A) are defined as the inserted residues in the Kabat-rule, and were named 100a–100 g. The Ka values against free E2 molecules and amino acid substitutions in this mutant are shown together with those of the first- and the second-generation scFv mutants (scFv#M1st and #M2nd, respectively) in Fig. 2A. Previously, we showed that scFv#M3rd(amb) was specific enough and applicable for use with clinical specimens14. The 11 amino acid substitutions in scFv#M3rd(amb) are located both in the VH and the VL domains (five and six substitutions, respectively), and both in CDRs and framework regions (FRs) (five and six substitutions, respectively).
(A) Summary of the process used for generating affinity-matured scFvs against estradiol-17β (Ε2)12,13,14. Three steps of genetic evolution, i.e., scFv#WT (the wild-type scFv combining the VH and VL domains derived from a mouse anti-E2 antibody)12 → scFv#M1st12,13,14 → scFv#M2nd13,14 → scFv#M3rd(amb)14, were performed, each of which involved random mutagenesis based on error-prone PCR and phage display-aided selection of improved species. The amino acid sequences of the wild-type VH and VL domains are shown in the purple box. The VH- and VL-CDRs, determined with the Kabat definition25 (H1, H2, H3, L1, L2, and L3), are shown with red and green, respectively. The Ka values of each scFv, determined by the Scatchard analysis28, are shown together with the increasing magnitudes observed with each step. The primary scFv structures are schematically illustrated, where the new amino acid substitution(s) introduced during the first, second, and third mutagenesis steps is represented with a red, dark blue, and purple star(s), respectively. The amino acids before and after each substitution are indicated with the one-letter code. (B,C) Schematic illustration of the primary structures of the scFvs introduced with a reverse mutation(s) [(shown with magenta cross(es)] for returning upstream (B) from scFv#M3rd(amb) to scFv#M2nd or (C) from scFv#M2nd to scFv#M1st. Two substitutions were simultaneously restored in scFv#R2-3. The downward double arrows (↓↓) mean >10-fold decrease in the affinity compared with the parent scFv before reverse mutation(s) was introduced.
In the scFv#M3rd(amb) gene variant, a T→A transversion introducing a nonsense mutation from TTG [encoding L] to TAG (amber termination codon) occurred in the VH-100g codon, which encodes the residue near the end of VH-CDR3. Considering that we used Escherichia coli (E. coli) XL1-Blue as host cells, which is an supE suppressor strain, this amber codon was expected to be readthrough and translated as Q, and this was confirmed by liquid chromatography/tandem mass spectrometry (LC/MS/MS) fingerprinting of the affinity-purified scFv#M3rd(amb) protein (Supplementary Fig. S2). As further confirmation, we modified the scFv gene variant replacing the TAG codon at the position with CAG (encoding Q) via oligonucleotide-directed mutagenesis. The product named scFv#M3rd(Q) exhibited almost the same Ka (1.23 ± 0.17 × 1010 M−1) [mean ± standard deviation (SD); (n = 3)] as that of scFv#M3rd(amb) (Ka = 1.19 ± 0.22 × 1010 M−1), as determined by the Scatchard analysis28 (Fig. 2A, Supplementary Fig. S3). By performing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis, we observed that both scFvs migrated as a single band at almost the same relative molecular mass (Mr), which was close to the expected Mr value (27133) (Supplementary Fig. S2). Chemical identity between scFv#M3rd(Q) and #M3rd(amb) was supported by the LC/MS/MS fingerprinting data of the newly generated scFv#M3rd(Q) (Supplementary Fig. S2).
Determining the substitutions responsible for enhanced affinity (the first stage)
In this study, we employed the “elimination approach” to discover a minimum and essential set of substitutions responsible for the increased E2-binding affinities of scFv#M3rd(amb) or (Q) [denoted as scFv#M3rd(amb/Q), hereafter]. We first evaluated the affinity of partial scFv revertants, where one of the multiple substitutions was restored to the original amino acid. The Ka values were determined by Scatchard analysis28 using tritium-labeled E2, taking care to avoid inadequate estimations due to any additional structures like “bridges” that link with signal-groups or proteins necessary for immobilization29,30.
We first analyzed the third mutagenesis step that generated scFv#M3rd(amb) with a 19-fold increased Ka as the consequence of only two amino acid substitutions, i.e., the VH-L100gQ (confirmed as described above) and the histidine (H) to asparagine (N) substitution at the VL50 position (Fig. 2A)14. The scFv#R1-1 revertant with a single amino acid restoration at the VH100g (Q → L) exhibited a 21-fold decreased Ka (5.6 × 108 M−1), which was quite similar to, and rather lower than that of the parent scFv (i.e., scFv#M2nd; Ka = 6.3 × 108 M−1) (Fig. 2B). In contrast, scFv#R1-2 with the VL50 N → H restoration maintained 47% of the affinity that observed for scFv#M3rd(amb). These results show that the substitution at the VH100g was essential whereas the mutation at the VL50 was not critical.
Then, we analyzed the second mutagenesis step involving eight amino acid substitutions that converted scFv#M1st into #M2nd, resulting in a 2.4-fold higher Ka13. We produced seven revertants (scFv#R2-1–#R2-7), each with a single restoration, except that scFv#R2-3 contained two restorations at the contiguous VH84 and VH85 positions (Fig. 2C). Considerably lowered affinity was found only for scFv#R2-5 and #R2-7 (i.e., 64-fold and 3.0-fold decreases, respectively), indicating that the methionine (M) at the VL36 and the G at the VL77 were responsible for the higher affinity. We note here that the Ka of scFv#R2-5 (9.9 × 106 M−1) was significantly lower than that of its parent scFv, scFv#M1st (2.6 × 108 M−1). This finding suggests a situation where the 10 substitutions excluding VL-L36M, found in scFv#M2nd, should have cooperatively reduced the affinity of its parent scFv (i.e., scFv#M1st) significantly (26-fold), but that the effect of the single VL-L36M substitution on increasing the affinity predominated.
The first mutagenesis step provided scFv#M1st with a 3.0-fold higher Ka over scFv#WT. This improvement should be due to a isoleucine (I) to valine (V) substitution at the VL29 position, which was solely introduced during this step12,13. Therefore, this mutation was estimated to be essentially responsible for the increased affinity of the third-generation mutant, which was confirmed later (see below).
Determining substitutions responsible for enhanced affinity (the second stage)
During the first-stage examination, we identified the following amino acid substitutions that must have been key for increasing the affinity: VH-L100gQ (introduced during the 3rd mutagenesis step), VL-L36M and VL-S77G (the 2nd mutagenesis step), and VL-I29V (the 1st mutagenesis step). However, substitutions selected in different mutagenesis stages do not always function cooperatively. Thus, we generated a new scFv containing these selected four substitutions, named scFv#4mut (Fig. 3A). This mutant showed an even slightly higher Ka [1.46 ± 0.35 × 1010 M−1; mean ± SD (n = 3)] than that of scFv#M3rd(amb/Q) (Ka = ~1.2 × 1010 M−1). Thus, these four substitutions conferred 170-fold greater Ka over the original scFv#WT, and some of the remaining seven substitutions prevented these desirable four substitutions from enhancing the affinity. To speculate the kinetic mechanism of this affinity enhancement, we compared the association and dissociation rate constants (ka and kd) of scFv#4mut and #WT, determined by the surface plasmon resonance (SPR) sensor. The parameters obtained were as follows: for scFv#4mut, ka = 2.97 × 104 M−1s−1, kd = 1.40 × 10−6 s−1 (ka/kd = Ka = 2.12 × 1010 M−1) and for scFv#WT, ka = 1.95 × 105 M−1s−1, kd = 6.52 × 10−3 s−1 (ka/kd = Ka = 2.99 × 107 M−1). These data demonstrate that the improvement in the affinity (706-fold based on the Ka determined by the SPR) was mainly attributed to the decreased kd value of scFv#4mut. In comparison with scFv#M3rd(Q) [ka = 1.13 × 105 M−1s−1, kd = 5.05 × 10−6 s−1 (ka/kd = Ka = 2.24 × 1010 M−1)], scFv#4mut is evaluated to exhibit almost similar affinity as a consequence of 3.6-fold less kd that compensates for 3.8-fold less ka.
Comparison of the affinities (Ka values determined by the Scatchard analysis28) between (A) scFv#M3rd(amb) and scFv#4mut having four substitutions, and between (B–D) scFv#4mut and its partial revertants retaining (B) three substitutions, (C) two substitutions, and (D) a single substitution. The restored substitutions are shown with magenta crosses. (E) The affinities (Ka) of 20 different scFv#M3rd(amb) variants, each of which had a different amino acid at the VH100g position (shown in abscissa with the one-letter code), were compared. ND means “not determined” because of too low affinities. The downward double arrows (↓↓) mean >30-fold decrease in the Ka value compared with scFv#4mut.
Determining substitutions responsible for enhanced affinity (the third stage)
Then, we examined whether even fewer substitutions could cause the increased affinity by analyzing revertants derived from scFv#4mut. Among the four revertants with a single restoration at VH100g (Q → L), VL29 (V → I), VL36 (M → L), or VL77 [G → S], named scFv#R3-1, #R3-2, #R3-3, and #R3-4, respectively, scFv#R3-1 exhibited a significantly (44-fold) decreased Ka compared to scFv#4mut, whereas the decreases of 1.6–3.4-fold were observed with the other three revertants retaining the VH-L100gQ substitution (Fig. 3B). These observations strongly suggested that the VH-L100gQ substitution was most important for the dramatically increased affinity. Comparison of the Ka values of scFv#R3-2, #R3-3, and #R3-4 suggested that the extent of cooperation with VH-L100gQ might be in the order of VL-I29V ≈ VL-L36M > VL-S77G. This was further analyzed using three revertants with double restorations (Fig. 3C). Regarding the revertants that retained the VH-L100gQ substitution, the order of the Ka was as follows: scFv#R4-2 (containing VL-L36M) > #R4-1 (VL-I29V) > #R4-3 (VL-S77G), all of which showed 109-order Ka values. However, the other three revertants that lacked the VH-L100gQ substitution showed obviously lower affinity (Ka = 1.3–4.1 × 108 M−1).
Finally, we directly evaluated the contribution of the VH-L100gQ substitution by determining the Ka of revertant scFv#R5-1 (Fig. 3D). The result (Ka = 1.5 × 109 M−1) indicated that the single VH-L100gQ substitution improved the affinity by 17-fold, suggesting the great potential of performing VH-CDR3-directed mutagenesis anew, which has commonly been performed (often with simultaneous VL-CDR3-randomization), particularly in early antibody-engineering studies31,32,33,34. Naturally, we became interested in the potential of substituting the other 18 amino acids (beside Q and the original L) at the VH100g position. Thus, we prepared 18 scFv additional mutants by replacing the VH100g amino acid of scFv#M3rd(amb) with one of the remaining 18 amino acids, and compared their Ka values with those of the already-evaluated VH-100gQ and VH-100gL mutants. As shown in Fig. 3E, the Q-substituted mutant [i.e., scFv#M3rd(Q)] exhibited the most improved affinity, and the next highest was the mutant substituted with N (~14-fold improvement over the VH-100gL mutant), a homolog of Q with an amide group but with one less carbon atom in the side chain. Moderate, but significant (>5-fold) improvements were observed for the G-, glutamic acid (E)-, H-, and arginine (R)-substituted mutants. Amino acids with aliphatic and hydrophobic side chains (I and V) or aromatic rings [phenylalanine (F), tryptophan (W), and tyrosine (Y)] contributed little to the improvement. It was surprising that substitution with S, though well-recognized as a residue (as well as Y) that often plays important roles in interactions with antigens35, deteriorated the affinity down to undetectable range, as also seen with the cysteine (C)-substituted mutant. We should also note that the Q substitution, which increases the binding affinity most, is not among the top 10 amino acids that frequently appear in the CDR sequences of mouse antibodies against haptens [i.e., Y, S, G, L, N, W, threonine (T), I, alanine (A), and R]27: this suggests that it may be difficult to use a prediction-based approach for improving amino acid sequences in this most diverse CDR25,26,27,36,37. Nonetheless, these data (Fig. 3E) suggested to us the possibility that every position in VH-CDR3 might be substitutable with much more potent amino acids that cause dramatically enhanced affinity, but that it should rarely be achieved via error-prone-PCR-based mutagenesis.
Summary of affinity-maturation results from scFv#WT to scFv#M3rd(amb/Q)
These findings enabled us to order the affinity-maturation process by focusing the importance of VH-L100gQ substitution, as shown in Fig. 4. The most improved mutant species with single, double, and triple substitutions were estimated to be scFv#R5-1, #R4-2, and #R3-4, respectively, the Ka values of which increased as the numbers of substitutions increased. Comparison of the Ka values between scFv#R3-1 and R3-3 (Fig. 3B), and between scFv#R4-3 and R4-6 (Fig. 3C), indicates a greater potential of substitution at VH-L100gQ than VL-L36M under the presence of one or two other substitution(s). These observations were compatible with the result where scFv#R5-2 with a single VL-L36M substitution exerted the Ka value that was only 2.2-fold greater than scFv#WT and much lower than scFv#R5-1. Quadruple substitutions in scFv#4mut were necessary to reach (and even exceed) the affinity of scFv#M3rd(amb/Q), the high-affinity mutant with 11-amino acid substitution. Therefore, assistance of VL-S77G substitution was necessary, although it was not as potent as that of VL-I29V and VL-L36M substitutions. After all, it was shown that, in scFv#4mut, the 4 substitutions functioned additively and showed a 170-fold higher affinity than that of scFv#WT.
Schematic illustration of the hierarchy of the anti-E2 scFvs in terms of the antigen-binding affinity (Ka determined by the Scatchard analysis28). The upward orange arrows indicate increases in the Ka, and the magnitudes are shown beside the arrows.
Analytical utility and structural aspects of the high-affinity scFvs
Immunoassay sensitivities basically correlate with the affinities of the antibodies used, and usually antibodies with a higher affinity enable immunoassays with higher sensitivity38. Indeed, scFv#M3rd(amb), #M3rd(Q), and #4mut (showing Ka values in the 1010-range), as well as scFv#R3-4, #R4-1, #R4-2, and #R5-1 (showing Ka values in the 109-range), displayed dramatically enhanced sensitivities in competitive ELISAs, as shown by 8.0–17-fold lower midpoint values (~13–28 pg/assay) than scFv#WT (220 pg/assay) in dose–response curves (Fig. 5). These dose–response curves with improved sensitivities cover a measurable range required for clinical applications14.
Dose–response curves of competitive ELISAs obtained with the scFvs shown in Fig. 4. The vertical bars indicate SD for intra-assay variance (n = 4). The midpoints of the curves (pg/assay) were as follows: scFv#4mut, 12.6 ± 0.12 [mean ± SD (n = 4)]; #M3rd(amb), 15.0 ± 0.76; #M3rd(Q), 16.8 ± 1.57; #R3-4, 14.3; #R4-2, 15.6; #R4-1, 19.0; #R5-1, 27.6; and #WT, 220 (average of determinations in duplicate). In these assays, the scFv concentrations were adjusted to give bound enzyme activities at B0 (the reaction without E2 standard) of approximately 1.0–1.5 absorbance after a 30-min enzyme reaction. The background absorbance (observed without addition of scFvs) was lower than 5.0% of the B0 absorbance. We should note that, in the original article where scFv#M3rd(amb) (denoted as scFv#m3-a18 therein) was generated14, we reported its Ka value as 1.3 × 1010 M−1, and the midpoint value in the ELISA using this scFv was determined to be 10.0 ± 1.2 pg/assay. In this study, we re-determined the Ka in triplicate. The midpoint values were also re-determined to perform equal and strict comparisons between the scFvs, because we had to use a newly prepared E2–BSA conjugate to coat the ELISA microplates. Difference in the quality of these conjugates (mainly in the hapten/protein molar ratio) influences on the ELISA sensitivity and often makes it difficult to strictly reproduce previous experimental data.
Protein modeling of the scFv#4mut and #WT, docked with E2, is shown in Fig. 6. Although such in silico approaches offer structural information with more or less limited reliability compared with X-ray crystallography, the modeling of scFv#4mut strongly suggested that none of the 4 substituted residues (Q at the VH100g position, or V, M, and G at the VL29, 36, and 77 positions, respectively) forms direct contacts with the E2 molecule in the immune complex. The Q residue was located near the C-terminus of VH-CDR3 and should function by raising the loop structure of this CDR. Substitution from the original L to Q is estimated to alter the steric conformation of VH-CDR3 loop, but the modeling did not suggest obvious interaction of VH-CDR3-related residues with E2 molecule. Instead, a possibility was shown where, in scFv#4mut, VH-CDR2 and VL-CDR1 might interact with E2. Thus, the hydroxy group of Y residue (at the VL32 in the CDR1) and the carboxy group of E residue (at the VH50 in the CDR2) might anchor E2 molecule via the hydrogen bonds with the hydroxy groups of the A- and D-ring of the steroid skeleton, respectively, resulting in drastic change of the orientation of the E2 molecule in the paratope compared with that in scFv#WT. The V (at the VL29) and/or M (at the VL36) residue(s) introduced in scFv#4mut might trigger such VL-CDR-dependent events.
Protein ribbon structures for (A) scFv#4mut and (B) scFv#WT were constructed using the SWISS-MODEL Protein Modelling Server50, and their conformations when docked to E2 were predicted using SwissDock51. Three different views observed from different angles are shown. In the ribbon representation of the scFv backbones, CDR H1 (yellow), H2 (orange), H3 (magenta), L1 (dark blue), L2 (light green), and L3 (light blue) are represented with β-sheet structures (bold gray arrows). The introduced amino acids after the substitutions are shown in orange. The backbone of the E2 molecule is shown in light purple. Image generated with PyMOL52.
We should note here that both the introduced M residue and original L residue are unusual amino acids as for the VL36 position: in fact, the residues at the VL35–41, which form the beginning of VL-FR2, are highly conserved in the sequence WYQQK(lysine)P(proline)G found in all VL subgroups25. The G residue at the VL77 position is a member of FR3: the original amino acid was S, which frequently appears at this position. The contribution of this G was not significant as that of the V or M residue, but was essential for increasing the affinity to that observed with scFv#3rd(amb/Q). The mechanism whereby the G residue contributed to the affinity, despite its considerably long-distance from the paratope, is of great interest.
Discussion
For those of us, working in the fields of analytical and diagnostic chemistry, antibodies with high affinity for target molecules are an essential tool. Because the higher affinity enables more sensitive analytical/diagnostic systems, “antibody-breeding” that generates mutant antibody fragments with improved affinity is an attractive methodology. However, the conventional approach combining random mutagenesis and panning-based selection has very often failed to provide satisfactorily improved mutants, despite much time consuming effort. To overcome such challenges, revolutionary strategies are required for efficiently introducing functional mutations without unnecessarily enlarging the diversity and for reliably selecting rare and improved mutants without overlooking them.
To achieve the former requirement, information is needed for designing “decisive mutations (substitutions)” essential for elevating the antigen-binding affinity. It would also be of great help if it were possible to discover “hot spots” in the VH and/or VL domains of antibodies, where a wide range of mutations introduced thereto significantly improves the affinity. Of course, it is more desirable if fewer mutations introduced in a narrower region facilitate isolating improved species with high probability.
Previously, we summarized the results of studies wherein antibody mutants were generated that bound to small molecules (haptens)14,38. Among the affinity-matured products reported, mutant scFvs or Fabs that showed Ka values greater than 1 × 109 M−1 (i.e., a standard value required for subpicomole-order analysis), owing to>10-fold enhancement, were selected and their structures are illustrated in Supplementary Fig. S1A. Four out of the six mutants generated, including the scFv with a Ka value greater than 1 × 1012 M−1 and the greatest improvement19, involved totally 10 or more substitutions and some of them were in VH-CDR3, similar to our scFv#M3rd(amb). Because two of these four mutants (i.e., anti-fluorescein-biotin and anti-fluorescein antibody fragments) were generated via error-prone PCR18 or related methods, only some of the multiple substitutions introduced might have driven the enhanced affinity. Seeking substitutions that do in fact contribute to the binding affinity is inevitable for developing simpler strategies that depend less on trial and error.
Thus, we analyzed our anti-E2 scFv#M3rd(amb): this mutant had 11 amino acid substitutions and showed an extremely high (1010-order) Ka against free E2 molecules, which was over 100-fold higher than that of the parent antibody (scFv#WT). To explore the hierarchy of these substitutions regarding contribution to the enhanced affinity, we employed a unique approach based on the comparison of the affinities of various partial scFv revertants. Although this case study was performed with a particular antibody mutant, we obtained several suggestive results that exceeded our initial expectations, as summarized below. First, a mutant with only four substitutions showed even higher affinity than scFv#M3rd(amb), i.e., the Ka was 1.46 × 1010 M−1, corresponding to a 170-fold improvement compared to scFv#WT. The incorporated amino acids (and positions) were Q (VH100g), V (VL29), M (VL36), and G (VL77). Second, the most influential residue was Q at VH100g (locates in VH-CDR3), which alone improved the affinity by 17-fold. Third, none of the substitutions at VH100g with the remaining 19 different amino acids resulted in an equivalently improved affinity. Fourth, the extent that these substitutions contributed to the enhanced affinity was in the order of Q ≫ M ≥ V ≫ G, and these four substitutions seemed to function in an additive manner. Consequently, the mutant with three substitutions (Q/M/V residues) showed 63% of the affinity observed with the mutant with four substitutions, and the mutant with two substitutions, Q/M or Q/V, exhibited 36% or 29% affinity, respectively. These mutants with Q/M/V, Q/M, and Q/V substitutions demonstrated 107-fold, 62-fold, and 49-fold higher affinity than the parent scFv#WT, maintaining 109-order Ka value.
These findings impressed upon us the importance of identifying a decisive substitution (“ace” substitution) like the VH-L100gQ substitution described above. Considering the mutation patterns found in other reported high-affinity mutants as well (Supplementary Fig. S1A), the VH-CDR3 should be focused on as a “hot region” where such an ace substitution might be discovered with high probability. VH-CDR3 often works as the “ace CDR” in antigen recognition36,37, and thus at the dawn of antibody engineering (~1990), “hard randomization”39 was frequently performed for multiple amino acid residues therein (often together with VL-CDR3) by the site-directed introduction of degenerated NNS [i.e., (A/C/G/T)(A/C/G/T)(C/G)] or NNK [i.e., (A/C/G/T)(A/C/G/T)(G/T)] codons encoding any of 20 different amino acids32,33. This was actually a potent strategy for generating new prototype antibody fragments that gained different specificities, but was unlikely to be suitable for improving the affinity while maintaining the original specificity. In fact, our previous affinity-maturated scFvs against cotinine, cortisol, and THC did not contain substitutions in VH-CDR3 (Fig. 1B–D).
Such outcomes should be mainly attributable to the highly diversified structure and nature of VH-CDR3, each with different binding specificity and affinity function to allow for “the best fitting” against only a limited antigen structures. Therefore, the “hardly” mutagenized libraries only rarely generate satisfactorily improved mutants, which, furthermore, should be buried by tremendously large excess of mutants with deteriorated binding performance. The most popular selection systems, i.e., those combining phage display and panning do not always facilitate successful isolation of such rare and hidden species, mainly due to the biased propagation of phage clones displaying antibody fragments and the competition with a large excess of the phage clones displaying antibodies with weaker or deteriorated affinities40. Moreover, the hard mutagenesis of more than seven amino acid residues, which should produce >207 (= 1.28 × 109) different amino acid sequences, generates libraries too large to deal with using standard experimental conditions. It is no wonder that these difficulties made us gradually avoid the VH-CDR3-focused strategies for the purpose of affinity-maturation.
However, the present findings suggest a much simpler strategy for mining a possible ace mutation(s) in VH-CDR3. The parent scFv (scFv#WT) has a VH-CDR3 composed of 15 amino acids (Fig. 2A), which also correspond to the definition by Chothia et al.41,42. We here assumed that 13 of these residues, avoiding the less-variable D and Y residues at the VH101 and VH102 positions, are important for generating affinity against E2. Searching a very small scFv library, i.e., a sum of 13 groups of sequences each containing 20 different sequences generated by the hard randomization of one of the 13 positions (therefore, theoretically composed of only 13 × 20 = 260 amino acid sequences), should have enabled discovery of the mutant with the ace VH-L100gQ substitution. Even in the cases where two (or more) ace-equivalent mutations were present in the CDR3 and cooperated together, each of them could be separately discovered. Parallel examinations following hard mutagenesis of two or three serial residues (each generates 12 × 202 = 4,800 or 11 × 203 = 88,000 sequences, respectively) might help in finding “ace-mutation motifs”. In our laboratory, development of a novel and efficient strategy for discovering affinity-matured scFv mutants, named “colony-array profiling”, is now progressing. We believe that this approach should be particularly suitable for examining such small libraries and will be of great help, the results of which will be reported in the near future. After finding such an ace mutation, some cooperating mutations (e.g., corresponding to the V and M residues in this study) should be searched for. Error-prone PCR might be still suitable for this purpose.
One of the motives for undertaking this study was an article published in 1990, where the author sought amino acid substitutions that caused a >200-fold difference in the Ka (determined by the fluorescence quenching method) between two hybridoma-derived antibodies against p-azophenylarsonate43. Although 19 amino acid differences were originally observed between these antibodies, the author finally showed that only three amino acid substitutions (two in VH-CDR2, one in VH-CDR3) were needed to produce this increased affinity by examining artificial antibodies produced with the aid of synthetic oligonucleotides (Supplementary Fig. S1B). A more striking example was shown by the affinities observed for anti-digoxin antibodies that were separately obtained from different hybridoma clones. Thus, the antibody named 26-10 exhibited 279-fold higher affinity (1010-range Ka as determined by the equilibrium saturation method) than that of the antibody named LB4, surprisingly due to only a single substitution at the VH52 position located in VH-CDR244 (Supplementary Fig. S1B). In both cases, substitutions in VH-CDR2 must have resulted in the dramatically enhanced affinity (with high probability for the former case, and absolutely for the latter case). These results are reasonable because, for antibodies against haptens, VH-CDR2 and VL-CDR1 tend to play important roles in forming the antigen-binding cavity45, and this speculation was supported by the successful affinity maturation of anti-tacrolimus scFvs due to randomizing VH-CDR2 and VL-CDR1 with NNS codons46. However, even in such cases, the approach for seeking ace substitutions mentioned above can easily applied by extending it to more than one CDR in a parallel manner.
We are greatly interested in identifying the greatest possible increase in the affinity with the fewest substitutions, because this suggests the potential of in vitro affinity maturation of antibodies. In this study, we achieved 170-fold higher affinity by introducing four substitutions (with scFv#4mut), 107-fold higher affinity by introducing three substitutions (with scFv#R3-4), and 62-fold higher affinity by two substitutions (with scFv#R4-2), and these improved scFv mutants showed >109-order Ka values. Considering the difficulty in performing extensive improvement from already-matured antibodies (e.g., whose Ka values exceed 108 M−1), our present results might be worthy of attention.
Materials and Methods
Buffers
The following buffers12,13,14,15,16,17 were used in this study. PB: 50 mM sodium phosphate buffer (pH 7.3); PBS: PB containing 9.0 g/L NaCl; G-PBS: PBS containing 1.0 g/L gelatin; T-PBS: PBS containing 0.050% (v/v) Tween 20; and PVG-PBS: G-PBS containing 1.0 g/L polyvinyl alcohol with an average polymerization degree of 500.
scFvs
Anti-E2 scFv#WT, scFv#M1st, scFv#M2nd, and scFv#M3rd(amb) (originally named as scFv#E4-412,13,14, scFv#m1-e712,13,14, scFv#m2-c413,14, and scFv#m3-a1814, respectively) were prepared as soluble proteins as we described previously12,13,14. Other scFvs having reverse mutation(s) (i.e., revertants) were produced by expressing the corresponding scFv genes in E.coli XL1-Blue cells as described previously12,13,14,15,16,17. These scFv genes were constructed by PCR using synthetic oligo-DNAs predesigned to introduce the targeted mutation(s) as shown below, whose nucleotide sequences were confirmed by the standard method. The scFv proteins were obtained as periplasmic extracts from mass-cultured transformants12,13,14,15,16,17, and used for Ka determinations and ELISAs. For SDS-PAGE analysis, the scFvs were affinity-purified with anti-FLAG M2 antibody-immobilized agarose gel (Sigma–Aldrich)14,15,16.
Preparation of gene fragments encoding the scFv revertants
Among the 21 scFv genes synthesized in this study, typical instances were selected and their preparations are shown below. PCR experiments were performed using an adequate scFv gene (subcloned into the pEXmide 5 vector47; 0.5–50 ng) as the template in a buffer (100 μL) containing Ex Taq (TaKaRa-Bio) (0.5 or 2.5 U) or KOD Fx DNA polymerase (TOYOBO) (2.5 U), 20 nmol of each dNTP, and a combination of reverse/forward primers (50–100 pmol each), unless otherwise specified. Usually, the following cycling condition was used: 94 °C(2 min); then 35 cycles of 98 °C for 10 sec, 55 °C for 30 sec, and 72 °C for 1 min, followed by a hold step at 72 °C for 10 min. The nucleotide sequences of the primers are shown in Supplementary Table S1. Every scFv gene fragment synthesized was digested with Nco I and Sal I, ligated with the similarly digested pEXmide 5 vector47, and introduced in the E. coli cells by electroporation12,13,14,15,16,17.
-
a)
scFv#R1-1. Three sets of PCRs were performed using the scFv#M3rd(amb) gene14 as the template and one of the following three combinations of primers: (i) R1 and F1, (ii) R2 and F2, or (iii) R3 and E2-VL-For-212 (Supplementary Figure S4A). Using the three kinds of PCR products (i–iii), two overlap-extension PCR steps were performed. First, the products i and ii (each 200 ng) were mixed and subjected to 10 cycles of PCR in a 25-μL buffer solution containing ExTaq polymerase. A portion of the reaction solution (10 μL) was mixed with R1 and F2 primers and re-amplified similarly, but for 15 cycles in a 100-μL buffer solution containing ExTaq polymerase. Second, the resulting product was gel-purified, and a portion (200 ng) was mixed with product iii (200 ng) and subjected to a similar serial two-step amplification to generate the desired gene fragment.
-
b)
scFv#R2-1. PCR was performed using the scFv#M2nd gene13 as template with R1 and F3 primers. The product obtained was used as reverse mega-primer (MP) in the next PCR, in combination with the E2-VL-For-2 primer12, to generate the desired gene fragment.
-
c)
scFv#4mut. PCRs were performed using the scFv#WT gene12,13 as template (Supplementary Figure S4B). The VH-portion gene was prepared by amplification with E2-VH-Rev and F4 primers. The VL-portion gene was prepared as follows. First, PCR was performed using R5 and F5 primers. The product obtained was gel-purified and used as reverse MP1 in the next PCR, in combination with the E2-VL-For-2 primer. The product was then used similarly as forward MP2 with the E2-VL-Rev primer12 to generate the VL-portion gene fragment with 5′-end sequence that was complementary to the 3′-end sequence of the VH-portion gene fragment. These VH- and VL-portion gene fragments (each 200 ng) were mixed and submitted to overlap-extension PCR as described above (see entry a) to generate the desired gene fragment.
-
d)
scFv#R3-1. PCRs were performed using the scFv#WT gene as template. The VL-portion gene was prepared as follows. First, PCR was performed using R4 and MP1 primers. The product obtained was gel-purified and used as MP3, which was submitted to overlap-extension PCR with MP2 as described above (see entry a) to generate the gene fragment containing whole the VL with a portion of the 3′-side of VH (extending over VH-CDR3). On the other hand, a gene fragment containing the VH-portion was amplified using E2-VH-Rev and E2-VH-For primers. These two gene fragments were combined by the overlap-extension PCR to generate the desired gene fragment.
-
e)
scFv#R4-3. PCR was performed using the scFv#WT gene as template with R2 and F5 primers. The product obtained was digested with BamH I and Sal I to generate a gene fragment covering the VL with the mutation that substitutes the VL77 residue, whereas the plasmid having scFv#4mut gene was digested with BamH I and Sal I and gel-purified to remove the corresponding gene fragment. The VL77-mutated gene fragment was ligated into the digested plasmid to construct the desirable gene as the form already incorporated in the plasmid.
-
f)
scFv#R5-1. The desired gene fragment was generated by PCR using the scFv#WT gene as template with an MP (the VH-portion gene fragment prepared in entry c) and the E2-VL-For-2 primer.
Determination of the scFv K a values
-
a)
Scatchard analysis28. Mixtures of [1, 2, 6, 7-3H]-E2 (3.53 TBq/mmol; PerkinElmer) (~250 Bq), varying amounts of standard E2, and a constant amount of each scFv (adjusted to bind to ~50% of the tritium-labeled E2) were incubated in G-PBS (500 μL) at 4 °C for 240 min. The bound (B) and free (F) fractions were separated using a dextran-coated charcoal method, and the radioactivity of the B fraction was measured.
-
b)
SPR analysis. The kinetic parameters of selected scFvs, which were affinity-purified with anti-FLAG-M2 agarose (Sigma–Aldrich)13, to the E2–bovine serum albumin (BSA) conjugate (prepared according to the previous method12: E2/BSA molar ratio was determined to be 7) immobilized on the CM5 sensor chip (using an Amine Coupling) were determined with Biacore T200 SPR system (GE Healthcare). Kinetic measurements were carried out at 25 °C in G-PBS with a constant flow rate of 30 μL/min. In the measurements, five different concentrations (0.16–10 μg/mL) of the purified scFvs were used. The kinetic evaluation of data was performed using Biacore T200 evaluation software (GE Healthcare).
ELISA
The 96-well microplates (#3590; Corning) coated with the E2–BSA conjugate (see above) were incubated at 4 °C for 240 min with a mixture of E2 standard (50.0 μL/well) and soluble scFv protein (100 μL/well), both diluted in PVG-PBS. The microplates were washed 3 times with T-PBS and probed with an anti-FLAG M2 antibody labeled with peroxidase (POD) (Sigma–Aldrich) diluted in G-PBS (0.20 μg/mL; 100 μL/well)12,13,14,15,16,17. After incubation at 37 °C for 30 min, the microplates were washed similarly and the captured POD activity was determined colorimetrically (490 nm), as described previously12,13,14,15,16,17. To construct the ELISA dose–response curves, Image J software48 (NIH) was used for curve fitting and determining the reaction parameters. The midpoint (i.e., IC50) values were derived from a four parametric logistic equation [log(analyte dose) vs. B/B0(%)] as the EC50 values. The unit “X g/assay” was used in the abscissa, which refers to the total mass (X g) of analyte that was added to each assay chamber (microwell) for the competitive antigen–antibody reactions.
Data availability
The data sets generated during the current study are available from the corresponding author upon request.
References
Gibney, E., Noorden, R. V., Ledford, H., Castelvecchi, D. & Warren, M. Nobel for test-tube evolution. Nature 562, 176 (2018).
Handbook of therapeutic antibodies (ed Dübel, S.) (Wiley-Blackwell, 2004).
Bradbury, A. R. M., Sidhu, S., Dübel, S. & McCafferty, J. Beyond natural antibodies: the power of in vitro display technologies. Nat. Biotechnol. 29, 245–254 (2011).
Chiu, M. L. & Gilliland, G. L. Engineering antibody therapeutics. Curr. Opin. Struct.Biol. 38, 163–173 (2016).
Kennedy, P. J., Oliveira, C., Granja, P. L. & Sarmento, B. Monoclonal antibodies: technologies for early discovery and engineering. Crit. Rev. Biotechnol. 38, 394–408 (2018).
Skerra, A. & Plückthun, A. Assembly of a functional immunoglobulin Fv fragment in Escherichia coli. Science 240, 1038–1041 (1988).
Bird, R. E. et al. Single-chain antigen-binding proteins. Science 242, 423–426 (1988).
Smith, G. P. & Petrenko, V. A. Phage display. Chem. Rev. 97, 391–410 (1997).
Phage display (eds Clackson, T. & Lowman, H. B.) (Oxford University Press, 2004).
Plückthun, A. Ribosome display: a perspective. Methods Mol. Biol. 805, 3–28 (2012).
Gai, S. A. & Wittrup, K. D. Yeast surface display for protein engineering and characterization. Curr. Opin. Struct. Biol. 17, 467–473 (2007).
Kobayashi, N. et al. Anti-estradiol-17β single-chain Fv fragments: generation, characterization, gene randomization, and optimized phage display. Steroids 73, 1485–1499 (2008).
Kobayashi, N. et al. Two-step in vitro antibody affinity maturation enables estradiol-17β assays with more than 10-fold higher sensitivity. Anal. Chem. 82, 1027–1038 (2010).
Oyama, H., Yamaguchi, S., Nakata, S., Niwa, T. & Kobayashi, N. “Breeding” diagnostic antibodies for higher assay performance: a 250-fold affinity-matured antibody mutant targeting a small biomarker. Anal. Chem. 85, 4930–4937 (2013).
Oyama, H. et al. One-shot in vitro evolution generated an antibody fragment for testing urinary cotinine with more than 40-fold enhanced affinity. Anal. Chem. 89, 988–995 (2017).
Oyama, H. et al. A single-step “breeding” generated a diagnostic anti-cortisol antibody fragment with over 30-fold enhanced affinity. Biol. Pharm. Bull. 40, 2191–2198 (2017).
Morita, I. et al. Antibody fragments for on-site testing of cannabinoids generated via in vitro affinity maturation. Biol. Pharm. Bull. 40, 174–181 (2017).
Leung, D. W., Chen, E. & Goeddel, D. V. A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. Technique 1, 11–15 (1989).
Boder, E. T., Midelfort, K. S. & Wittrup, K. D. Directed evolution of antibody fragments with monovalent femtomolar antigen-binding affinity. Proc. Natl. Acad. Sci. USA 97, 10701–10705 (2000).
Barderas, R., Desmet, J., Timmerman, P., Meloen, R. & Casal, J. I. Affinity maturation of antibodies assisted by in silico modeling. Proc. Natl. Acad. Sci. USA 105, 9029–9034 (2008).
Lippow, S. M., Wittrup, K. D. & Tidor, B. Computational design of antibody affinity improvement beyond in vivo maturation. Nat. Biotechnol. 25, 1171–1176 (2007).
Sormanni, P., Aprile, F. A. & Vendruscolo, M. Third generation antibody discovery methods: in silico rational design. Chem. Soc. Rev. 47, 9137–9157 (2018).
Cannon, D. A. et al. Experimentally guided computational antibody affinity maturation with de novo docking, modelling and rational design. PLoS Comput. Biol. 15, e1006980 (2019).
Knappik, A. & Plückthun, A. An improved affinity tag based on the FLAG® peptide for the detection and purification of recombinant antibody fragments. Biotechniques 17, 754–761 (1994).
Kabat, E. A., Wu, T. T., Perry, H. M., Gottesman, K. S. & Foeller, C. Sequences of Proteins of Immunological Interest. U.S. Department of Health and Human Services, National Institutes of Health (U.S. Government Printing Office, 1991).
Wu, T. T., Johnson, G. & Kabat, E. A. Length distribution of CDRH3 in antibodies. Proteins: Struct. Funct. Genet. 16, 1–7 (1993).
Collis, A. V. J., Brouwer, A. P. & Martin, A. C. R. Analysis of the antigen combining site: correlations between length and sequence composition of the hypervariable loops and the nature of the antigen. J. Mol. Biol. 325, 337–354 (2003).
Scatchard, G. The attractions of proteins for small molecules and ions. Ann. N. Y. Acad. Sci. 51, 660–672 (1949).
Hosoda, H., Kobayashi, N. & Nambara, T. Effect of bridge heterologous combination on sensitivity in enzyme immunoassay for 11-deoxycortisol. Chem. Pharm. Bull. 31, 953–958 (1983).
Hosoda, H., Kobayashi, N., Ishii, N. & Nambara, T. Bridging phenomena in steroid immunoassays. The effect of bridge length on sensitivity in enzyme immunoassay. Chem. Pharm. Bull. 34, 2105–2111 (1986).
Hoogenboom, H. R., Marks, J. D., Griffiths, A. D. & Winter, G. Building antibodies from their genes. Immunol. Rev. 130, 41–68 (1992).
Barbas, C. F. III, Bain, J. D., Hoekstra, D. M. & Lerner, R. A. Semisynthetic combinatorial antibody libraries: a chemical solution to the diversity problem. Proc. Natl. Acad. Sci. USA 89, 4457–4461 (1992).
Griffiths, A. D. et al. Isolation of high affinity human antibodies directly from large synthetic repertoires. EMBO J. 13, 3245–3260 (1994).
Valjakka, J. et al. Crystal structure of an in vitro affinity- and specificity-matured anti-testosterone Fab in complex with testosterone. J. Biol. Chem. 277, 44021–44027 (2002).
Fellouse, F. A. et al. Molecular recognition by a binary code. J. Mol. Biol. 348, 1153–1162 (2005).
Oliva, B., Bates, P. A., Querol, E., Avilés, F. X. & Sternberg, M. J. E. Automated classification of antibody complementarity determining region 3 of the heavy chain (H3) loops into canonical forms and its application to protein structure prediction. J. Mol. Biol. 279, 1193–1210 (1998).
Angelo, S. D. et al. Many routes to an antibody heavy-chain CDR3: necessary, yet insufficient, for specific binding. Front. Immunol. 9, 395 (2018).
Kobayashi, N. & Oyama, H. Antibody engineering toward high-sensitivity high-throughput immunosensing of small molecules. Analyst 136, 642–651 (2011).
Dennis, M. S. & Lowman, H. B. Phage selection strategies for improved affinity and specificity of proteins and peptides. In Phage display (Ref. 9).
Derda, R. et al. Diversity of phage-displayed libraries of peptides during panning and amplification. Molecules 16, 1776–1803 (2011).
Chothia, C. et al. Conformations of immunoglobulin hypervariable regions. Nature 342, 877–883 (1989).
Al-Lazikani, B., Lesk, A. M. & Chothia, C. Standard conformations for the canonical structures of immunoglobulins. J. Mol. Biol. 273, 927–948 (1997).
Sharon, J. Structural correlates of high antibody affinity: three engineered amino acid substitutions can increase the affinity of an anti-p-azophenylarsonate antibody 200-fold. Proc. Natl. Acad. Sci. USA 87, 4814–4817 (1990).
Schildbach, J. F. et al. Altered hapten recognition by two anti-digoxin hybridoma variants due to variable region point mutations. J. Biol. Chem. 266, 4640–4647 (1991).
Vargas-Madrazo, E., Lara-Ochoa, F. & Almagro, J. C. Canonical structure repertoire of the antigen-binding site of immunoglobulins suggests strong geometrical restrictions associated to the mechanism of immune recognition. J. Mol. Biol. 254, 497–504 (1995).
Siegel, R. W., Baugher, W., Rahn, T., Drengler, S. & Tyner, J. Affinity maturation of tacrolimus antibody for improved immunoassay performance. Clin. Chem. 54, 1008–1017 (2008).
Jirholt, P., Ohlin, M., Borrebaeck, C. A. K. & Söderlind, E. Exploiting sequence space: shuffling in vivo formed complementarity determining regions into a master framework. Gene 215, 471–476 (1998).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Oyama, H., Yamaguchi, S., Nakata, S., Niwa, T. & Kobayashi, N. Correction to “Breeding” diagnostic antibodies for higher assay performance: a 250-fold affinity-matured antibody mutant targeting a small biomarker. Anal. Chem. 91, 15325 (2019).
Guex, N., Diemand, A. & Peitsch, M. C. Protein modelling for all. Trends Biochem. Sci. 24, 364–367 (1999).
Grosdidier, A., Zoete, V. & Michielin, O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 39, W270–W277 (2011).
DeLano, W. The PyMOL Molecular Graphics System; DeLano Scientific LLC: Palo Alto, CA, USA, 2008. Available at, http://www.pymol.org.
Acknowledgements
We thank Dr. Eskil Söderlind (Avena Partners AB, Sweden) and Professor Carl A.K. Borrebaeck (Lund University, Sweden) for providing the pEXmide 5 vector.
Author information
Authors and Affiliations
Contributions
N.K. conceived the project; N.K. and H.O. designed the research; H.O., Y. K., I.M., C.Y., Y.H., M.T., T.T., Y.E., Y.T., H.H. and A.T. performed the research and analyzed the data; N.K. and H.O. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Oyama, H., Kiguchi, Y., Morita, I. et al. Seeking high-priority mutations enabling successful antibody-breeding: systematic analysis of a mutant that gained over 100-fold enhanced affinity. Sci Rep 10, 4807 (2020). https://doi.org/10.1038/s41598-020-61529-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-61529-7
This article is cited by
-
Development of an inhibiting antibody against equine interleukin 5 to treat insect bite hypersensitivity of horses
Scientific Reports (2023)
-
The VH framework region 1 as a target of efficient mutagenesis for generating a variety of affinity-matured scFv mutants
Scientific Reports (2021)
-
Clonal array profiling of scFv-displaying phages for high-throughput discovery of affinity-matured antibody mutants
Scientific Reports (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.