Abstract
The bronze bug, Thaumastocoris peregrinus, an Australian native insect, has become a nearly worldwide invasive pest in the last 16 years and has been causing significant damage to eucalypts (Myrtaceae), including Eucalyptus spp. and Corymbia spp. Its rapid expansion leads to new questions about pathways and routes that T. peregrinus used to invade other continents and countries. We used mtDNA to characterize specimens of T. peregrinus collected from 10 countries where this species has become established, including six recently invaded countries: Chile, Israel, Mexico, Paraguay, Portugal, and the United States of America. We then combined our mtDNA data with previous data available from South Africa, Australia, and Europe to construct a world mtDNA network of haplotypes. Haplotype A was the most common present in all specimens of sites sampled in the New World, Europe, and Israel, however from Australia second more frequently. Haplotype D was the most common one from native populations in Australia. Haplotype A differs from the two major haplotypes found in South Africa (D and G), confirming that at least two independent invasions occurred, one from Australia to South Africa, and the other one from Australia to South America (A). In conclusion, Haplotype A has an invasion success over many countries in the World. Additionally, analyzing data from our work and previous reports, it is possible to suggest some invasive routes of T. peregrinus to predict such events and support preventive control measures.
Similar content being viewed by others
Introduction
The eucalypts represent over 20% of commercial tree plantations worldwide1 and are also common in many landscapes. However, in recent years the significant expansion of planted areas has also increased the risk of introduction of invasive pest species2,3. From 1870 to 2014, at least 42 eucalypt insect pests were introduced outside their native ecosystems, notably of the order Hemiptera (17 species)1. Among the various invasive pests of eucalypts, the bronze bug, [Thaumastocoris peregrinus Carpintero & Dellapé 2006 (Hemiptera: Thaumastocoridae)]4, has become the most widespread. In areas of recent colonization, T. peregrinus caused significant economic and esthetic damage to eucalypt trees. Symptoms of the insect's attack include leaf discoloration ranging from chlorosis to silver, bronze, and finally brown at defoliation, decreasing photosynthetic area and leading to the death of young trees5,6,7,8.
Thaumastocoris peregrinus was firstly detected as invasive in South Africa in the year 20039 and shortly thereafter in Argentina in 20054,10. A few years later, it was found in other African and South American countries, and later also in North America, Europe, and Israel (Table 1). The EPPO (European Plant Protection Organization) added T. peregrinus to the EPPO alert list in 2012, due to its invasive behavior and its potential damage to eucalypt trees11.
Increasing economic globalization has facilitated the dispersal of T. peregrinus and other invasive pests1. The intensification in intercontinental goods and passenger transport has significantly contributed to the introduction of invasive insects12,13,14. DNA barcoding can support studies of invasive pests and its possible routes of invasion as well as provide accurate species identification. Furthermore, analysis of the variability within a standard DNA barcode region can also aid determining potential origin and pattern of dispersal of an invasive pest15,16,17,18. Previous analysis of mtDNA (partial COI region) from South African and South American populations of T. peregrinus revealed the presence of three haplotypes and more than one route of dispersal from Australia to Africa and South America were proposed16. The molecular characterization of T. peregrinus populations in the newly invaded areas was lacking. Thus, we sequenced part of the mitochondrial gene of Cytochrome C Oxidase subunit I (COI) of T. peregrinus collected in different countries. Possible invasion routes for each country, based on data from the literature and collected in the present work, are analyzed and discussed.
Material and Methods
Sampling procedure
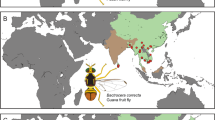
Specimens of T. peregrinus were randomly collected from Eucalyptus trees in five countries in South America (Argentina, Brazil, Chile, Paraguay, and Uruguay), two in North America (the USA and Mexico), one in the Middle East (Israel) and two in Europe (Italy and Portugal) (Fig. 1). At each site, adults of T. peregrinus were manually detached from branches and/or leaves using a paintbrush or aspirator and placed into 1.5 mL vials containing 98% ethanol to preserve their DNA. Approximately 300 specimens of T. peregrinus were collected at 28 sampling sites from November 2016 to May 2018. Each vial was labeled with the municipality, date, and coordinates of the collection site. Vials from all sample collection sites were sent to the Biotechnology Laboratory of the Federal University of Santa Maria, Santa Maria, RS, Brazil, where they were stored at −20 °C until genomic DNA extraction.
DNA extraction, PCR amplification, and COI-gene sequencing
The identification of all T. peregrinus specimens was confirmed based on morphological characters4. DNA extraction was performed individually for each specimen using the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Depending on the availability of insects and the proximity of sampling sites, we used 1 to 10 insects per location for DNA extraction (Table 2). Each adult insect was removed from the vial with ethanol and left to air dry on a paper towel. The entire body was individually macerated in a 1.5 mL tube containing 180 µL of buffer ATL and 20 µL of proteinase K and incubated at 56 °C for 12 hours. Subsequently, genomic DNA was purified in a silica-based matrix and eluted in 35 µL of buffer AE. The concentration of DNA was assessed in a spectrophotometer (NanoDrop™ 1000, Thermo Scientific, Wilmington, DE, USA).
A fragment of the mitochondrial COI gene (468 bp) was amplified from 116 individuals through polymerase chain reaction (PCR) using the primers Tp2390F (5′ACCCGAGCATACTTTACTTC) and Tp2937R (5′ATTGTGGCTCGTTTTGATA)16. Each PCR reaction was performed with a final volume of 12 µL composed by 1.25 µL of JumpStartTM 10X reaction buffer; 0.625 µL de dNTP mix (10 nM of each); 1.0 µL of each primer (10 pM), 0.220 µL of JumpStartTM DNA Polymerase (2.5 U/µL) (Sigma-Aldrich, St. Louis, MO, USA); 1 µL de template DNA (05–100 ng/µL) and 7.0 µL of ultra-pure water.
PCR amplification consisted of an initial denaturation step at 98 °C for 30 s, followed by 30 cycles at 95 °C for 30 s, 48 °C for 30 s and 72 °C for 1.5 min, and a final extension at 72 °C for 10 min16. Amplified products were resolved on 1.0% agarose electrophoresis gel, pre-stained with Nancy-520 DNA gel stain (Sigma-Aldrich) and visualized using a gel documentation system. Successfully amplified PCR products were sequenced by ACTGene Molecular Analyses (Alvorada, RS, BR), using the BigDye Terminator method on an ABI 3500 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA).
Data analysis
Quality assessment, trimming, editing, and analysis of each DNA sequence were performed using the software Pregap and Gap4 within the Staden package19. CLC Sequence Viewer (Version 7.8.1-QIAGEN Aarhus A/S) was used to retrieve and align sequences with a length of 468 bp. A pBLAST analysis (amino acid homology confirmation) with T. peregrinus partial mtDNA COI genes deposited on the NCBI (National Center for Biotechnology Information, USA) database was also performed.
The 116 COI sequences generated in this study were combined with 197 sequences of 16 populations from Australia16,20. We also used 32 sequences from South America, 62 from South Africa16, 1 from Italy (KF437485)21, 10 from Spain (MN401749)22, and 5 from Reunion Island (France) (GenBank − KT273623, KT273624, KT273625, KT273626, and KT273627)23. The inference and visualization of genetic relationships among intraspecific sequences used to generate a haplotype network were conducted using TCS network24 within the program PopART25.
Results
Fragments of mtDNA from 116 individuals of T. peregrinus from 28 populations were amplified and sequenced, resulting in a trimmed sequence of 468 pb of the COI gene. Samples of T. peregrinus from six countries (Chile, Paraguay, Mexico, Portugal, Israel, and the USA) were sequenced for the first time. For all the sampled sites from South America (Chile, Paraguay, Argentina, Brazil, and Uruguay), North America (Mexico and the USA) and the Mediterranean basin (Italy, Portugal, and Israel), we confirmed the presence of a unique haplotype (haplotype A).
Combining our data with sequences not deposited in GenBank20 and with others previously deposited in this database (Table 2), we obtained a total of 423 T. peregrinus COI sequences and 45 unique haplotypes were identified. The proportions of the different haplotypes among the countries are presented on a haplotype network (Fig. 2). This network reveals that the greatest diversity of haplotypes is from the native region of T. peregrinus in Australia. Haplotype D is the ancestral one and all the other haplotypes radiate from it (Fig. 2). Haplotype D was the most frequent haplotype in Australia and found in 10 sites20. Haplotype A was present in three of 10 sites where haplotype D was also reported: Sydney, Coonabarabran, and Cootamundra20. The difference between the two haplotypes is only one base pair in the partial mtDNA COI fragment (468 bp)16 and possibly represents a silent mutation.
Mitochondrial haplotype network. Circle sizes are in approximate proportion to haplotype frequencies. Each mark represents one nucleotide difference. Countries: Australia (AU); Reunion Island (RE); Argentina (AR); Brazil (BR); Chile (CL); Paraguay (PY); Uruguay (UY); Israel (IL); Mexico (MX); United States of America (US); Portugal (PT); Spain (ES); Italy (IT); South Africa (SA).
Outside of Australia, only four haplotypes were found: D and G in South Africa16, IT in Italy21, and A in South America16, North America, Europe, and Asia. From our work, haplotype A was the most frequent and widespread, found at 28 sites (116 sequences), in three populations from Australia (48 sequences)20, six populations in South America (32 sequences)16 and one population in Spain (10 sequences)22, comprising a total of 206 sequences (Fig. 2). Based on the combined sequences, haplotype D was found in Australia (67 sequences)16,20, Reunion Island (five sequences)23, and in nearly all the sites sampled in South Africa (46 sequences)16, making a total of 118 sequences. The third and fourth most frequent haplotypes were G and H, 31 sequences (16 for South Africa and 15 for Australia) of G and 14 sequences (all in Australia) of H, totaling 45 sequences16,20. All the remaining haplotypes were represented by three or fewer individuals. We note that all haplotypes found in the invaded regions were also found in their naturally occurring regions in Australia, except for the IT haplotype, which was found only in Italy (Genbank KF437485)21 (Fig. 2). To understand and to have a wide view of these results properly, the Fig. 3 shows the haplotype A distribution at our sampling sites in Americas, Europe and Israel (10 countries). In addition, the Fig. 4 represent a world haplotype distribution from 14 countries and 423 sequences.
Haplotype distribution of T. peregrinus in 14 countries. Australia (AU); Reunion Island (RE); Argentina (AR); Brazil (BR); Chile (CL); Paraguay (PY); Uruguay (UY); Israel (IL); Mexico (MX); United States of America (US); Portugal (PT); Spain (ES); Italy (IT); South Africa (SA). The size of pie charts is relative to the number of individuals sequenced at each locality.
Discussion
Genetic diversity
Our study updates information on the genetic diversity and mtDNA haplotype distribution of T. peregrinus worldwide. Samples from several locations in seven countries, including newly invaded countries in South America (Chile and Paraguay), North America (the USA and Mexico) and the Mediterranean Basin (Portugal, Italy, and Israel) were collected and analyzed. We also combined our mtDNA data with previously available sequences of T. peregrinus16,20,21,22,23 (Table 1). Our results demonstrate that haplotype A is predominant in South America, North America, Europe, and Israel, indicating a low genetic diversity of T. peregrinus outside its native region.
In contrast with the low genetic diversity found in the invaded areas, the Australian populations of T. peregrinus (the native range of the species) revealed a high mtDNA COI haplotype diversity16,20. Genetic diversity is expected to be higher in a species native range in comparison with invasive populations26, possibly because of bottleneck effects during the invasion process. The newly introduced species might be reduced to a few individuals because of environmental events (e.g. hurricane, forest fires) and anthropic actions, resulting in low genetic diversity27. Furthermore, genetic drift and the founder effect might also account for the low genetic diversity of the new invasions28. Selection pressure on a given population of the invasive species, that allow the survival of the fittest insects, might result in low genetic diversity during the first decades of species establishment29. These evolutionary forces associated with the adaptability of haplotype A to different environments seem to have driven the expansion of T. peregrinus worldwide, compared to the haplotypes D and G.
Low genetic diversity has also been reported for many other forest invasive pests. For example, populations of Anoplophora glabripennis (Motschulsky) (Coleoptera: Cerambycidae)30, Leptocybe invasa Fisher & La Salle, 2004 (Hymenoptera: Eulophidae)31,32, Solenopsis invicta Buren, 1972 (Hymenoptera: Formicidae)33, Solenopsis geminata (Fabricius, 1804) (Hymenoptera: Formicidae)34, and Sirex noctilio Fabricius, 1793 (Hymenoptera: Siricidae)35 demonstrated low genetic diversity in areas where these species are considered invasive. In a new environment, invasive populations can take several years to increase their genetic diversity36,37.
Possible invasion routes of T. peregrinus in South America, Europe, North America, and Asia
Knowledge of possible ways by which an insect can invade a new area and about its biology is essential to identify routes of invasion and to develop and adopt effective strategies to prevent and/or manage additional introductions1,38. Possible routes of invasion of eucalypts pests include active dispersion, passive transport by biotic and abiotic agents, or accidental transport39. Even though our data are limited to a partial mtDNA COI fragment (468 bp) from T. peregrinus, an invasive range can be projected using mtDNA which possesses a unique haplotype signature of a population as stated by Goldstein et al.40. The mtDNA is inherited most often only from the mother and does not undergo recombination41, enabling to assess the number of genetically unique founder females42,43,44,45. Studies of invasive species used similar methodologies, for example of T. peregrinus16, wood-boring beetles46, Melanagromyza sojae (Zehntner, 1900) (Diptera: Agromyzidae)47 and Helicoverpa armigera (Hübner, 1808) (Lepidoptera: Noctuidae)48.
Using data collected in our work and from the literature, we suggest nine possible invasion routes of T. peregrinus (Fig. 5). These conclusions are based on (1) spatio-temporal events; (2) previous report of invasion routes of each country; (3) movement of people, goods, and timber through motorways and waterways between countries; and (4) our findings that analyzed 116 sequences from specimens collected in countries in South and North Americas, Europe, and Asia.
Routes 1 and 2: In 2003, T. peregrinus4 was firstly reported in South Africa, erroneously identified as Thaumastocoris australicus Kirkaldy9. Years later, a study using DNA barcoding in specimens of T. peregrinus from South Africa revealed the presence of two haplotypes, D (route 1) and G (route 2), indicating that the introduction happened in two distinct invasion events, both aided by transport of people and goods primarily through airline with flights from Australia (Sydney)16. Further studies should update the T. peregrinus haplotypes present in Africa because its occurrence has increased throughout the continent, possibly as a consequence of new invasions or further dispersal of populations already present in the invaded area.
Route 3: T. peregrinus was first reported in the Americas in Argentina in 20054. This invasion was an independent event of haplotype A directly from Australia and possibly from Sydney since the haplotype was different from that of the South African invasion16. Geographical proximity, land border, surface transport of people and goods, and the presence of extensive eucalypt plantations alongside highways margins in Argentina, Brazil, Paraguay, and Uruguay facilitated the spread of this invasive pest through South America. The monitoring data of T. peregrinus in Brazil suggest its entrance via airport8. In Chile, the invasion of T. peregrinus was thought to be passive and was spread by different means, including trucks, buses and ships, and other vectors, mainly birds49,50. Natural dispersion by flight is also possible due to Argentina's proximity to other countries.
The geographical position of a country can influence and help to predict the number of invasive insects, which is related to the length and nature of borders with neighboring countries and correlations among land area, population, and gross domestic product1,51. However, some factors may be more likely than others, as is the case of eucalypt pest introductions into New Zealand from Australia, probably due to the proximity and frequent trade between the two countries1,52.
Route 4: In Europe, the first reported occurrence of T. peregrinus was in Italy in 2011, possibly introduced through the transport of timber from South America and South Africa53. However, this hypothesis was not based on the population genetic signatures. In 2014, mtDNA molecular characterization of T. peregrinus specimens from Italy was performed, and a haplotype called IT was identified21. Strangely, haplotype IT has not been reported from Australia and several factors might offer an explanation, including a not yet detected haplotype with unusual low frequency in Australia, or a new haplotype restricted to Italy or from an undetermined population along the invasive routes between Australia and Italy. Additionally, our results confirm the presence of haplotype A in Italy, suggesting an invasive pathway from South America because of wood transport between the two countries. This scenario is more likely than an introduction from South Africa as previously reported53. Nonetheless, based on the presence of haplotype A in Portugal and recently in Spain22, we also hypothesize dispersion and incursions from many of these countries into Italy because of the short geographical distance, the land border, and the motorway connections between countries (Portugal, Spain, France, and Italy), and intense movement of tourists and goods.
Route 5: The second country in Europe to report an invasion of T. peregrinus was Portugal in 201254. Based on South America's status as the main supplier of eucalypts wood to Europe55,56, South America has been suggested as a route of invasion of T. peregrinus into Europe54. Also, the genetic signature of the sampled population in Portugal revealed that it is most likely an arrival from South America. The latest European countries to report T. peregrinus were Spain (2014)57, Albania (2016)58, and Greece (2017)59. The genetic diversity in these countries remains unknown, except for Spain, where haplotype A was reported in Madrid, with two main invasion routes being suggested: either an invasion from South America to Portugal and then an expansion to Spain; or that T. peregrinus invaded Spain from South America22.
For other European countries, we suggest that the bronze bug spread within the Mediterranean countries similarly to other invasive species that feed on eucalyptus trees. Possible invasion routes of G. brimblecombei into Europe were suggested from South or North America, considering the Spatio-temporal events and the presence of this insect in the New World since the 20th century60. Subsequent events demonstrated the spread of G. brimblecombei to Mediterranean countries61. The connections between countries, mainly motorways and waterways linking Portugal, Spain, France, Italy, Greece, Algeria, and Morocco, were suggested as the main invasion routes of this pest62.
Route 6: The invasion of T. peregrinus to Reunion Island (2013)23 may have resulted from the flow of people and goods and the island's proximity to South Africa, where haplotype D is predominant16. However, a direct introduction from Australia is also possible based on distance and the presence of haplotype D in the site of origin of this species.
Route 7: The invasion of T. peregrinus into Israel possibly occurred from a country with the bordering to the Mediterranean Sea, where this species was already present, based on connections of motorways and waterways and the intense movement of people and goods between countries60,62. The recent expansion of eucalypts forestry in Asia could also influence the occurrence and detection of pests1, including the spread of T. peregrinus soon into new countries in Asia.
Routes 8 and 9: In North America, the first report of T. peregrinus was in Mexico in 201563, ten years after its detection in Argentina. In 2016, this pest was found in the USA, in California in the greater Los Angeles area, specifically north Hollywood in the San Fernando Valley64,65, and Heartwell Park in Long Beach65. The first detection event might not be the actual first incursion and the establishment of a pest. Therefore, because of the short distance and the short period of this pest’s detection between Mexico and California (USA), it is impossible to know definitely in which country of North America T. peregrinus first entered. However, the presence of the same haplotype A in all areas of the Americas, suggests that a single invasion starting from South America and spreading northward to California might have occurred. Nevertheless, as before, Australia should also be considered as a source of T. peregrinus in North America because of the presence of haplotype A.
In general, in the USA, two invasion routes are considered dominant in the introduction of insects and diseases. Most harmful, non-native species (62%) likely entered North America with live plants and 30% probably arrived with wood packaging material (WPM) or other wood products66. In 2017, several specimens of T. peregrinus were found on Eucalyptus twigs in a shipment of cut flowers from Mexico that was intercepted at the port of Brownsville (Texas). Because T. peregrinus had quarantine status, the cut flowers returned to Mexico as a safety measure67. The twin ports of Los Angeles and Long Beach are the busiest in the USA and the international airports of San Francisco and Los Angeles are main entry points for passengers and air cargo68; all serve as gateways for pest introductions.
Our mtDNA analysis, including temporal and geographical sequence data, provides information that suggested scenarios of invasion routes of T. peregrinus in South America, North America, Europe, and Asia. These scenarios are an important source of information that assists in the planning and management of biosafety measures to prevent pests like T. peregrinus, from invading and colonizing new sites. Further studies using nuclear markers (e.g. microsatellites) or other mtDNA regions can aid to postulate new scenarios.
Factors involved in the successful invasion and establishment of T. peregrinus
Several insect pests of eucalypts have undergone dispersal to two or more continents, for example, L. invasa, Gonipterus spp., and Glycaspis brimblecombei (Moore, 1964) (Hemiptera: Aphalaridae). Evidence exists that these invasive species possess and exploit mechanisms to aid in their dispersal1. In the specific case of T. peregrinus, adults do not show a strong natural ability to disperse except when aided by human activities20. However, nymphs are particularly mobile8 and could be more easily dispersed. Besides, biological factors account for the success of many invasive insects into a new location69, and we believe that certain biological characteristics of T. peregrinus represent an important portion of its invasion success throughout more than 20 countries. The life cycle of T. peregrinus is about 60 days and has a rather high potential for breeding (fecundity of 60 eggs/female) conditioning to reproduce more than twice a year and increase its population density50,70. Once this pest invades a new location, the lack of natural enemies facilitates its establishment and dispersion71 into the wide range of host trees of Eucalyptus and Corymbia (over 52 host species) found in many countries in the world72. T. peregrinus also thrives under a wide range of temperatures, varying from 4 °C to 34 °C, which explains its wide distribution and possible invasion into new areas in the future73. Its invasion success is also related to the small body size (± 3–4 mm)9 that facilitates its dispersal over long distances, whether actively or passively by the wind (draft)74 or by the movement of people or any kind of goods, especially timber, where is very difficult to be detected due to its small size.
Successful invaders are those featuring a small body size, several generations per year, long flight displacement and low incidence of diapause69. All these traits account for the rapid adaptation to new environmental conditions75. For example, the successful invasion of L. invasa into new areas was suggested to be a consequence of this pest's high dispersal capabilities32, strong resistance to low temperatures, the presence of eucalypt plantations worldwide76, and a lack of natural enemies in the invaded areas77. Moreover, synergic interactions among invasive species have been reported as a “meltdown” process that accelerates the impact of these species in the invaded area78. In line with this theory, both the eucalyptus lerp psyllid G. brimblecombei and T. peregrinus display a positive interaction that benefits from the preference of bronze bug females to oviposit on leaves bearing lerps79.
Considering the invasion success of haplotype A worldwide reported in this work and that the most common maternal lineages of T. peregrinus sampled in Australia, specifically in Sydney from 2001 to 2009, were haplotypes A, D, G and H16,20, the most likely source of T. peregrinus seems to be Sydney16,20 or Chelmsford20 for haplotypes A and D. The most commons haplotypes in a given population are more likely to be sampled and eventually to spread16,43,80,81, as it seems to have happened to haplotype A of T. peregrinus. On the other hand, haplotypes D, G and H were found restricted to some areas or not found outside its place of origin, although individuals might have dispersed to new areas but did not adapted and successfully establish as haplotype A.
The rate of invasion by insect pests in eucalypt plantations has increased approximately fivefold since the 1980s1. During our sampling of T. peregrinus specimens in over 28 locations nearly worldwide, we found that this insect was present in eucalypt trees along highways, and in parks, urban forests and landscapes, and commercial plantations. The growing flow of people among countries using diverse ways of transport and the unfamiliarity concerning the risks of transporting plant materials, either within or outside their natural areas, favor this invasive process.
References
Hurley, B. P. et al. Increasing numbers and intercontinental spread of invasive insects on eucalypts. Biol. Invasions. 18, 921–933, https://doi.org/10.1007/s10530-016-1081-x (2016).
Paine, T. D., Steinbauer, M. J. & Lawson, S. A. Native and exotic pests of Eucalyptus: a worldwide perspective. Annu. Rev. Entomol. 56, 11–20, https://doi.org/10.1146/annurev-ento-120709-144817 (2011).
Branco, S., Videira, N., Branco, M. & Paiva, M. R. A review of invasive alien species impacts on eucalypt stands and citrus orchards ecosystem services: towards an integrated management approach. J. Environ. Manag. 149, 17–26, https://doi.org/10.1016/j.jenvman.2014.09.026 (2015).
Carpintero, D. L. & Dellapé, P. M. A new species of Thaumastocoris Kirkaldy from Argentina (Heteroptera: Thaumastocoridae: Thaumastocorinae). Zootaxa 1228, 61–68 (2006).
Button, G. Thaumastocoris peregrinus. NCT Forestry Co-operative Limited. http://www.nctforest.com/home.php?nav=pubs&pubtypeid=1&archive=1 (2007).
Soliman, E. P. et al. Biology of Thaumastocoris peregrinus in different eucalyptus species and hybrids. Phytoparasitica 40, 223–230, https://doi.org/10.1007/s12600-012-0226-4 (2012).
Wilcken, C. F. Percevejo bronzeado do eucalipto (Thaumastocoris peregrinus) (Hemiptera: Thaumastocoridae): ameaça às florestas de eucalipto brasileiras. IPEF Programa de proteção florestal - PROTEF/IPEF. http://www.ipef.br/protecao/alerta-percevejo.pdf (2008).
Wilcken, C. F. et al. Bronze bug Thaumastocoris peregrines Carpintero and Dellapé (Hemiptera: Thaumastocoridae) on Eucalyptus in Brazil and its distribution. J. Plant Protect. Resear. 50, 201–205, https://doi.org/10.2478/v10045-010-0034-0 (2010).
Jacobs, D. H. & Neser, S. Thaumastocoris australicus Kirkaldy (Heteroptera: Thaumastocoridae): a new insect arrival in South Africa, damaging to Eucalyptus trees: research in action. S. Afr. J. Sci. 101, 233–236 (2005).
Noack, A. E. & Coviella, C. E. Thaumastocoris australicus Kirkaldy (Hemiptera: Thaumastocoridae): first record of this invasive pest of Eucalyptus in the Americas. Gen. Appl. Entomol. 35, 13–14 (2006).
EPPO. Global database. Addition of Thaumastocoris peregrinus to the EPPO Alert List, https://gd.eppo.int/reporting/article-1974 (2012).
Lesieur, V. et al. The rapid spread of Leptoglossus occidentalis in Europe: a bridgehead invasion. J. Pest Sci. 92, 189–200, https://doi.org/10.1007/s10340-018-0993-x (2019).
Seebens, H. et al. No saturation in the accumulation of alien species worldwide. Nat. Communications 8, 14435, https://doi.org/10.1038/ncomms14435 (2017).
Westphal, M. I., Browne, M., MacKinnon, K. & Noble, I. The link between international trade and the global distribution of invasive alien species. Biol. Invasions 10, 391–398, https://doi.org/10.1007/s10530-007-9138-5 (2008).
Madden, M. J. L. et al. Using DNA barcoding to improve invasive pest identification at U.S. ports-of-entry. Plos One 14, e0222291, https://doi.org/10.1371/journal.pone.0222291 (2019).
Nadel, R. L. et al. DNA bar-coding reveals source and patterns of Thaumastocoris peregrinus invasions in South Africa and South America. Biol. Invasions 12, 1067–1077, https://doi.org/10.1007/s10530-009-9524-2 (2010).
Kang, T. H., Kim, S., Hong, K. J. & Lee, H. S. DNA barcoding in quarantine inspection: a case study on quarantine insect monitoring for Lepidoptera obtained through quarantine inspection on foreign vessels. Mitochondrial DNA Part B 4, 43–48, https://doi.org/10.1080/23802359.2018.1536447 (2018).
Piper, A. M. et al. Prospects and challenges of implementing DNA metabarcoding for high-throughput insect surveillance. GigaScience 8, 1–22, https://doi.org/10.1093/gigascience/giz092 (2019).
Staden, R. B. K. F. & Bonfield, J. K. The Staden package, 1998. Methods in Molecular Biology 132, 115–130, https://doi.org/10.1385/1-59259-192-2:115 (2000).
Lo, N. et al. Population genetics of the Australian eucalypt pest Thaumastocoris peregrinus: evidence for a recent invasion of Sydney. J. Pest Science 92, 201–212, https://doi.org/10.1007/s10340-018-0995-8 (2019).
Nugnes, F., Caprio, E., di Prisco, G., Laudonia, S. & Sasso, R. Thaumastocoris peregrinus Carpintero & Dellapé (Heteroptera, Thaumastocoridae): a new haplotype in Italy. In: XXIV Italian National Congress of Entomology, Orosei, 2014, p. 98. https://doi.org/10.13140/2.1.3217.5045 (2014).
Machado, D. N. et al. The ongoing dispersion of the Eucalyptus bronze bug (Thaumastocoris peregrinus) in Spain. Forest Systems 28; https://doi.org/10.5424/fs/2019283-15353 (2019).
Streito, J. C. et al. Report of the invasive species Thaumastocoris peregrinus Carpintero & Dellapé, 2006, on Réunion Island (Hemiptera, Heteroptera, Thaumastocoridae). Bul. Soc. Entomol. France 12, 65–72 (2016).
Clement, M., Posada, D. C. & Crandall, K. A. TCS: a computer program to estimate gene genealogies. Molecular Ecology 9, 657–1659, https://doi.org/10.1046/j.1365-294x.2000.01020.x (2000).
Leigh, J. W. & Bryant, D. Popart: full-feature software for haplotype network construction. Methods in Ecology and Evolution 6, 1110–1116, https://doi.org/10.1111/2041-210X.12410 (2015).
Puillandre, N. et al. Genetic bottleneck in invasive species: the potato tuber moth adds to the list. Biol. Invasions 10, 319–333 (2008).
Caron, V., Norgate, M., Ede, F. J., Nyman, T. & Sunnucks, P. Novel microsatellite DNA markets indicate strict parthenogenesis and few genotypes in the invasive willow saw fly Nematus oligospilus. Bull. Entomol. Res. 103, 74–88 (2013).
Rollins, L. A. et al. Invasive species can’t cover their tracks: using microsatellites to assist management of starling (Sturnus vulgaris) populations in Western Australia. Mol. Ecol. 18, 1560–1573 (2009).
Nei, M., Maruyama, T. & Chakraborty, R. The bottleneck effect and genetic variability in populations. Evolution 29, 1–10 (1975).
Carter, M., Smith, M. & Harrison, R. Genetic analyses of the Asian longhorn beetle (Coleoptera, Cerambycidae, Anoplophora glabripennis), in North. Biol. Invasions 12, 165–1182, https://doi.org/10.1007/s10530-009-9538-9 (2010).
Nugnes, F. et al. Genetic Diversity of the Invasive Gall Wasp Leptocybe Invasa (Hymenoptera: Eulophidae) and of its Rickettsia endosymbiont, and associated sex-ratio differences. PloS One 10, e0124660, https://doi.org/10.1371/journal.pone.0124660 (2015).
Dittrich-Schröder, G. et al. Population genetic analyses of complex global insect invasions in managed landscapes: a Leptocybe invasa (Hymenoptera) case study. Biol. Invasion 20, 2395–2420, https://doi.org/10.1007/s10530-018-1709-0 (2018).
Ascunce, M. S. et al. Global invasion history of the fire ant Solenopsis invicta. Science 331, 1066–1068 (2011).
Gotzek, D. et al. Global invasion history of the tropical fire ant: a stowaway on the first global trade routes. Mol. Ecol. 24, 374–388 (2015).
Boissin, E. et al. Retracing the routes of introduction of invasive species: the case of the Sirex noctilio woodwasp. Mol. Ecol. 21, 5728–5744 (2012).
Dlugosch, K. M. & Parker, I. M. Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Mol. Ecology 17, 431–449 (2008).
Garnas, J. R. et al. Complex patterns of global spread in invasive insects: eco-evolutionary and management consequences. Biol. Invasions 18, 935–952, https://doi.org/10.1007/s10530-016-1082-9 (2016).
Essl, F. et al. Crossing frontiers in tackling pathways of biological invasions. Biosci. 65, 769–782 (2015).
Lopes da Silva, M. et al. The Role of Natural and Human-Mediated Pathways for Invasive Agricultural Pests: A Historical Analysis of Cases from Brazil. Agricul. Sci. 5, 634–646 (2014).
Goldstien, S. J. et al. Global Phylogeography of the Widely Introduced North West Pacific Ascidian Styela clava. PLoS One 6, https://doi.org/10.1371/journal.pone.0016755 (2011).
Townsend, C. R., Begon, M. & Harper, J. L. Fundamentos em Ecologia. 3 Ed. Artmed, Porto Alegre (2010).
Vincek, V. et al. How large was the founding population of Darwin's finches? Proc. Biol. Sci. 264, 111–18 (1997).
Ficetola, G. F., Bonin, A. & Miaud, C. Population genetics reveals origin and number of founders in a biological invasion. Mol. Ecol. 17, 773–782 (2008).
Dickey, A. M. et al. Population Genetics of Invasive Bemisia tabaci (Hemiptera: Aleyrodidae) Cryptic Species in the United States Based on Microsatellite Markers. J. Econ. Entomol. 106, 1355–1364 (2013).
Tay, T. W. et al. Mitochondrial DNA and trade data support multiple origins of Helicoverpa armigera (Lepidoptera, Noctuidae) in Brazil. Scientific Reports 7, 45302 (2017).
Yunke, W. et al. Identification of wood-boring beetles (Cerambycidae and Buprestidae) intercepted in tradeassociated solid wood packaging material using DNA barcoding and morphology. Scientific Reports 7, 40316, https://doi.org/10.1038/srep40316 (2017).
Arnemann, J. A. et al. Complete mitochondrial genome of the soybean stem fly Melanagromyza sojae (Diptera: Agromyzidae). Mitochondrial DNA Part A 27, 4534–45335 (2016).
Arnemann, J. A. et al. Multiple incursion pathways for Helicoverpa armigera in Brazil show its genetic diversity spreading in a connected world. Scientific Reports 9, https://doi.org/10.1101/762229 (2019).
Mayorga, S. I., Ruiz, C. G., Sandoval, A. C. & Valenzuela, J. E. Detection of Thaumastocoris peregrinus (Hemiptera: Thaumastocoridae) associated to Eucalyptus spp. in Chile. (Detección de Thaumastocoris peregrinus (Hemiptera: Thaumastocoridae) asociado a Eucalyptus spp. en Chile. Bosque 32, 309–313, https://doi.org/10.4067/S0717-92002011000300012 (2011).
Noack, A. E. & Rose, H. A. Life-history of Thaumastocoris peregrinus and Thaumastocoris sp. In the laboratory with some observations on behavior. Gen. App. Entomol. 36, 27–34 (2007).
Roy, B. A. et al. Increasing forest loss worldwide from invasive pests requires new trade regulations. Front. Ecol. Environ. 12, 457–465 (2014).
Withers, T. M. Colonization of eucalypts in New Zealand by Australian insects. Aust. Ecol. 26, 467–476 (2001).
Laudonia, S. & Sasso, R. The bronze bug Thaumastocoris peregrinus: a new insect recorded in Italy, damaging to Eucalyptus trees. Bul. Insectology 65, 89–93 (2012).
Garcia, A., Figueiredo, E., Valente, C., Monserrat, V. J. & Branco, M. First record of Thaumastocoris peregrinus in Portugal and of the Neotropical predator Hemerobius bolivari in. Europe. Bul. Insectology 66, 251–256 (2013).
UNECE, FAO. Forest products: annual market review 2004-2005, Timber Bulletin, volume 58 (2005). United Nations Economic Comission for Europe, Food and Agriculture Organization, Geneva, Switzerland. http://www.ipef.br/estatisticas/relatorios/FAO-forest_products-2005.pdf (2005).
UNECE, FAO. Forest products: annual market review 2011-2012. United Nations Economic Comission for Europe, Food and Agriculture Organization, Geneva, Switzerland. http://www.unece.org/fileadmin/DAM/timber/publications/FPAMR_2012.pdf (2012).
Vivas, L., Crespo, J. & Jacinto, V. Primer registro de la especie invasora Thaumastocoris peregrinus Carpintero & Dellapé, 2006 en España y nuevos datos para Portugal (Hemiptera: Thaumastocoridae). BV News 4, 30–35 (2015).
Heyden, T. V. D. The first record of Thaumastocoris peregrinus Carpintero & Dellapé, 2006 (Hemiptera: Heteroptera: Thaumastocoridae) for Albania. Rev. Gad. Entomol. VIII, 133–135 (2017).
Petrakis, P. V. First record of the bug Thaumastocoris peregrinus in Greece. Entomologia Hellenica 27, 1–9, https://doi.org/10.12681/eh.18703 (2018).
Brennan, E. B., Gill, R. J., Hrusa, G. F. & Weinbaum, S. A. First record of Glycaspis brimblecombei (Moore) (Homoptera: Psyllidae) in North America: initial observations and predator associations of a potentially serious new pest of Eucalyptus in California. Pan-Pacific. Entomol 75, 55–57 (1999).
Dhahri, S., Ben Jamaa, M. L., Garcia, A., Boavida, C. & Branco, M. Presence of Glycaspis brimblecombei and its Parasitoid Psyllaephagus bliteus in Tunisia and Portugal. Silva Lusit. 22, 99–115 (2014).
Reguia, K. & Peris-Felipo, F. J. Glycaspis brimblecombei Moore,1964 (Hemiptera Psyllidae) invasion and new records in the Mediterranean area. Biodiversity J. 4, 501–506 (2013).
Jiménez-Quiroz, E., Vanegas-Rico, J. M., Morales-Martínez, O., Lomeli-Flores, J. R. & Rodríguez-Leyva, E. First Record of the Bronze Bug, Thaumastocoris peregrinus Carpintero & Dellapé 2006 (Hemiptera: Thaumastocoridae), in Mexico. J. Agric. Urban Entomol. 32, 35–39, https://doi.org/10.3954/1523-5475-32.1.35 (2016).
BugGuide. 2016. Thaumastocoris peregrinus. On-line, http://bugguide.net/node/view/1236781 (2019).
Hodel, D. R., Arakelian, G. & Ohara, L. M. The Bronze Bug Another New Threat to Eucalypts in California. PalmArbor 5, 1–11, https://ucanr.edu/sites/HodelPalmsTrees/files/248430.pdf (2016).
Liebhold, A. M., Brockerhoff, E. G., Garrett, L. J., Parke, J. L. & Britton, K. O. Live plant imports: the major pathway for forest insect and pathogen invasions of the US. Front. Ecol. Environ. 10, 135–143 (2012).
U. S. Department of Homeland Security. Brownsville Port of Entry CBP Agriculture Specialist Intercepts First in Port Pest in Eucalyptus Branches. https://www.cbp.gov/newsroom/local-media-release/brownsville-port-entry-cbp-agriculture-specialist-intercepts-first-1 (2017).
Paine, T. D., Millar, J. G. & Daane, K. M. Accumulation of Pest Insects on Eucalyptus in California: random Process or Smoking Gun. J. Econ. Entomol. 103, 1943–1949 (2010).
EPPO. Global Database. Distribution Thaumastocoris peregrinus, https://gd.eppo.int/taxon/THMCPE/distribution (2018).
CABI. Invasive Species Compendium. Thaumastocoris peregrinus (bronze bug). https://www.cabi.org/isc/datasheet/109741#20123208401 (2018).
Chilima, C. Z. New Eucalyptus Pest Recorded from Zimbabwe. Forest Invasive Species Network for Africa (FAO) [WWW document]. http://www.fao.org/forestry/fisna/26061/en/ (2007).
Nahrung, H. F. & Swain, A. J. Strangers in a strange land: do life history traits differ for alien and native colonisers of novel environments? Biol. Invasions 17, 699–709 (2015).
Nadel, R. L. et al. Population dynamics of Thaumastocoris peregrinus in Eucalyptus plantations of South Africa. J. Pest Sci. 88 (97–106) (2015).
Corallo, B. et al. Entomopathogenic fungi naturally infecting the eucalypt bronze bug, Thaumastocoris peregrinus (Heteroptera: Thaumastocoridae), in Uruguay. J. Appl. Entomol. 1–14; https://doi.org/10.1111/jen.12624 (2019).
Saavedra, M. C., Withers, T. M. & Holwell, G. I. Susceptibility of four Eucalyptus host species for the development of Thaumastocoris peregrinus Carpintero and Dellapé (Hemiptera: Thaumastocoridae). Forest Ecol. Manag. 336, 210–2016 (2015a).
Saavedra, M. C., Avila, G. A., Withers, T. M. & Holwell, G. I. The potential global distribution of the Bronze bug Thaumastocoris peregrinus Carpintero and Dellapé (Hemiptera: Thaumastocoridae). Agr. Forest Entomol. 17, 375–388 (2015b).
Osborne, J. L.; Loxdale, H. D. & Woiwod, I. P. Monitoring insect dispersal: methods and approaches. In: Bullock, J. M., Kenward, R. E and Hails, R. S. (Eds.). Dispersal Ecology: 42nd Symposium of the British Ecological Society, P. 24–49 (Cambridge University Press, 2002).
Whitney, K. D. & Gabler, C. A. Rapid evolution in introduced species, ‘invasive traits’ and recipient communities: challenges for predicting invasive potential. Diversity distrib. 14, 569–580 (2008).
Zheng, X. L. et al. A review of invasive biology, prevalence and management of Leptocybe invasa Fisher & La Salle (Hymenoptera: Eulophidae: Tetrastichinae). Afr. Entomol. 22, 68–79 (2014).
Protasov, A., Doganlar, M., Salle, J. L. & Mendel, Z. Occurrence of two local Megastigmus species parasitic on the eucalyptus gallwasp Leptocybe invasa in Israel and Turkey. Phytoparasitica 36, 449–459 (2008).
Simberloff, D. & Holle, B. V. Positive interactions of nonindigenous species: invasional meltdown? Biol. Invasions 1, 21–32 (1999).
Martínez, G., González, A. & Dicke, M. Effect of the eucalypt lerp psyllid Glycaspis brimblecombei on adult feeding, oviposition‐site selection, and offspring performance of the bronze bug, Thaumastocoris peregrinus. Entomol. Experim. Appl. 166, 395–401 (2018).
Sakai, A. K. et al. The population biology of invasive species. Ann. Rev. Ecol. Syst. 32, 305–332 (2001).
Roderick, G. K. Geographic structure of insect populations: gene flow, phylogeography, and their uses. Ann. Rev. Entomol. 41, 325–352 (1996).
Martínez, G. & Bianchi, M. Primer registro para Uruguay de la chinche del eucalipto, Thaumastocoris peregrinus Carpintero y Dellappé, 2006 (Heteroptera: Thaumastocoridae). Agrociencia XIV, 15–18 (2010).
Díaz, E. A. B., Coronel, R. S. & Godziewski, D. Consideraciones sobre dos nuevas plagas del eucalipto em Paraguay. El psílido de la concha o escudo Glycaspis brimblecombei (Hemiptera: psyllidae) y La chinche marrón Thaumastocoris peregrinus (Hemiptera: Thaumastocoridae). Bol. Museo Nac. Hist. Nat. Parag. 17, 72–75 (2013).
Sopow, S., George, S. & Ward, N. Bronze bug, Thaumastocoris peregrinus: a new Eucalyptus pest in New Zealand. Surveillance 39, 43–46 (2012).
Carapezza, A. The arrival of one more eucalyptus pest in Sicily: Thaumastocoris peregrinus Carpintero & Dellapé, 2006 (Hemiptera Heteroptera Thaumastocoridae). Naturalista Sicil. XXXVIII, 127–129 (2014).
Suma, P., Nucifora, S. & Bella, S. New distribution record of the invasive bronze bug Thaumastocoris peregrinus Carpintero and Dellapé (Heteroptera, Thaumastocoridae) in Italy. Bull. OEPP/EPPO 44, 79–182, https://doi.org/10.1111/epp.12122 (2014).
Novoselsky, T. & Freideberg, A. First record of Thaumastocoris peregrinus (Hemiptera: Thaumastocoridae) in the Middle East, with biological notes on its relations with eucalyptus trees. Isr. J. Entomol. 46, 43–55 (2016).
Acknowledgements
Dayanna Machado would like to thank the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for granting the doctoral scholarship. We also thank Nathan Lo for providing us with T. peregrinus mtDNA data from the Sydney, Australia.
Author information
Authors and Affiliations
Contributions
D.N.M., E.C.C., J.V.C.G., C.R.P. and J.A.A. conceived this research and designed the experiments; D.N.M., I.V., G.A.U., C.R.P. and J.A.A. analyzed the data; L.R.B., G.M., S.I.M., S.O.R., M.B., A.G., J.M.V.-R., E.J.-Q., S.L., T.N., D.R.H., G.A. and H.S. assisted with sample collection; D.N.M., I.V., M.B., A.G., C.R.P., G.M. and J.A.A. wrote the paper. All authors read and commented on the information provided in the manuscript and approved the final draft.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Machado, D.d.N., Costa, E.C., Guedes, J.V.C. et al. One maternal lineage leads the expansion of Thaumastocoris peregrinus (Hemiptera: Thaumastocoridae) in the New and Old Worlds. Sci Rep 10, 3487 (2020). https://doi.org/10.1038/s41598-020-60236-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-60236-7
This article is cited by
-
Coconut rhinoceros beetle in Solomon Islands: a tale of two invasions
Biological Invasions (2023)
-
Highly diverse and rapidly spreading: Melanagromyza sojae threatens the soybean belt of South America
Biological Invasions (2021)
-
Global incursion pathways of Thaumastocoris peregrinus, an invasive Australian pest of eucalypts
Biological Invasions (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.