Abstract
Three new CoII-coordination polymers (Co-CPs) containing glutarates and bipyridyl ligands, formulated as [Co2(Glu)2(µ-bpa)2]·(H2O)4 (1), [Co4(Glu)4(µ-bpp)2] (2), and [Co2(Glu)2(µ-bpe)2]·(H2O)0.5 (3), were prepared, and their structures were determined by X-ray crystallography. Glutarates bridge CoII ions to form 2D sheets, and the sheets are connected either by bpa or by bpp ligands to form 3D networks 1 and 2, respectively. Both frameworks 1 and 2 are two-fold interpenetrated, and there is no significant void volume in either network. Four glutarates bridge two CoII ions to form chains, and these chains are connected by bpe ligands to form the 2D sheet 3. The antifungal properties of these new Co-CPs were tested against two model fungal pathogens, Candida albicans and Aspergillus niger. Under the maximum concentration of Co-CPs, 2.0 mg mL−1, the inhibition rates of Co-CPs against A. niger were much lower (44–62%) than those (90–99.98%) observed in C. albicans. The results indicate that 1–3 can inactivate C. albicans cells more efficiently than A. niger spores in the same treatment time, and the greater inactivation of C. albicans can be explained by dramatic changes in the morphology of C. albicans cells. We also found that Co-CPs could generate the reactive species NO and H2O2, and these species might play a role in inactivating fungal cells. Additionally, degradation tests confirmed that the leaching of CoII ions from Co-CPs was not significant. The small amount of leached CoII ions and the robust Co-CPs themselves as well as the reactive species generated by Co-CPs can actively participate in fungal inactivation.
Similar content being viewed by others
Introduction
Metal-organic frameworks (MOFs) containing large void volumes after the removal of captured solvent molecules1,2,3,4 can be valuable for a wide range of applications, including separation5,6,7, CO2 capture8,9,10, H2 storage11,12,13, heterogeneous catalysis14,15,16, and biomedical usage17,18. The properties of MOFs can depend on many factors, such as the metal ion, rigidity/flexibility, chemical functionality, and geometry of the framework. MOFs can be tuned by changing bridging ligands. The flexible α,ω–alkane (or alkene) dicarboxylates have been utilized as bridging ligands by our group and others for constructing new functional MOFs19,20,21. A systematic study of MOFs containing these α,ω-alkane (or alkene)-dicarboxylate ligands has broadened the scope of MOF materials.
Glutarate is a flexible dicarboxylate ligand that can bridge CuII ions to form two-dimensional (2D) sheets, and bipyridyl ligands can be used to connect these sheets to form three-dimensional (3D) frameworks, Cu-MOFs19,22,23,24,25,26. We and other groups have studied Cu-MOFs containing these glutarates and bipyridyl ligands. Our group has reported 3D Cu-MOFs containing glutarates and the bipyridyl ligands (bpa = 1,2-bis(4-pyridyl)ethane and bpp = 1,3-bis(4-pyridyl)propane). Both MOFs contain Cu2 paddle-wheel units connected by glutarates to form 2D sheets, and the sheets were also bridged by either bpa or bpp to form 3D frameworks19. They showed good selectivity for CO2 over N2 and H2. Esterhuysen, Barbour and coworkers performed a systematic study of three closely related microporous Cu-MOFs containing glutarates and bipyridyl linkers, namely, bpy, bpe, and bpymh (bpy = 4,4′-bipyridine, bpe = 1,2-bis(4-pyridyl)ethylene, bpymh = N,N′-bis(pyridine-4-ylmethylene)hydrazine)22. These linkers increased the pore dimensions for the gas sorption behaviour of the frameworks. Solvent- and pressure-induced phase changes in the same 3D Cu-MOFs via glutarate conformational isomerism have also been reported23. Recently, these water-stable robust Cu-MOFs containing glutarates and bipyridyl linkers (bpy, bpa, bpe, and bpp) were tested for antibacterial activities against five bacteria27. They exhibited excellent antibacterial activities against five kinds of bacteria tested, including both Gram-positive bacteria (Staphylococcus aureus and MRSA) and Gram-negative bacteria (Escherichia coli, Klebsiella pneumonia, and Pseudomonas aeruginosa), with very low MBCs (minimal bactericidal concentration).
As an extension of our previous work, we present here two new 3D CoII-coordination polymers (Co-CPs) containing glutarates and bipyridyl ligands, formulated as [Co2(Glu)2(µ-bpa)2]·(H2O)4 (1) and [Co4(Glu)4(µ-bpp)2] (2) (Glu = glutarate) for a systematic study of MOFs and CPs containing glutarates and bipyrdyl ligands. We also present a new 2D Co-CP containing bipyridyl ligands with double bonds, formulated as [Co2(Glu)2(µ-bpe)2]·(H2O)0.5 (3). They were prepared by a solvothermal reaction in a DMF solution of the mixture of Co(NO3)2, glutaric acid and bpa, bpp, or bpe (Scheme 1). The antifungal properties of these new Co-CPs were tested against two model fungal pathogens, Candida albicans and Aspergillus niger. C. albicans is an opportunistic fungal pathogen that can infect human tissues, causing candidiasis28. C. albicans occurs mostly in yeast form but becomes filamentous during infection29. A. niger is a filamentous Ascomycete fungus implicated in human and plant infection as well as food contamination and fermentation29. This fungus is often found in bakery products, intermediate-moisture food products, cheeses, preserved fruits and grains. Although it has been reported that the water-stable Cu-BTC MOF (BTC = 1,3,5-benzenetricarboxylate) can inhibit the growth rate of C. albicans30, and other Cu-based HKUST-1 MOF can inactivate Saccharomyces cerevisiae and its antifungal property is due to releasing CuII ions from its own structure31, Co-based MOFs and CPs have rarely been tested for their antifungal properties. A Co-based MOF (Co-TDM, TDM8− = [(3,5-dicarboxyphenyl)-oxamethyl]methane) was shown to be highly effective at inactivating E. coli32. However, there are few reports on the antifungal activity of Co-based MOFs and CPs.
Results and Discussion
Structure description
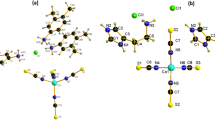
Co-CPs 1 and 2 crystallize in the monoclinic P21/n space group, and single-crystal X-ray analysis (Table 1) revealed that the glutarates bridge CoII ions to form two-dimensional (2D) sheets, and the sheets are also connected either by bpa ligands or by bpp ligands to form 3D networks 1 (Fig. 1) and 2 (Fig. 2), respectively. In 1, two glutarates bridge two CoII ions in bridging mode and chelating/bridging mode to form one-dimensional (1D) chains, and these chains are connected by other glutarates in chelating and monodentate coordination modes to form a 2D sheet. The bpa ligands connect these 2D sheets to form a 3D network. The 3D networks are two-fold interpenetrated (Fig. S1a), and there is no significant void volume for solvents (4.9%, 161.5 Å3/3297.9 Å3 on PLATON analysis33). The coordination geometry of one CoII ion is distorted octahedral constructed by one bridging carboxylate O atom (η1:η1:μ2), two chelating O atoms (η1:η1), one monodentate carboxylate O atom (η1:η0), and two pyridyl N atoms. That of the other CoII ions is also distorted octahedral constructed by one bridging O atom (η1:η1:μ2), two chelating O atoms (η1:η1), one chelating/bridging O atom (η2:η1:μ2), and two pyridyl N atoms. The Co-O bond distances range from 1.994(4) to 2.285(4) Å, and the Co-N distances range from 2.150(3) to 2.184(3) Å (Table 2). The Co···Co distance in a Co2 unit is 3.804 Å.
For 2, two CoII ions are bridged by three glutarates in two bridging and one chelating/bridging modes, and one of the CoII ions is chelated by the other glutarate to form 2D sheets. These 2D sheets are connected by bpp ligands to form a 3D network. The 3D networks are also two-fold interpenetrated (Fig. S1b), and there is no void volume for solvents. The coordination geometry of one CoII ion is distorted square pyramidal was constructed by two bridging O atoms, two chelating O atoms, and one pyridyl N atom and that of the other CoII ion is distorted octahedral constructed by two bridging O atoms, two chelating O atoms, one chelating/bridging O atom, and one pyridyl N atom. The Co-O bond distances range from 1.952(3) to 2.239(3) Å, and the Co-N distances range from 2.048(3) to 2.154(3) Å (Table 2). The Co···Co distances in Co2 units are 3.49 and 3.49 Å.
The 3D networks of both 1 and 2 indicate a 6-connected uninodal net (pcu alpha-Po primitive cubic) with a Schläfli symbol of 412·63, assuming that the Co2 unit acts as a node by ToposPro analysis34.
Co-CP 3 crystallizes in a triclinic space group, and X-ray analysis (Table 1) shows that four glutarates bridge two CoII ions to form chains, and these chains are connected by bpe ligands to form a 2D sheet (Fig. 3). The coordination geometry of a CoII ion is distorted octahedral constructed by two bridging O atoms from the different glutarates, two chelating O atoms from glutarate, and two pyridyl N atoms. The C=C double bonds of the bpe ligands are close enough to produce photoreaction [2 + 2] addition products, but no photoaddition reaction occurred in Co-CP 3. The distances between double bonds are 3.81 and 3.85 Å (Fig. 3). The Co-O bond distances range from 2.029(2) to 2.260(2) Å, and the Co-N distances range from 2.153(2) to 2.168(2) Å (Table 2). The Co···Co distances in Co2 units are 4.011 and 4.018 Å.
The coordination modes of two the carboxylates in a glutarate are shown in Scheme 2. For 3D networks, two types of coordination modes are used: (η1:η0)(η1:η1) and (η2:η1:μ2)(η1:η1:μ2) for 1, (η1:η1:μ2)(η1:η1) and (η1:η1:μ2)(η2:η1:μ2) for 2. For 2D sheets, one coordination mode (η1:η1)(η1:η1:μ2) is used for 3. The flexible glutarate can have different conformations in coordination compounds23, 1 and 3 have a gauche-trans conformation, and 2 has a trans-trans conformation.
The bulk purities of 1–3 were confirmed by powder X-ray diffraction (PXRD) (Figs S2–S4), elemental analysis (see experimental section), and thermogravimetric analysis (TGA) of the as-prepared samples (Figs S5–S7). The scanning electron microscopy (SEM) images of three Co-CPs are shown in Fig. S8. 1 and 3 have rod type crystals, 2 has aggregated crystals. The crystals of all three compounds were used as prepared without grinding. The structural stabilities of 1–3 in water or 1x PBS (Phosphate Buffered Saline) solution have been tested by PXRD, and the PXRD patterns of 1 and 2 indicate that their structures are stable after 4 days in water or PBS solution (Fig. S9). For 3 after 4 days in water or PBS, the intensities of diffraction planes of (011), (101), (112), and (1-1-1) at 2θ = 8.63, 9.54, 13.677, and 14.484°, respectively, were reduced compared with those of the as-prepared sample. Contrarily, the diffraction planes of (110) and (1–10) at 2θ = 11.457 and 11.962° were maintained. These differences of diffraction patterns between the as-prepared sample and samples after 4 days in water or PBS imply that partial degradation of the framework. This also consists with the small amount of CoII ions released from Co-CPs in PBS which will be discussed in the later part.
Antifungal activities
The antifungal properties of the three Co-CPs against C. albicans and A. niger were assessed by using the plate counting method. C. albicans cells were incubated in different concentrations of each compound (2, 1, 0.5, 0.25, and 0.125 mg mL−1) for 4 days. When yeasts were incubated with compounds for 1 day, no significant antifungal effects were observed (Fig. S10a). However, after 4 days, the difference in CFU numbers between the control and treatments became more dramatic (Figs 4 and S11). Representative images of C. albicans colonies on PDA (Potato Dextrose Agar) plates are shown in Fig. 5. 1 inactivated C. albicans by approximately 91% at 2 mg mL−1, 2 by 99.98% at 0.25 mg mL−1, and 3 by 99.93% at 0.25 mg mL−1 (Figs 4 and S11). 2 and 3 showed relatively higher antifungal activity against C. albicans than 1 (Figs 4 and 5). The result indicates that the viability of yeast cells is reduced by increasing the concentration of Co-CPs, although a relatively longer incubation time is required to obtain a similar level of antifungal effect to that of the previously reported Cu-BTC, in which C. albicans was effectively inactivated with a 500 ppm (0.5 mg mL−1) concentration and a short incubation time (60 min)30.
Antifungal activities of 1 (a), 2 (b), and 3 (c) against Candida albicans at different concentrations. Graphs on the left show log scale CFU (colony forming unit) numbers of C. albicans in each treatment. The percentage in the right graphs was calculated as follows: (CFU number in each compound treatment/CFU number of control) × 100. Each bar represents the average and standard deviation of 9 replicates. Student’s t-test was performed between the control and each treatment; **p < 0.01.
A. niger spores were also incubated in different concentrations of each compound (2, 1, 0.5, 0.25, and 0.125 mg mL−1) for 4 days, and spore germination was assessed as an indicator of spore viability. As observed in C. albicans, treatment of A. niger spores with compounds for 1 day did not produce any significant antifungal effects (Fig. S10b). In contrast to C. albicans cells, A. niger spores are pigmented, and this pigmentation may affect the responses to Co-CPs. Inactivation of A. niger spores by Co-CPs measured as number and percentage of spore germination was not dramatic compared to that observed in C. albicans (Figs 6 and S12). Representative images of A. niger colonies on PDA plates are shown in Fig. 7. The germination percentage of A. niger spores was more slowly reduced over increased concentrations of compounds compared to the CFU percentage of C. albicans cells (Figs 4 and 6 and S11, S12). At the maximum concentration of compounds, 2.0 mg mL−1, the inhibitory effect of 1, 2, and 3 against A. niger were approximately 44%, 46%, and 62%, respectively, after incubation for 4 days (Fig. 6). These inhibition rates were much lower than those (90–99.98%) observed in C. albicans (Figs 4 and 6).
Antifungal activities of 1 (a), 2 (b), and 3 (c) against Aspergillus niger at different concentrations. Graphs on the left show log scale CFU (colony forming unit) numbers of A. niger in each treatment. The percentage in the right graphs was calculated as follows: (CFU number in each compound treatment/CFU number of control) × 100. Each bar represents the average and standard deviation of 9 replicates. Student’s t-test was performed between the control and each treatment; *p < 0.05, **p < 0.01.
Our results indicate that Co-CPs can inactivate C. albicans cells more efficiently than A. niger spores in the same treatment time. The antifungal activity of Co-CPs is likely to be variable depending on fungal species. The differential sensitivity of fungal species to MOFs was also observed in a previous study in which Cu-BTC inhibited spores of F. oxysporum and A. oryzae up to 30% but was not affected by A. niger spores even after 7 days of incubation30. Our results can be compared with those of silver(I) CPs and MOFs showing a notable antimicrobial efficiency against a panel of bacteria (E. coli, P. Aeruginosa and S. aureus) and yeast (C. albicans) with the minimum inhibitory concentrations (MIC) of 30–60 μg mL−1 (normalized MIC of 11–68 nmol mL−1), but their antimicrobial action is due to release AgI ions from Ag-MOFs or Ag-CPs35,36,37,38. The antifungal activities of Co-CPs can also be compared with those of other nanomaterials: Ag nanoparticles on C. albicans biofilms39, ZnO nanoparticles against E. coli40, and reduced graphene oxide nanosheets against A. niger, A. oryzae, and F. oxysporum (Fusarium oxysporum) with half-maximal inhibitory concentrations (IC50) of 50, 100, and 100 μg mL−1, respectively41.
Our SEM analysis demonstrated that the morphology of C. albicans cells and A. niger spores was differentially changed after treatment with Co-CPs (Fig. 8). Loosened spore surfaces (probably destruction of the fungal cell wall) without severe changes in cell shape were observed in many A. niger spores, while a large proportion of C. albicans cells were severely crushed (Fig. 8). In A. niger treated with Co-CPs, the spore surface was wrinkled, and the surface layers peeled off (Fig. 8b). This result indicates damage to the spore cell wall, most likely making the spores more vulnerable to mechanical and other environmental stresses. However, we could not see wrinkles or peel-off of the surface in C. albicans cells treated with Co-CPs. This may be because of the difference in the composition and structure of the cell wall between C. albicans and A. niger, although both are fungi. More dramatic changes in morphology were observed in C. albicans than in A. niger, which might explain the greater inactivation by Co-CPs in C. albicans. The morphological changes observed after treatment with Co-CPs were not significantly different among the 3 types of Co-CPs (Fig. 8), where 1 and 2 are two-fold-interpenetrated 3D frameworks and 3 is a 2D sheet.
To find the mechanism for antifungal effects, ROS (reactive oxygen species) and RNS (reactive nitrogen species) levels in PBS solution were analysed after treatment with Co-CPs. These Co-CPs can generate ROS/RNS that can interact with biomolecules in fungal spores, producing an inhibitory effect on fungal growth42. Due to the limitations of the available methods, we could not measure many reactive species. The concentrations of nitric oxide (NO) and hydrogen peroxide (H2O2) were measured. The levels of both reactive species increased more dramatically with compound concentration in 1 and 2 than in 3 at day 0 (Fig. 9). After 4 days, NO concentration was generally elevated in all treatments without a significant change in trend among Co-CPs (Fig. 9). However, a similar level of H2O2 (approximately 12 µM) was observed in all treatment concentrations of 2, while the H2O2 level was decreased in the treatment with higher concentrations of 1 and 3 (Fig. 9). Our data demonstrate that Co-CPs can generate NO and H2O2, and these species may play a role in inactivating fungal cells. Recently, it is reported that ROS can be generated by photocatalysis of MOFs43,44. Photoinduced metal ion activates O2, producing ROS such as superoxide or singlet oxygen via electron transfer. Since our experiments were performed in the presence of light, light-induced electron transfer from Co metal to O2 might produce ROS such as superoxide or singlet oxygen. H2O2 and NO can be further generated through reaction with dissolved oxygen and nitrogen in PBS background solution. However, the relatively lower levels of NO and H2O2 observed in the treatment with 3 are not sufficient to explain the high antifungal activity of 3. Moreover, the H2O2 level was maintained at approximately 12 µM in all treatment concentrations of 2 and reduced with the increased concentration of 1 and 3. These results are not well matched with the antifungal effects. In addition to reactive species, other factors, such as mechanical damage to fungal cells by contact with the surface of Co-CPs or toxicity of glutarate (ligand chemical in Co-CPs) and CoII ion, should also be considered.
Additionally, degradation tests were carried out in the 1x PBS solution used for antifungal testing at room temperature to confirm that the leaching of CoII ions from Co-CPs was not significant. The quantities of CoII ions released from Co-CPs were monitored by ICP-MS (ICP-MS measurements were performed in Seoul Center, Korea Basic Science Institute) after 1 day, 2 days, 3 days and 4 days (Fig. S13). The concentration of released CoII ions from 1 mg of a compound in 1 mL of 1x PBS after 4 days was approximately 0.013 mg mL−1 for all three Co-CPs. The amount of CoII ions released from 3 was calculated to be 0.0033 mg mL−1, which is negligible compared to 0.25 mg mL−1 of the 99.93% inactivating property. Co-CPs cannot easily release CoII ions and are stable in 1x PBS. From these results, we infer that reactive species from Co-CPs, the small amount of leached CoII ions, and Co-CPs themselves actively participate in fungal inactivation.
To find the toxicity of glutarate and CoII ion, we incubated C. albicans cells and A. niger spores in PBS solutions containing glutarate (2 mg/ml, equivalent to Co-CPs concentration showing the highest antifungal activity) and Co salt (0.02 mg/ml, equivalent to the released CoII concentration after 4 days) for 4 days and measured the fungal viability. Glutarate did not affect significantly the viability of both fungal cells (Fig. S14). However, treatment with Co salt reduced the viability of C. albicans cells up to around 50% while no significant effect on A. niger spores was observed (Fig. S14). Although reduction in C. albicans cell viability by Co-CPs (over 90%) was much higher than that by Co salt (around 50%), CoII ion released from Co-CPs could partially contribute to fungal inactivation, probably through the redox activity of CoII ion on biomolecules leading to the fungal cell damage.
In summary, three new Co-CPs containing glutarates and bipyridyl ligands, formulated as [Co2(Glu)2(µ-bpa)2]·(H2O)4 (1), [Co4(Glu)4(µ-bpp)2] (2), and [Co2(Glu)2(µ-bpe)2]·(H2O)0.5 (3), were prepared and characterized by X-ray structure determination. Glutarates bridge CoII ions to form 2D sheets, and the sheets were also bridged either by bpa or by bpp ligands to form 3D networks 1 and 2, respectively. Both 1 and 2 are two-fold interpenetrated, and there is no significant void volume in both networks. Four glutarates bridge two CoII ions to form chains, and these chains are connected by bpe ligands to form the 2D sheet 3. The antifungal properties of these new Co-CPs were tested against two model fungal pathogens, C. albicans and A. niger. Under the maximum concentration of Co-CPs, 2.0 mg mL−1, the inhibition rates of Co-CPs against A. niger were much lower (44–62%) than those (90–99.98%) observed in C. albicans. The results indicate that Co-CPs can inactivate C. albicans cells more efficiently than A. niger spores in the same treatment time, and the greater inactivation of C. albicans by Co-CPs may result from dramatic changes in the morphology of C. albicans. Co-CPs can generate NO and H2O2, and these species may play a role in inactivating fungal cells. Additionally, degradation tests showed that the leaching of CoII ions from Co-CPs was not significant. These results imply that the small amount of leached CoII ions and Co-CPs themselves as well as reactive species generated by Co-CPs can participate in fungal inactivation. Further investigation into the antifungal mechanisms of Co-CPs and other MOFs will be conducted to obtain a clear conclusion.
Experimental Section
Preparation of [Co2(Glu)2(µ-bpa)2]·(H2O)4 (1)
A mixture of Co(NO3)2·6H2O (0.116 g, 0.4 mmol), glutaric acid (0.053 g, 0.4 mmol), and 1,2-bis(4-pyridyl)ethane (0.147 g, 0.8 mmol) in 10 mL of N,N-dimethylformamide (DMF) was placed in a Teflon-lined high pressure vessel. The vessel was placed in an oven at 100 °C for 72 h. After cooling to room temperature, the plum purple crystalline rods were retrieved by filtration, washed with DMF, and dried in air overnight. The yield was 0.080 g. IR (KBr): ν(cm−1) = 3067(w), 2966(w), 2904(w), 1613(s), 1581(s), 1541(s), 1413(s), 1390(s), 1301(s), 1273(m), 1223(m), 1145(w), 1072(w), 1044(w), 1022(m), 838(s), 765(w) (Fig. S15). Anal. Calcd. for [Co2(C5H6O4)2(C12H12N2)2]·(H2O)4 (818.60), 1: C, 49.89; H, 5.42; N, 6.84%. Found; C, 49.85; H, 4.71; N, 7.28%.
Preparation of [Co4(Glu)4(µ-bpp)2] (2)
A mixture of Co(NO3)2·6H2O (0.116 g, 0.4 mmol), glutaric acid (0.053 g, 0.4 mmol), and 1,2-bis(4-pyridyl)propane (0.159 g, 0.8 mmol) in 10 mL of DMF was placed in a Teflon-lined high pressure vessel. The vessel was placed in an oven at 100 °C for 72 h. After cooling to room temperature, the purple crystalline rods were retrieved by filtration, washed with DMF, and dried in air overnight. The yield was 0.060 g. IR (KBr): ν(cm−1) = 3072(w), 2943(w), 2865(w), 1602(s), 1541(s), 1407(s), 1312(m), 1273(m), 1223(m), 1161(w), 1072(m), 1027(m), 1022(m), 882(w), 815(m) (Fig. S15). Anal. Calcd. for [Co4(C5H6O4)4(C13H14N2)2] (1152.66), 3: C, 47.93; H, 4.55; N, 4.86%. Found; C, 47.41; H, 4.59; N, 5.15%.
Preparation of [Co2(Glu)2(µ-bpe)2]·(H2O)0.5 (3)
A mixture of Co(NO3)2·6H2O (0.116 g, 0.4 mmol), glutaric acid (0.053 g, 0.4 mmol), and 1,2-bis(4-pyridyl)ethylene (0.146 g, 0.8 mmol) in 10 mL of DMF and distilled water (7:3 vol/vol%) was placed in a Teflon-lined high pressure vessel. The vessel was placed in an oven at 100 °C for 72 h. After cooling to room temperature, the burgundy crystalline rods were retrieved by filtration, washed with DMF, and dried in air overnight. The yield was 0.062 g. IR (KBr): ν(cm−1) = 3066(w), 3021(w), 2971(w), 2949(w), 2927(w), 2888(w), 1592(s), 1564(s), 1418(s), 1351(m), 1312(m), 1206(m), 1156(m), 1056(w), 1022(m), 988(m), 882(m), 838(s), 798(m), 737(w), 664(m) (Fig. S15). Anal. Calcd. for [Co2(C5H6O4)2(C12H10N2)2]·(H2O)0.5 (751.51), 3: C, 54.34; H, 4.43; N, 7.46%. Found; C, 54.17; H, 4.26; N, 7.36%.
X-ray crystallography
The X-ray diffraction data for all three Co-CPs were collected on a Bruker APX-II diffractometer equipped with a monochromator in the Mo Kα (λ = 0.71073 Å) incident beam. Each crystal was mounted on a glass fibre. The CCD data were integrated and scaled using the Bruker-SAINT software package, and the structure was solved and refined using SHEXL-201445. All hydrogen atoms were placed in the calculated positions. The crystallographic data for the three compounds are listed in Table 1. The selected bond distances and angles of compounds are listed in Table 2. Structural information was deposited at the Cambridge Crystallographic Data Centre. The CCDC reference numbers are 1561941 for 1, 1561902 for 2, and 1561903 for 3.
Instrumentation
Thermogravimetric analysis was carried out on a TGA Q5000 (TA Instruments) under a nitrogen atmosphere. The elemental compositions of Co-CP samples were analysed at the Organic Chemistry Research Center, Sogang University (Seoul, Korea) by using EA1112 (CE Instruments, Italy). Powder X-ray diffraction patterns were recorded on a Bruker D8 Focus diffractometer (40 kV, 30 mA, step size = 0.02°). IR spectra were measured on a BIO RAD FTS 135 spectrometer as KBr pellets.
Antifungal assays
We treated C. albicans (KCTC7270) cells and A. niger (130708) spores with 3 types of Co-CPs to assess the antifungal activity of Co-CPs. Both C. albicans and A. niger were maintained and propagated in PDA medium. To obtain C. albicans cells for the assay, 1–2 colonies from PDA plates were suspended in 1 mL of PDB (potato dextrose broth). Then, 10 µL of suspension was inoculated into 15 mL of PDB placed in a 50 ml conical tube. The tube was incubated at 30 °C with shaking for 16–20 h and then centrifuged at 3,134 × g for 5 min. After the liquid part was discarded, the C. albicans cell pellet was washed with 1x PBS. The washed pellet was resuspended in new PBS, and the cell concentration was adjusted to 108 per mL.
Spores of A. niger were harvested from 2- to 3-week-old PDA culture plates. Approximately 20 mL of sterile deionized (DI) water was added to each PDA culture plate, and fungal materials were scraped using a plastic scraper. The resulting suspension was filtered through 3 layers of sterile Miracloth (Millipore, Burlington, MA, USA) to remove fungal mycelia. The spore suspension was centrifuged at 3,134 × g for 5 min, and the liquid was discarded. The spore pellet was washed with 1x PBS and then resuspended in fresh 1x PBS, and the spore concentration was adjusted to 108 per ml.
For treatment with Co-CPs, a suspension of C. albicans cells or A. niger spores was placed in a 24-well plate (1 mL per well). Then, the Co-CP (suspended in PBS) was added to each well to the indicated concentrations (2, 1, 0.5, 0.25, 0.125 mg mL−1). The well plate was shaken at room temperature for 4 days. The treated suspension was serially diluted, and then 100 µL of diluted suspension was spread on PDA plates. The plates were incubated at 30 °C for 2 days, and the number of colonies was counted. Three replicates of treatment were performed at each compound concentration, and colonies were counted from 9 replicate plates.
Analysis of morphology by scanning electron microscopy (SEM)
Morphological changes in fungal cells and spores were analysed using SEM following Co-CP treatment. After treatment with Co-CPs (2 mg mL−1) for 4 days, control (no Co-CPs) and treated C. albicans cells and A. niger spores were washed twice with PBS and fixed in Karnovsky’s fixative [2% (v/v) paraformaldehyde and 2% (v/v) glutaraldehyde in 1x PBS] overnight at 4 °C. After primary fixation, the fungal cells and spores were washed three times with PBS. The washed cells and spores were mixed with approximately 1 ml of 1% (v/v) osmium tetroxide (Ted Pella Inc., Redding, CA, USA) and incubated for 2 h at room temperature in the dark. The cells and spores were then washed twice with PBS and dehydrated via serial incubations in 30, 50, 70, 80, 90, and 100% (twice) ethanol. In each dehydration step, the cells and spores were mixed with 1 ml of ethanol and incubated for 5 min at 4 °C. After dehydration, 500 µL of hexamethyldisilazane (HMDS) was applied to the samples, and the samples were incubated for 15 min for drying. This drying step was repeated once. Then, the dried cells and spores were mounted on carbon tape, coated with platinum, and examined under a scanning electron microscope (SEM) (JEOL, Tokyo, Japan).
Measurement of NO (nitric oxide) and H2O2 concentration
To analyse NO and H2O2 concentrations in 1x PBS solution treated with Co-CPs, 1 mL of PBS solution was placed in each well of a plate, and the Co-CPs (suspended in PBS) were added at the indicated concentrations (2, 1, 0.5, 0.25, 0.125 mg mL−1). The well plate was shaken at room temperature for 4 days. The concentrations of NO and H2O2 were measured immediately after Co-CPs were added (0 day) and after 4 days. The NO and H2O2 levels were measured using a QuantiChromTM Nitric Oxide Assay Kit (BioAssay Systems, Hayward, CA, USA) and an Amplex™ Red Hydrogen Peroxide/Peroxidase Assay Kit (Molecular Probes, Eugene, OR, USA), respectively, following the manufacturer’s protocols.
Co-CP degradation and release of CoII ion
To assess whether Co-CPs will be able to release their constituent CoII ions, 1 mL of 1x PBS solution was added to 1 mg of each compound and stirred for 1 day–4 days at r.t. After different times, each sample was centrifuged, and the supernatant was removed from the reaction tube. The amount of CoII released in the sample was quantified by ICP-MS (ICP-MS measurements were performed in Seoul Center, Korea Basic Science Institute). The degree of degradation is represented as the concentration of CoII ions released into the medium.
References
Yaghi, O. M. et al. Reticular synthesis and the design of new materials. Nature 423, 705–714 (2003).
Kim, J. et al. Assembly of metal-organic frameworks from large organic and inorganic secondary building units: new examples and simplifying principles for complex structures. J. Am. Chem. Soc. 123, 8239–8247 (2001).
Foo, M. L., Matsuda, R. & Kitagawa, S. Functional hybrid porous coordination polymers. Chem. Mater. 26, 310–322 (2014).
Eddaoudi, M. et al. Zeolite-like metal–organic frameworks (ZMOFs): design, synthesis, and properties. Chem. Soc. Rev. 44, 228–249 (2015).
He, Y., Zhou, W., Krishnad, R. & Chen, B. Microporous metal–organic frameworks for storage and separation of small hydrocarbons. Chem. Commun. 48, 11813–11831 (2012).
Cadiau, A., Adil, K., Bhatt, P. M., Belmabkhout, Y. & Eddaoudi, M. A metal-organic framework–based splitter for separating propylene from propane. Science 353, 137–140 (2016).
Cui, X. et al. Pore chemistry and size control in hybrid porous materials for acetylene capture from ethylene. Science 353, 141–144 (2016).
Li, J.-R. et al. Carbon dioxide capture-related gas adsorption and separation in metal-organic frameworks. Coord. Chem. Rev. 255, 1791–1823 (2011).
Sumida, K. et al. Carbon dioxide capture in metal-organic frameworks. Chem. Rev. 112, 724–781 (2012).
Bae, Y.-S. & Snurr, R. Q. Development and evaluation of porous materials for carbon dioxide separation and capture. Angew. Chem., Int. Ed. 50, 11586–11596 (2011).
Eberle, U., Felderhoff, M. & Schüth, F. Chemical and physical solutions for hydrogen storage. Angew. Chem., Int. Ed. 48, 6608–6690 (2009).
Seayad, A. M. & Antonelli, D. M. Recent advances in hydrogen storage in metal-containing inorganic nanostructures and related materials. Adv. Mater. 16, 765–777 (2004).
Suh, M. P., Park, H. J., Prasad, T. K. & Lim, D.-W. Hydrogen storage in metal-organic frameworks. Chem. Rev. 112, 782–835 (2012).
Farrusseng, D., Aguado, S. & Pinel, C. Metal-organic frameworks: opportunities for catalysis. Angew. Chem., Int. Ed. 48, 7502–7513 (2009).
Dhakshinamoorthy, A., Opanasenko, M., Čejka, J. & Garcia, H. Metal organic frameworks as heterogeneous catalysts for the production of fine chemicals. Catal. Sci. Technol. 3, 2509–2540 (2013).
Gu, J.-M., Kim, W.-S. & Huh, S. Size-dependent catalysis by DABCO-functionalized Zn-MOF with one-dimensional channels. Dalton Trans. 40, 10826–10829 (2011).
Horcajada, P. et al. Metal-organic frameworks in biomedicine. Chem. Rev. 112, 1232–1268 (2012).
Cunha, D. et al. Rationale of drug encapsulation and release from biocompatible porous metal−organic frameworks. Chem. Mater. 25, 2767–2776 (2013).
Hwang, I. H. et al. Bifunctional 3D Cu-MOFs containing glutarates and bipyridyl ligands: selective CO2 sorption and heterogeneous catalysis. Dalton Trans. 41, 12759–12765 (2012).
Hwang, I. H. et al. Zn-MOFs containing flexible α,ω-alkane (or alkene)-dicarboxylates and 1,2-bis(4-pyridyl)ethane ligands: CO2 sorption and photoluminescence. Cryst. Growth Des. 13, 4815–4823 (2013).
Kim, H.-C. et al. Zn-MOFs containing flexible α,ω-alkane (or alkene)-dicarboxylates with 1,2-bis(4-pyridyl)ethylene: comparison with Zn-MOFs containing 1,2-bis(4-pyridyl)ethane ligands. Cryst. Eng. Comm, 19, 99–109 (2017) and references there in.
Bezuidenhout, C. X., Smith, V. J., Bhatt, P. M., Esterhuysen, C. & Barbour, L. J. Extreme carbon dioxide sorption hysteresis in open-channel rigid metal–organic frameworks. Angew. Chem. Int. Ed. 54, 2079–2083 (2015).
Bezuidenhout, C. X., Smith, V. J., Esterhuysen, C. & Barbour, L. J. Solvent- and pressure-induced phase changes in two 3D copper glutarate-based metal–organic frameworks via glutarate (+gauche ⇄ −gauche) conformational Isomerism. J. Am. Chem. Soc. 139, 5923–5929 (2017).
Rather, B. & Zaworotko, M. J. A 3D metal-organic network, [Cu2(glutarate)2(4,4’-bipyridine)], that exhibits single-crystal to single-crystal dehydration and rehydration, Chem. Commun. 830–831 (2003).
Chen, B. et al. Metal-organic framework with rationally tuned micropores for selective adsorption of water over methanol. Inorg. Chem. 47, 5543–5545 (2008).
Seco, J. M. et al. Modular structure of a robust microporous MOF based on Cu2 paddle-wheels with high CO2 selectivity. Chem. Commun. 49, 11329–11331 (2013).
Jo, J. H., Kim, H.-C., Huh, S., Kim, Y. & Lee, D. N. Antibacterial activities of Cu-MOFs containing glutarates and bipyridyl ligands. Dalton Trans. 47, 8084–8093 (2019).
Kabir, M. A., Hussain, M. A. & Ahmad, Z. Candida albicans: a model organism for studying fungal pathogens. ISRN Microbiol. 2012, 538694 (2012).
Baker, S. E. Aspergillus niger genomics: past, present and into the future. Med. Mycol. 44, S17–S21 (2006).
Bouson, S., Krittayavathananon, A., Phattharasupakun, N., Siwayaprahm, P. & Sawangphruk, M. Antifungal activity of water-stable copper-containing metal-organic frameworks. R. Soc. open sci. 4, 170654 (2017).
Chiericatti, C., Basillico, J. C., Zapata, M. L. & Zamaro, J. M. Novel application of HKUST-1 metal-organic framework as antifungal: Biologicla tests and physicochemical characterizations. Micropor. Mesopor. Mater. 162, 60–63 (2012).
Zhuang, W. et al. Highly potent bactericidal activity of porous metal-organic frameworks. Adv. Healthcare Mater. 1, 225–238 (2012).
Spek, A. L. PLATON-A multipurpose Crystallographic Tool, Utrecht University, Utrecht, The Netherlands, (2004).
Blatov, V. A., Carlucci, L., Ciani, G. & Proserpio, D. M. Interpenetrating metal–organic and inorganic 3D networks: a computer-aided systematic investigation. Part I. Analysis of the Cambridge structural database. CrystEngComm 6, 377–395 (2004).
Jaros, S. W. et al. Silver(I) 1,3,5-Triaza-7-phosphaadamantane Coordination Polymers Driven by Substituted Glutarate and Malonate Building Blocks: Self-Assembly Synthesis, Structural Features, and Antimicrobial Properties. Inorg. Chem. 55, 5886–5894 (2016).
Jaros, S. W. et al. Bioactive Silver−Organic Networks Assembled from 1,3,5-Triaza-7-phosphaadamantane and Flexible Cyclohexanecarboxylate Blocks. Inorg. Chem. 55, 1486–1496 (2016).
Jaros, S. W. et al. Aliphatic Dicarboxylate Directed Assembly of Silver(I) 1,3,5-Triaza-7-phosphaadamantane Coordination Networks: Topological Versatility and Antimicrobial Activity. Cryst. Growth Des. 14, 5408–5417 (2014).
Jaros, S. W. et al. New silver BioMOFs driven by 1,3,5-triaza-7-phosphaadamantane-7-sulfide (PTALS): synthesis, topological analysis and antimicrobial activity. CrystEngComm 15, 8060–8064 (2013).
Lara, H. H. et al. Effect of silver nanoparticles on Candida albicans biofilms: an ultrastructural study. J. Nanobiotechnol 13, 91 (2015).
Dutta, R. K., Nenavathu, B. P., Gangishetty, M. K. & Reddy, A. V. Studies on antibacterial activity of ZnO nanoparticles by ROS incuced lipid peroxidation. Colloid. Surface. B: Biointerface. 94, 143–150 (2012).
Sawangphruk, M., Srimuk, P., Chiochan, P., Sangsri, T. & Siwayaprahm, P. Synthesis and antifungal activity of reduced graphene oxide nanosheets. Carbon 50, 5156–5161 (2012).
Fu, P. P., Xia, Q., Hwang, H.-M., Ray, P. C. & Yu, H. Mechanisms of nanotoxicity: generation of reactive oxygen species. J. Food Drug Anal. 22, 64–75 (2014).
Liu, Y., Howarth, A. J., Hupp, J. T. & Farha, O. K. Selective photooxidation of a mustard-gas simulant catalyzed by a porphyrinic metal-organic framework. Angew. Chem. Int. Ed. 54, 9001–9005 (2015).
Li, P. et al. Metal-organic frameworks with photocatalytic bactericidal activity for integrated air cleaning. Nat. Comm. 10, 2177 (2019).
Sheldrick, G. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. C: Struct. Chem. 71, 3–8 (2015).
Acknowledgements
This work was supported by the Basic Science Research Program of the National Research Foundation of Korea (NRF), which was funded by the Ministry of Education, Science and Technology (2016R1D1A1B03934922, 2018R1D1A1B07043017, and 2018R1D1A1B07045327) and by RP-Grant 2019 of Ewha Womans University.
Author information
Authors and Affiliations
Contributions
The original idea was conceived by Y.K. and G.P.; experiments and data analysis were performed by S.H., H.-C.K., S.M., M.V., J.-S.L. and H.-R.J.; the crystal structures of the MOFs were analysed by Y.K.; and the manuscript was drafted by Y.K., G.P. and S.H. All authors analysed the data and reviewed and contributed to the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kim, HC., Mitra, S., Veerana, M. et al. Cobalt(II)-coordination polymers containing glutarates and bipyridyl ligands and their antifungal potential. Sci Rep 9, 14983 (2019). https://doi.org/10.1038/s41598-019-50258-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-50258-1
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.