Abstract
The pathogenesis of heart failure associated with dilated cardiomyopathy (DCM) may result in part from adenosine triphosphate (ATP) dysregulation in the myocardium. Under these conditions, diabetes-associated protein in insulin-sensitive tissue (DAPIT), which is encoded by the upregulated during skeletal muscle growth 5 (USMG5) gene, plays a crucial role in energy production by mitochondrial ATP synthase. To determine whether USMG5 is related to the development of heart failure, we performed clinical and experimental studies. Microarray analysis showed that the expression levels of USMG5 were positively correlated with those of natriuretic peptide precursor A in the human failed myocardium. When endogenous z-usmg5 in zebrafish was disrupted using morpholino (MO) oligonucleotides, the pericardial sac and atrial areas were larger and ventricular fractional shortening was reduced compared to in the control MO group. The expression levels of natriuretic peptides were upregulated in the z-usmg5 MO group compared to in controls. Further, microarray analysis revealed that genes in the calcium signalling pathway were downregulated in the z-usmg5 MO group. These results demonstrate that DAPIT plays a crucial role in the development of heart failure associated with DCM and thus may be a therapeutic target for heart failure.
Similar content being viewed by others
Introduction
Heart failure is a leading cause of mortality, even in developed countries. The prevalence of heart failure among the elderly population (>70 years old) is approximately 10% and is estimated to be increasing1,2. Despite improvements in pharmacological and mechanical therapies, the prognosis of heart failure patients remains poor, with a 1-year mortality of 25–30%3. Therefore, dissecting the molecular mechanisms of heart failure progression is necessary to develop novel therapeutics.
Dilated cardiomyopathy (DCM) is an idiopathic cardiomyopathy condition causing progressive left ventricular enlargement and systolic dysfunction related to advanced heart failure and cardiac transplantation. Although gene mutations are evident in 30–35% of DCM patients4,5, the mechanism of disease progression remains unclear. As the heart consumes more oxygen than all other human organs, myocardial energy metabolism is an important factor affecting disease progression in DCM5. Furthermore, impaired adenosine triphosphate (ATP) utilization in the myocardium has been associated with the severity and mortality of DCM6.
Mitochondria are organelles that produce most of the cellular ATP in cardiomyocytes. In this organelle, ATP synthase is an important enzyme complex that generates ATP, functioning in the last step of the mitochondrial oxidative phosphorylation system7,8. Diabetes-associated protein in insulin-sensitive tissue (DAPIT) is encoded by the upregulated during skeletal muscle growth 5 (USMG5), which is a component of the Fo subunit of the ATP synthase9,10 and plays a crucial role in energy production during ATP synthase11. However, the role of DAPIT in the myocardium has not been investigated. In this study, we hypothesized that DAPIT, a component of ATP synthase, plays a crucial role in the pathogenesis of heart failure associated with DCM.
Results
Expression levels of USMG5 and natriuretic peptide precursor A in failed myocardium of NICM patients
We first investigated whether the expression levels of USMG5 are associated with the severity of heart failure in patients with non-ischemic cardiomyopathy (NICM) by microarray analysis. The relative expression levels of USMG5 were generally positively correlated with those of natriuretic peptide precursor A (NPPA) in failed myocardium (R2 = 0.40, p = 0.08, Fig. 1). There were no significant differences in the relative expression levels of NPPA and USMG5 between control subjects (n = 2) and NICM patients (n = 8) (Suppl. Figure S1)
Phenotypes recapitulating heart failure induced by z-usmg5 MO injection in zebrafish embryos
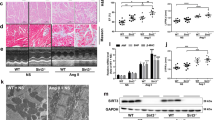
We then examined whether knockdown of z-usmg5 induced phenotypes of heart failure in zebrafish embryos. Compared to control MO-injected embryos, the percentage of embryos with a swollen pericardial sac (>2 SD from wild-type embryos) was significantly greater in z-usmg5 MO-injected embryos at both 48 and 72 hours post-fertilization (hpf) (Suppl. Figure S2). Evaluation at 72 hpf revealed no differences in heart rates between control MO (161 ± 12 bpm), z-usmg5 MO (158 ± 14 bpm), and z-usmg5 MO with its wild-type mRNA (152 ± 12 bpm) groups (Suppl. Figure S3). However, the z-usmg5 MO group (n = 19) showed a larger pericardial sac area (60,520 ± 16,872 μm2 vs. 32,962 ± 6,295 μm2, p < 0.0001) and atrial area (11,190 ± 1,370 μm2 vs. 9,052 ± 1,361 μm2, p < 0.0001) than the control MO group (n = 21) (Figs 2 and 3A,B). Further, ventricular fractional shortening was significantly reduced in the z-usmg5 MO group compared to in the control MO group (16.5 ± 4.7% vs. 22.0 ± 6.0%, p = 0.003) (Figs 2 and 3D). Importantly, there were no differences in cardiac morphology and function, such as pericardial area, atrial area, ventricular diameter, and ventricular fractional shortening between the wild-type embryos and control MO group (Suppl. Table S1).
Representative images of zebrafish hearts at 72 hpf. Brightfield and fluorescent microscopy images of zebrafish embryos injected control MO (A,B) and z-usmg5 MO (C,D), respectively. z-usmg5 MO injected embryos showed swollen pericardial sacs (C) and reduced ventricular contraction (D,G,H) compared to the control MO embryos (B,E,F). hpf: hours post fertilization, MO: morpholino oligonucleotide, Dd: diastolic diameter, Ds: systolic diameter.
Quantitative measurements of cardiac dimensions and functions in the zebrafish embryos injected with control MO (control MO group, n = 21), z-usmg5 MO (z-usmg5 MO group, n = 19) and z-usmg5 MO with its wild-type mRNA (z-usmg5 MO with mRNA group, n = 20). Compared to the control, the MO group showed that the pericardial sac area (60,520 ± 16,872 μm2 vs. 32,962 ± 6,295 μm2, p < 0.0001) (A) and the atrial area (11,190 ± 1,370 μm2 vs. 9,052 ± 1,361 μm2, p < 0.0001) (B) were significantly increased. Although there were no differences in ventricular diastolic diameter (C), fractional shortening was significantly reduced in the z-usmg5 MO group compared to in the control MO group (16.5 ± 4.7% vs. 22.0 ± 6.0%, p = 0.003) (D). These heart failure phenotypes caused by z-usmg5 knockdown were rescued by co-injection of its wild-type mRNA ((A–D), z-usmg5 MO with mRNA group). Statistical analyses were performed using Mann-Whitney U test.
To confirm that the observed phenotypes were specific to z-usmg5 deficiency, we conducted rescue experiments. Co-injection of wild-type z-usmg5 mRNA with its MO (n = 20) resulted in a significant reduction in the pericardial sac and atrial areas and improvement in ventricular fractional shortening compared to injection of only z-usmg5 MO (Fig. 3A−D).
Changes in foetal cardiac genes and ATP synthase-related genes in z-usmg5 MO-injected zebrafish embryos
We next performed microarray analysis to examine the differences in gene expression between z-usmg5 MO-injected, control MO-injected, and wild-type embryos. Hierarchical clustering analysis revealed that differences in gene expression profiles were more evident between wild-type embryos and z-usmg5 MO embryos than between wild-type embryos and control MO embryos (Fig. 4A). Compared to that in wild-type, 2,096 genes were upregulated (Fig. 4B) and 4,298 genes were downregulated (Fig. 4C) in z-usmg5 MO embryos when a fold-change of 1.5 was set as the minimum.
Hierarchical clustering analysis revealed that differences in gene expression profiles were more evident between wild-type embryos and z-usmg5 MO embryos than between wild-type embryos and control MO embryos (A). A Venn diagram showing that 2,096 genes were upregulated > 1.5-fold (B) and 4,298 genes were downregulated by > 1.5-fold (C) in z-usmg5 MO embryos compared to wild-type embryos.
Gene Ontology (GO) analysis revealed that 58 GO terms were upregulated and 133 were downregulated in z-usmg5 MO embryos compared to in wild-type embryos. Importantly, GO terms related to ATP driving intracellular ions transfer, such as ATPase activity coupled to the transmembrane movement of ions (GO: 0042625), substrate-specific transmembrane transporter activity (GO: 0022891), and ion transmembrane transporter activity (GO: 0015075), were significantly downregulated (Table 1).
Pathway enrichment analysis revealed that 7 pathways were downregulated and 11 pathways were upregulated in embryos injected with z-usmg5 MO compared to in wild-type embryos (Table 2). Among the 7 downregulated pathways, major factors involved in calcium signalling, such as atp2a2 encoding sarcoplasmic/endoplasmic reticulum Ca2+-ATPase (SERCA), slc8a1b encoding a member of proteins constituting the sodium/calcium exchanger (NCX), ryr3 encoding a ryanodine receptor (RyR), and atp2b3 encoding a plasma membrane calcium ATPase, were significantly downregulated (p < 0.0001) in z-usmg5 MO embryos compared to in wild-type embryos (http://www.wikipathways.org/index.php/Pathway:WP1365; Fig. 5). Furthermore, the transforming growth factor β signalling pathway was significantly upregulated (p < 0.0001) in z-usmg5 MO embryos compared in wild-type embryos.
Pathway analysis using WikiPathways revealed that the factors related to the calcium signalling pathway (Kelder, T., Chichester, C., Hanspers, K.: Calcium Regulation in the Cardiac Cell (Danio rerio). http://www.wikipathways.org/instance/WP1365_r71502) were significantly downregulated (p < 0.0001) in z-usmg5 MO embryos compared to in wild-type embryos. Downregulated genes in z-usmg5 MO embryos with a fold change > 1.5 compared to wild-type embryos are highlighted in yellow boxes.
Quantitative real-time PCR confirmed that z-usmg5 knockdown did not affect other components of ATP synthase such as atp5a, atp5b, and atp5h (Fig. 6A). Additionally, we confirmed that the expression levels of major factors involved in calcium signalling including atp2a2, slc8a1b, ryr3, and atp2b3 were significantly downregulated (Fig. 6B). The expression levels of cardiac-specific sarcomere genes including vmhc (cardiac myosin heavy chain), myl2 (cardiac myosin light chain), and mybpc3 (cardiac myosin binding protein c) were significantly downregulated in z-usmg5 MO embryos compared to the control MO embryos (Fig. 6C). Furthermore, the expression levels of natriuretic peptides, such as nppa (NM_198800) and nppb (NM_001327776), were significantly upregulated in z-usmg5 MO embryos compared to in control MO embryos (Fig. 6D).
Quantitative real-time PCR confirmed that the expression level of z-usmg5 was significantly downregulated in z-usmg5 MO-injected embryos. However, the expression levels of other components of ATP synthase such as atp5a, atp5b, and atp5h were not significantly changed (A). The expression levels of major factors in the calcium signalling pathway, such as atp2a2a, atp2b3a, ryr3, and slc8a1b, were significantly downregulated (B) and cardiac-specific sarcomere-associated genes, such as vmhc, mybpc3, and myl2, were downregulated (C) in z-usmg5 MO embryos compared to in control MO embryos. Furthermore, the expression levels of the natriuretic peptide a (nppa) and b (nppb) were significantly upregulated (D) in z-usmg5 MO embryos compared to in control MO embryos. *P < 0.05 compared to the respective control. Each bar indicates the mean ± SD of three independent experiments. White and black bars indicate control MO embryos and z-usmg5 MO embryos, respectively. The results are representative of three independent experiments. Statistical analyses were performed using Student’s t test.
Discussion
In this study, we demonstrated that z-usmg5 MO-injected zebrafish embryos showed (1) reduced ventricular contraction and pericardial sac enlargement, (2) downregulated expression levels of major factors in the calcium signalling pathway, and (3) gene expression fluctuations of natriuretic peptides and cardiac sarcomere-associated peptides. These findings indicate that z-usmg5 knockdown can cause heart failure phenotypes associated with DCM in zebrafish embryos.
DAPIT, encoded by USMG5, consists of 58 amino acids12 and is known to be a Fo subunit component of mitochondrial ATP synthase13,14. Although whether DAPIT contributes to ATP production and maintaining ATP concentrations in cardiomyocytes is unclear, several studies have revealed the functions of this protein. Ohsakaya et al. showed that mitochondrial ATP production was significantly reduced in DAPIT-depleted HeLa cells11, indicating that DAPIT plays a critical role in ATP production by mitochondrial ATP synthase.
ATP synthase is the major generator of ATP in the mitochondrial respiratory chain and maintains ATP concentrations in cardiomyocytes7. As the heart requires ATP synthesis than any other organ, cardiac dysfunction in z-usmg5 knockdown zebrafish embryos may result from mitochondrial ATP synthase dysfunction and subsequent intracellular ATP depletion (Suppl. Figure S4)8. Indeed, our data demonstrate that GO terms related to ATP driving intracellular ion transfer, such as ATPase activity, were significantly downregulated in z-usmg5 knockdown zebrafish embryos (Table 1), supporting this hypothesis. Additionally, we demonstrated that factors in the calcium signalling pathway, a major pathway activated in the failing heart, were significantly downregulated in z-usmg5 knockdown embryos (Fig. 5). ATP synthase dysfunction resulted in excessive reactive oxygen species (ROS) accumulation in the cytosol, which can cause calcium overload in cardiomyocytes (Fig. 5)15,16. Previous studies demonstrated that intracellular accumulation of ROS leads to the deterioration of the calcium regulator pumps in the sarcoplasmic reticulum (SR) membrane, such as RyR and SERCA, and in the cell membrane, such as NCX17,18,19,20. Subsequently, calcium overload may lead to cardiomyocyte apoptosis21 and myocardial fibrosis22, resulting in contractile dysfunction of cardiomyocytes (Suppl. Figure S4)23,24. Further, upregulation of the TGF-β signalling pathway (Table 2) and foetal cardiac genes (Fig. 6D) in z-usmg5 knockdown embryos may contribute to the development of pathological cardiac remodelling25,26. As the activity of the mitochondrial respiratory chain including the ATP synthase is reduced in the failing myocardium8,27, the intervention of ATP synthase-interacting proteins modulating ATP synthase function may be a novel therapeutic target for heart failure in DCM patients. To date, cyclophilin D28, ATPase inhibitor factor 129, and protein kinase C delta30 have been reported to interact with and modulate ATP synthase function. For DAPIT, Kontro et al. showed that overexpression of DAPIT saturated the respiratory chain by decreasing H+-ATP synthase activity, leading to increased mitochondrial membrane potential and superoxide levels in human embryonic kidney 293 cells14. Taken together with our results, in which DCM phenotypes were detected in z-usmg5 knockdown zebrafish, these observations reported by Kontro et al. indicate that the modulation of DAPIT may be important for maintaining ATP synthase activity and mitochondrial respiratory function and thus improving the cardiac function in heart failure patients. Indeed, co-injection of wild-type z-usmg5 mRNA with z-usmg5 MO into the embryos rescued the DCM phenotypes induced by z-usmg5 MO alone (Fig. 3), suggesting that modulation of USMG5 mRNA levels can improve cardiac function. Further, expression levels of USMG5 were generally positively correlated with the severity of heart failure in NICM patients (Fig. 1), indicating that DAPIT expression is compensatory to preserve ATP production during the process of heart failure progression. Together, these findings suggest that DAPIT plays a crucial role in preserving ATP production in the failing heart and thus is a novel target molecule for DCM treatment.
There are several limitations to our study. First, our microarray data was not validated at the protein level by western blot analysis. However, we confirmed the major results of microarray analysis by quantitative real-time PCR. Second, DCM phenotypes in z-usmg5 knockdown zebrafish embryos should be carefully interpreted because generalized DAPIT knockdown in zebrafish may increase myocardial stress to meet the oxygen demand of peripheral tissues. However, systemic oxygen demand may not be significantly increased in z-usmg5 knockdown zebrafish embryos compared to in control zebrafish embryos because heart rates were similar between the 2 groups (Suppl. Figure S3). Third, although we suggest that z-usmg5 knockdown can induce intracellular ATP depletion leading to the heart failure phenotype in zebrafish embryos, further investigations are needed to confirm our results and reveal the role of DAPIT in mitochondrial function. Finally, the statistical evaluations of NPPA and USMG5 expression levels between human control subjects and NICM patients were difficult because of the small sample size (control = 2). Additional study will be necessary to confirm these differences.
Our data demonstrate that z-usmg5 knockdown induced phenotypes recapitulating heart failure in human with reduced ventricular contraction in zebrafish embryos. DAPIT, a component of ATP synthase, may play a crucial role in the pathogenesis of heart failure associated with DCM and thus could be a therapeutic target for heart failure.
Methods
Ethics
This study complied with the Declaration of Helsinki. All zebrafish experimental protocols were approved by the Animal Care and Use Committee of Kanazawa University. In addition, all the experimental protocols used in this study were approved by the Bioethical Committee of Medical Researches of Kanazawa University.
Microarray analysis using human failed myocardium
Eight RNA samples extracted from human failed myocardium and two samples of normal control subjects were used. Failed myocardium samples were obtained from patients with non-ischemic cardiomyopathy (NICM) who underwent cardiac reconstructive surgery, such as a Batista or Dor procedure, after obtaining written informed consent31. The use of myocardial samples was approved by the Bioethical Committee of Medical Researches of the Osaka University (Suita, Japan). Gene expression levels were evaluated using a HG-U95 Affymetrix GeneChip. All expression data were normalized by global scaling and were analysed using GeneSpring software (Agilent Technologies, Santa Clara, CA, USA)31.
Animals
All zebrafish experiments were performed in accordance with the Guide for the Care and Use of Laboratory Animals (National Institutes of Health Publication, 8th Edition, 2011). Zebrafish were raised and maintained at 28 °C on a 14:10 h light-dark cycle. We used the hspGFF3A strain of zebrafish, which expresses green fluorescent protein in the heart32, to analyse the size and function of the cardiac ventricle.
In vitro synthesis of zebrafish usmg5 mRNA
The cDNA of the wild-type zebrafish usmg5 orthologue, z-usmg5 (NM_001200033), was subcloned into a PCS2P + plasmid expression vector. Capped mRNA was synthesized using mMESSAGE mMACHINE SP6 Transcription Kit (Thermo Scientific, Waltham, MA, USA) according to the manufacturer’s protocol.
Design of z-usmg5 morpholino oligonucleotides and injection
The morpholino (MO) oligonucleotides were synthesized using Gene-Tools (Philomath, OR, USA). To knockdown z-usmg5, we designed a splice site blocking MO to disrupt the translation of z-usmg5. The sequence of the acceptor site MO was 5′-GTATGCAATCTGTTAATAAAGGAGA-3′ and that of the donor site MO was 5′-TAATGTCGACTTACATTCCTCCTGC-3′. We also designed control MOs with 5-base pair mismatches corresponding to each of the z-usmg5 MOs. The control MO sequences for the donor and acceptor MOs were 5′-GTATaCAATaTaTTAATAAAaGAaA-3′ and 5′-TAATaTCGAaTTAaATTCaTCCTaC-3′, respectively. Both z-usmg5 MOs (2.5 ng for the acceptor and donor site MO) or equal amounts of control MOs were injected into the hspGFF3A zebrafish strain embryos at the 1-cell stage using a microinjector. For rescue experiments, we co-injected synthesized wild-type z-usmg5 mRNA (400 pg) with z-usmg5 MO into the embryos.
Quantification of cardiac morphology and function in zebrafish embryos
Zebrafish hearts at 72 h post fertilization (hpf) were recorded using a Leica digital camera (DFC 310 for colour images and DFC 365 FX for fluorescence images) on a fluorescence stereomicroscope M205A (Leica, Wetzlar, Germany). Cardiac morphologies such as ventricular area, atrial size, and pericardial sac area were quantified with LAS AF software (version 3.1.0; Leica). The end-diastolic ventricular dimension was measured at its largest point. Ventricular fractional shortening was evaluated with recorded movies converted to M-mode images using the original software31,33.
RNA extraction, cDNA preparation, and microarray analysis of zebrafish embryos
Total RNA was isolated from the whole bodies of 72 hpf zebrafish embryos homogenized in 500 μL of RNA BEE reagent (Tel-Test, Friendswood, CA, USA). Subsequently, cDNA was synthesized from extracted RNA using the One-Step reverse transcription PCR Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s protocol. The integrity and concentration of extracted RNA and cDNA were evaluated using the Nano Drop 2000 (Thermo Scientific). The aliquots were stored at −80 °C. We performed microarray analyses to evaluate the influence of z-usmg5 knockdown on gene expression of zebrafish embryos. RNA samples isolated from wild-type embryos, control MO, and z-usmg5 MO-injected embryos were compared. Gene expression was evaluated using the GeneChip Zebrafish Genome array containing the probes for 15,509 genes provided by Affymetrix (Santa Clara, CA, USA). Microarray data were analysed using GeneSpring GX software (version 12.1; Agilent Technologies). Gene Ontology (GO) annotation of genes was obtained from the NCBI gene database. Pathway enrichment analysis was performed using the Gene Spring GX software importing WikiPathways (http://www.wikipathways.org)34.
Quantitative real-time PCR analysis
The primer sequences of z-usmg5 were designed using Primer3Plus (primer3plus.com) and additional primer sequences were taken from the literature35,36,37,38 (Suppl. Table S2). Quantitative real-time PCR was performed in a total volume of 25 μL, comprised of 12.5 μL of SYBR Green Master Mix (Thermo Fisher Scientific), 50 ng of cDNA, and 200 nM of each primer using an Stratagene Mx3000P qPCR system (Agilent Technologies). Duplicate reactions were performed for each cDNA sample. Forty amplification cycles were performed, with each cycle consisting of 95 °C for 30 s followed by 60 °C for 1 min. Amplification and dissociation curves generated by the MxPro QPCR software (version 4.10; Agilent Technologies) were used for gene expression analysis. The relative quantification of expression of each gene was normalized to the gene expression of gapdh.
Statistics
All analyses were performed using commercially available software (JMP 9.0, SAS Institute, Cary, NC, USA). Data are presented as the mean ± SD. Demographic data were expressed as continuous and categorical variables. Comparisons between continuous variables were performed with the Student’s t test or Mann-Whitney U test. Categorical variables were compared using the chi-square test. P values < 0.05 were considered statistically significant.
Change history
14 May 2018
A correction to this article has been published and is linked from the HTML and PDF versions of this paper. The error has been fixed in the paper.
References
Bleumink, G. S. et al. Quantifying the heart failure epidemic: prevalence, incidence rate, lifetime risk and prognosis of heart failure The Rotterdam Study. Eur. Heart J. 25, 1614–1619 (2004).
Roger, V. L. The heart failure epidemic. Int. J. Environ. Res. Public Health 7, 1807–1830 (2010).
Mosterd, A. & Hoes, A. W. Clinical epidemiology of heart failure. Heart 93, 1137–1146 (2007).
Petretta, M., Pirozzi, F., Sasso, L., Paglia, A. & Bonaduce, D. Review and metaanalysis of the frequency of familial dilated cardiomyopathy. Am. J. Cardiol. 108, 1171–1176 (2011).
Japp, A. G., Gulati, A., Cook, S. A., Cowie, M. R. & Prasad, S. K. The diagnosis and evaluation of dilated cardiomyopathy. J. Am. Coll. Cardiol. 67, 2996–3010 (2016).
Neubauer, S. et al. Myocardial phosphocreatine-to-ATP ratio is a predictor of mortality in patients with dilated cardiomyopathy. Circulation 96, 2190–2196 (1997).
Tsutsui, H., Kinugawa, S. & Matsushima, S. Oxidative stress and mitochondrial DNA damage in heart failure. Circ. J. 72(Suppl A), A31–A37 (2008).
Long, Q., Yang, K. & Yang, Q. Regulation of mitochondrial ATP synthase in cardiac pathophysiology. Am. J. Cardiovasc. Dis. 5, 19–32 (2015).
Meyer, B., Wittig, I., Trifilieff, E., Karas, M. & Schagger, H. Identification of two proteins associated with mammalian ATP synthase. Mol Cell Proteomics 6, 1690–1699 (2007).
Chen, R., Runswick, M. J., Carroll, J., Fearnley, M. & Walker, J. E. Association of two proteolopids of unknown function with ATP synthase from bovine heart mitochondria. FEBS Lett. 581, 3145–3148 (2007).
Ohsakaya, S., Fujikawa, M., Hisabori, T. & Yoshida, M. Knockdown of DAPIT (Diabetes-associated Protein in Insulin-sensitive Tissue) results in loss of ATP synthase in mitochondria. J. Biol. Chem. 286, 20292–20296 (2011).
Paivarinne, H. & Kainulainen, H. DAPIT, a novel protein down-regulated in insulin-sensitive tissues in streptozotocin-induced diabetes. Acta Diabetol. 38, 83–86 (2001).
Lee, J. et al. Organisation of subunits in the membrane domain of the bovine F-ATPase revealed by covalent cross-linking. J. Biol. Chem. 290, 13308–13320 (2015).
Kontro, H., Cannino, G., Rustin, P., Dufour, E. & Kainulainen, H. DAPIT over-expression modulates glucose metabolism and cell behaviour in HEK293T cells. PLos One 10, e0131990 (2015).
Gorlach, A., Bertram, K., Hudecova, S. & Krizanova, O. Calcium and ROS: A mutual interplay. Redox Biol. 6, 260–271 (2015).
Nakamura, K. et al. Beta-blockers and oxidative stress in patients with heart failure. Pharmaceuticals 4, 1088–1100 (2011).
Xu, K. Y., Zweier, J. L. & Becker, L. C. Hydroxyl radical inhibits sarcoplasmic reticulum Ca(2+)-ATPase function by direct attack on the ATP binding site. Circ. Res. 80, 76–81 (1997).
Yano, M. et al. Correction of defective interdomain interaction within ryanodine receptor by antioxidant is a new therapeutic strategy against heart failure. Circulation 112, 3633–3643 (2005).
Rothstein, E. C., Byron, K. L., Reed, R. E., Fliegel, L. & Lucchesi, P. A. H(2)O(2)-induced Ca(2+) overload in NRVM involves ERK 1/2 MAP kinases: role for an NHE-1-dependent pathway. Am. J. Physiol. Heart Circ. Physiol. 283, H598–H605 (2002).
Orrenius, S., Zhivotovsky, B. & Nicotera, P. Regulation of cell death: the calcium-apoptosis link. Nat. Rev. Mol. Cell Biol. 4, 552–565 (2003).
Von Harsdorf, R., Li, P. F. & Dietz, R. Signaling pathways in reactive oxygen species-induced cardiomyocyte apoptosis. Circulation 99, 2934–2941 (1999).
Swick, D. A., Pagano, P. J. & Colucci, W. S. Oxidative stress regulates collagen synthesis and matrix metalloproteinase activity in cardiac fibroblasts. Am. J. Physiol. Cell Physiol. 280, C53–C60 (2001).
Vassalle, M. & Lin, C. I. Calcium overload and cardiac function. J. Biomed. Sci. 11, 542–565 (2004).
Josephson, R. A., Silverman, H. S., Lakatta, E. G., Stern, M. D. & Zweier, J. L. Study of the mechanisms of hydrogen peroxide and hydroxyl free radical-induced cellular injury and calcium overload in cardiac myocytes. J. Biol. Chem. 266, 2354–2361 (1991).
Koitabashi, N. et al. A. Pivotal role of cardiomyocyte TGF-β signaling in the murine pathological response to sustained pressure overload. J. Clin. Invest. 121, 2301–2312 (2011).
Dobaczewski, M., Chen, W. & Frangogiannis, N. G. Transforming Growth Factor (TGF)-β signaling in cardidac remodeling. J. Mol. Cell Cardiol. 51, 600–606 (2011).
El-Hattab, A. W. & Scaglia, F. Mitochondrial cardiomyopathies. Front Cardiovasc. Med. 3, 25 (2016).
Halestrap, A. P. & Richardson, A. P. The mitochondrial permeability transition: a current perspective on its identity and role in ischaemia/reperfusion injury. J. Mol. Cell Cardiol. 78, 129–141 (2015).
Garcia-Bermudez, J. & Cuezva, J. M. The ATPase Inhibitory Factor 1 (IF1): a master regulator of energy metabolism and of cell survival. Biochem. Biophys. Acta 1857, 1167–1182 (2016).
Nguyen, T. T., Ogbi, M., Yu, Q. & Johnson, J. A. Attenuation of the hypoxia-induced protein kinase Cdelta interaction with the ‘d’ subunit of F1Fo-ATP synthase in neonatal cardiac myocytes: implications for energy preservation and survival. Biochem. J. 429, 335–345 (2010).
Seguchi, O. et al. A cardiac myosin light chain kinase regulates sarcomere assembly in the vertebrate heart. J. Clin. Invest. 117, 2812–2824 (2007).
Asakawa, K. et al. Genetic dissection of neural circuits by Tol2 transposon-mediated Gal4 gene and enhancer trapping in zebrafish. Proc. Natl. Acad. Sci. USA 105, 1255–1260 (2008).
Hodatsu, A. et al. Compound heterozygosity deteriorates phenotypes of hypertrophic cardiomyopathy with founder MYBPC3 muation: evidence from patients and zebrafish models. Am. J. Physiol. Heart Circ. Physiol. 307, H1594–H1604 (2014).
Kutmon, M. et al. WikiPathways: capturing the full diversity of pathway knowledge. Nucl. Acids Res. 44, D488–D494 (2016).
Shin, Y. H. et al. Cardiac transcriptome and dilated cardiomyopathy genes in zebrafish. Circ. Cardiovasc. Genet. 8, 261–269 (2015).
Li, M., Andersson-Lendahl, M., Sejersen, T. & Arner, A. Knockdown of fast skeletal myosin-binding protein C in zebrafish results in a severe skeletal myopathy. J. Gen. Physiol. 147, 309–322 (2016).
Ling, X. P., Lu, Y. H. & Huang, H. Q. Differential protein in zebrafish (Danio rerio) brain under the joint exposure of methyl parathion and cadmium. Environ. Sci. Pollut. Res. Int. 19, 3925–3941 (2012).
Wang, Z. et al. Zebrafish beta-adrenergic receptor mRNA expression and control of pigmentation. Gene 446, 18–27 (2009).
Acknowledgements
The authors are grateful to Takako Obayashi, BS, Yasuko Tamura, BS, for excellent technical assistance. This work was supported by a grant from JSPS KAKENHI (50377389).
Author information
Authors and Affiliations
Contributions
Y.N., C.N. and K.H. contributed to the analysis and interpretation of the data from the zebrafish experiments. M.Y. designed the study and wrote the manuscript. S.I., Y.N., O.S., and Y.A. contributed to analysis and interpretation of the microarray data in human myocardium., T.K., N.F. and M.K. contributed to the collection and assembly of the data. S.T. and M.K. contributed to the critical revision of the article and provided important intellectual content.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nagata, Y., Yamagishi, M., Konno, T. et al. Heart Failure Phenotypes Induced by Knockdown of DAPIT in Zebrafish: A New Insight into Mechanism of Dilated Cardiomyopathy. Sci Rep 7, 17417 (2017). https://doi.org/10.1038/s41598-017-17572-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-17572-y
This article is cited by
-
NP protein and F protein of pigeon paramyxovirus type 1 are associated with its low pathogenicity in chickens
Archives of Virology (2019)
-
Zebrafish heart failure models: opportunities and challenges
Amino Acids (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.