Abstract
Truncating variants in TTN (TTNtv), coding for the largest structural protein in the sarcomere, contribute to the largest portion of familial and ambulatory dilated cardiomyopathy (DCM). TTN haploinsufficiency caused by TTNtv is suggested as the disease mechanism. However, it is unclear whether TTN insufficiency causes DCM. Moreover, it is unknown whether modulation of downstream pathways serves as a therapeutic strategy for DCM caused by TTN insufficiency. Here, we show that reduction of cardiac Ttn expression by adeno-associated virus mediated shRNA (Ttn shRNA) generated DCM in mouse, demonstrating impaired cardiac performance, enlarged left ventricle (LV) and reduced LV wall thickness. A screen of 10 dysregulated and selected genes identified that Yin Yang 1 (Yy1) significantly suppressed DCM caused by Ttn shRNA. Gene profiling by RNAseq showed Yy1 modulated cell growth related genes. Ttn insufficiency activated cardiomyocyte cell cycle reentry by upregulating of Ccnd1 and Ccnd2. Cardiomyocytes activated by Ttn insufficiency did not advance to S phase by EdU incorporation assay. Yy1 promoted cardiomyocyte cell cycle by further enhancing Ccnd1 and Ccnd2 and increasing DNA replication without undergoing cell division. Importantly, upregulation of Ccnd1 and Ccnd2 suppressed DCM caused by Ttn insufficiency. Our findings demonstrate that DCM caused by Ttn insufficiency can be treated by therapeutically promoting cardiac cell cycle.
Similar content being viewed by others
Introduction
Dilated cardiomyopathy (DCM) occurs as many as 1 in 250 people1. There are currently no approved therapeutic products indicated for DCM treatment. Typical treatments are those indicated for broader cardiovascular disease. As the disease progresses, patients have limited treatment options, such as surgical or other invasive interventions and heart transplant2. DCM will result in heart failure with reduced ejection fraction (EF), usually without prior ischemic. The walls of the left ventricle are thin and over-expanded, leading to improper contraction and insufficient blood being pumped by the dilated heart. DCM results from a variety of external factors, such as viral infection, alcohol abuse, exposure to cardiotoxic medications and pregnancy, as well as from genetic variants in a number of causal genes including TTN, LMNA, ACTC1, MHY7 and PLN3,4,5,6,7,8,9. Titin (coded by TTN) plays an important role in the contraction and relaxation of cardiac muscles by connecting Z-disc to the M-line in the sarcomere. TTN truncating variants (TTNtv) contribute up to 15% ambulatory DCM and 25% end-stage or familial DCM3,8,9. In DCM, TTNtv are significantly enriched most in A band as well as other regions including I-band, Z-disc or M-line with variable position-related odds ratios3,10. TTN haploinsufficiency caused by TTNtv is emerging as the potential disease mechanism. Rat models with TTNtv in Z-disc and A band did not result in change in titin protein levels and obvious cardiac performance under normal physiological conditions. It was not known whether Ttn insufficiency causes DCM in mouse.
Mammalian cardiomyocytes exit their cell cycle shortly after birth, preventing heart repair through cardiac regeneration11. Cell cycle reactivation is observed in a limited cardiomyocytes under physiological conditions12,13. As an emerging strategy for cardiac therapeutic regeneration, we and others showed that enhancing this process by removing cell cycle brakes or augmenting accelerators is beneficial for heart recovery from heart failure models caused by ischemia or pressure overload14,15,16,17,18. We previously observed that cardiomyocytes undergo an extra round of cell cycle in Mybpc3 deficient mice, suggesting cell cycle reactivation could compensate sarcomere insufficiency19. It was not known whether Ttn insufficiency could induce cardiac cell cycle reactivation. Moreover, it was unknown whether enhancing this process could be a therapeutic strategy for DCM caused by Ttn insufficiency. Here, we address these major gaps and identify therapeutic candidates that are vital for advancing potential hits into a therapeutic approach for DCM.
Results
Ttn shRNA induces DCM in mice
To produce a set of Ttn shRNA sequences targeting Ttn gene expression, we designed 4 unique shRNA constructs, designated Ttn shRNA-1, -2, -3 or -4 (Fig. 1A). To assess their knockdown efficacy, we developed an EGFP/Cherry two-color system. Each shRNA (co-expressed with Cherry) was co-transfected with a plasmid carrying its respective target region (~500 bp) of Ttn gene (co-expressed with EGFP) into 293 T human embryonic kidney cells. Two shRNA constructs, Ttn shRNA-1 and -2, significantly reduced EGFP signal (Fig. 1B). To quantify shRNA knockdown efficacy, EGFP expression was measured by qPCR (Table 1). Ttn shRNA-1 and -2 significantly reduced Ttn target expression by ~ 90% (Fig. 1C). Both shRNA sequences were sub-cloned into AAV vectors (AAV-shRNA) to generate AAV-Ttn shRNA-1 and -2. To assess the efficacy of Ttn shRNA in vivo, we injected variable doses of AAV viruses encoding Ttn shRNA-1 and -2 (0.8E + 13, 2.5E + 13, and 5.0E + 13 vg/kg) into the thoracic cavity of 1.5 week old male neonates, as described20,21. Three weeks after viral transduction, all mice injected with medium or high dose of Ttn shRNA-1or -2 viruses (2.5E + 13 vg/kg, n = 4 and 5.0E + 13 vg/kg, n = 4) died due to severe heart failure. Mice treated with low dose (0.8E + 13 vg/kg, n = 8) were evaluated by echocardiography. Ttn shRNA-1 or -2 transduced mice reached the endpoint of showing impaired cardiac performance, enlarged left ventricle (LV) and reduced LV wall thickness (Fig. 1D). The fractional shortening (FS) of Ttn shRNA transduced mice (shRNA-1, 6.22 ± 1.81%, n = 8, P = 1.02E-13; shRNA-2, 4.12 ± 1.27%, n = 8, P = 2.90E-14) was significantly reduced compared to that of mice transduced with control shRNA (28.45 ± 3.80%, n = 10) (Table 2). Correspondingly, the left ventricular diastolic dimension (LVDD) (shRNA-1, 4.25 ± 0.11 mm, P = 1.75E-07; shRNA-2, 4.41 ± 0.18 mm, P = 1.67E-09, n = 8) was significantly enlarged compared to control (3.70 ± 0.12 mm, n = 10). Left ventricular wall thickness (LVWT) of Ttn shRNA-transduced mice (shRNA-1, 0.44 ± 0.06 mm, P = 1.60E-05, n = 8; shRNA-2, 0.41 ± 0.04 mm, P = 1.03E-06, n = 8) was significantly reduced compared to control (0.58 ± 0.05 mm, n = 10). Histological analysis did not reveal myocardial disarray in both Ttn shRNA-transduced mice. Moreover, cardiac fibrosis, assessed by image analysis of Masson’s trichrome (MT)-stained heart specimens and commonly observed in other DCM models including PLN (R9C) and Lmna−/−, was not significantly induced in Ttn shRNA transduced mice (shRNA-1, MT = 1.08 ± 0.21%, n = 7, P = ns; shRNA-2, MT = 1.20 ± 0.61%, n = 7, P = ns) relative to control shRNA-transduced mice (MS = 1.00 ± 0.24%, n = 9) (Fig. 1E)6,19,22. Instead, the expression of myocardial stress gene markers including Nppa, Nppb and Myh7 was significantly upregulated in mice transduced with Ttn shRNA compared to mice transduced with control shRNA (Fig. 1F). Ttn shRNA in vivo resulted in a significant ~50% reduction of Ttn transcripts in Ttn shRNA-1 or -2 transduced mice compared to control shRNA-transduced mice. We selected Ttn shRNA-2 (designated as Ttn shRNA) for subsequent assessment. Cardiomyocytes with anti-Troponin I staining was EGFP positive after AAV-EGFP transduction, indicating the virus was distributed to most cardiomyocytes (Supplementary Fig. S1A). Ttn shRNA significantly reduced Ttn protein level (n = 5, P = 4.36E-06) (Supplementary Fig. S1B,C). These results suggest Ttn expression level is critical for cardiac structure and contractile function of heart and reduction of Ttn expression causes DCM in mice. Blast of Ttn shRNA targeting sequence identified potential off-target genes with at least 4 mismatch such as Wdr95, Sptbn1 and Tnrc6b. We did not detect a significant reduction in these gene expression after Ttn shRNA by qPCR (Supplementary Fig. S1D). To extend the cardiac assessment window, we further titrated virus dose of Ttn shRNA to 0.2E + 13 vg/kg. Thus, we could evaluate cardiac performance by additional timepoints (Table 3).
Ttn shRNA induces dilated cardiomyopathy in mice. (A) Schematic of two main isoforms of Ttn and the targeting sites of 4 Ttn shRNA candidates. Image was adapted by genome assembly of UCSC (University of California, Santa Cruz) mouse genome browser (Dec 2011)43. (B) HEK293 cells co-transfected with Ttn targets (Green) and corresponding shRNAs or control shRNA (Red). Magnification = 4×, scale bar = 500 µm. (C) Quantitative real-time PCR analysis of EGFP expression in HEK293 cells with two-color system. Transcription level were normalized to targets with control shRNA. n = 3. (D) H&E and Masson Trichrome (MT) of paraffin sections of mice 3 weeks after control shRNA, Ttn shRNA-1 and -2 transduction respectively. Virus does, 0.8E + 13 vg/kg, n ≥ 8. For completed images: magnification = 4×, scale bar = 1000 µm; for enlarged images: magnification = 20 × , scale bar = 100 µm. (E) Quantification of myocardial fibrosis of MT sections in (d), n ≥ 7. (F) Quantitative real-time PCR analysis of Ttn, Nppa, Nppb and Myh7 expression, n ≥ 8.
Upregulation of Yin Yang 1 (Yy1) suppresses DCM induced by Ttn shRNA
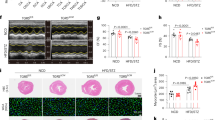
To dissect the molecular mechanisms of DCM induced by Ttn shRNA, we profiled control (n = 3) and Ttn shRNA (n = 3) treated hearts (dose = 0.2E + 13 vg/kg) by RNAseq, and the significantly changed were analyzed by Gene Ontology (Supplementary Fig. S2A). We selected 8 candidates involved in downregulated categories upon Ttn shRNA, including Atp2a2, Esrra, Sod2, Sirt3, Pdgfb, Gja1, Cav1 and Tmsb4x (Supplementary Table S1, Supplementary Fig. S2B)23,24,25,26,27,28. Anti-apoptosis gene Bcl2 was included because cardiac apoptosis exhibits in DCM models29,30. In addition, we added Yy1 which significantly enhanced TTN gene expression when co-transduced together with 5 factors (GATA4, MEF2C, TBX5, HAND2 and MYOCD, designated as 5 F) in human dermal fibroblasts (Supplementary Fig. S3). All 10 candidates were constructed into an AAV cis-vector (AAV-cTnT) transcriptionally controlled by cardiac Troponin T promoter. After each candidate (0.2E + 13 vg/kg) was co-introduced with Ttn shRNA (0.2E + 13 vg/kg), cardiac performance was assessed by echocardiography. When treated with most selected candidates, Ttn shRNA transduced mice developed a similar or even more severe DCM compared to those treated with control cDNA (0.2E + 13 vg/kg, expressing EGFP under cardiac troponin T promoter, designated as EGFP control), indicating that re-introducing these pathway-associated candidates could not suppress Ttn shRNA-induced DCM (Supplementary Table S2). Conversely, Yy1 (FS = 16.39 ± 1.44%, n = 5, P = 1.26E-03) significantly improved cardiac performance of Ttn transduced mice compared to those treated with EGFP control (FS = 9.91 ± 4.32%, n = 20).We further assessed the suppressive effect of Yy1 on DCM induced by Ttn shRNA for three additional timepoints (Fig. 2A,B, Supplementary Table S3). Three weeks after virus transduction, cardiac chamber dilation (LVDD) of Ttn shRNA transduced mice treated by Yy1 (3.90 ± 0.18 mm, n = 8, P = 4.20E-02) was significantly suppressed compared to that treated by EGFP control (4.20 ± 0.34 mm, n = 12). Remarkably, Yy1 treated mice continued to maintain significantly rescue of the cardiac chamber dilation compared to Ttn shRNA transduced mice treated by EGFP control. Cardiac function (FS) of Ttn shRNA mice treated with Yy1 was significantly improved compared to Ttn shRNA mice treated by EGFP control, suggesting that Yy1 suppresses DCM induced by Ttn shRNA. At dose of 0.2E + 13 vg/kg, candidate Yy1 was induced ~ 4.5 fold by qPCR analysis. The expression of heart failure markers including Nppa, Nppb and Myh7 in mice treated with Yy1 was significantly reduced compared to that in DCM mice treated with EGFP control (Fig. 2C). Ttn shRNA significantly reduced Ttn expression and protein level which were not induced by upregulation of Yy1, suggest Yy1 does not regulate Ttn in mouse hearts (Fig. 2C, Supplementary Fig. S4). Importantly, we did not observe increased fibrosis by Yy1 (Fig. 2D). Upregulation of Yy1 in wildtype mice did not alter cardiac performance at least 5 months post virus transduction, when ~3.0 fold increase of Yy1 expression was detected by qPCR analysis (Supplementary Table S4, Fig. 2E).
Upregulation of Yy1 suppresses dilated cardiomyopathy induced by Ttn shRNA. (A) Experimental timeline showing timepoints of virus injection and echocardiogram. (B) Ejection fraction (EF), Fractional shortening (FS), left ventricular diastolic dimension (LVDD) and left ventricular wall thickness (LVWT) of mice from control group (control shRNA + EGPF), Ttn shRNA group (Ttn shRNA + EGFP) and Yy1 treated group (Ttn shRNA + Yy1). Virus dose, 0.2E + 13 vg/kg. Cardiac performance was assessed by echocardiogram once a week from 4.5 to 7.5 weeks old. Data were presented as mean and SD, with associated P value, calculated by ANOVA and Tukey’s multiple comparisons test, representing comparison to Ttn shRNA group at each time point, n ≥ 8. (C) Quantitative real-time PCR analysis of Yy1, Nppa, Nppb, Myh7 and Ttn expression in mice from control, Ttn shRNA and Yy1 treated groups at 5.5 weeks old, n ≥ 4. (D) MT of Paraffin sections (left) of mice at 7.5 weeks old, magnification = 4 × , scale bar = 1000 µm, and quantification of myocardial fibrosis (right), n ≥ 4. (E) Quantitative real-time PCR analysis of Yy1 expression in mice injected with control (EGFP) and Yy1 respectively at 21.5 weeks old, virus dose, 0.2E + 13 vg/kg, n = 5.
Upregulation of Yy1 modulates cardiac structural contractile and cell growth related gene expression
To dissect the molecular mechanisms of Yy1 suppressing Ttn shRNA-induced DCM, we profiled control shRNA + EGFP (designated as control shRNA group, male, n = 4, dose = 0.2E + 13 vg/kg), Ttn shRNA + EGFP (designated as Ttn shRNA group, male, n = 4, dose = 0.2E + 13 vg/kg) and Ttn shRNA + Yy1 animals (designated as Yy1 treated group, male, n = 3, dose = 0.2E + 13 vg/kg) four weeks after virus transduction by RNAseq. To uncover signaling pathways regulated by Yy1, we found 134 genes significantly changed in Yy1 treated group compared to Ttn shRNA group (Fig. 3A). Among them, 80 genes were elevated, and 54 genes were reduced by Yy1. Hierarchical clustering of control shRNA, Ttn shRNA and Yy1 groups built significantly dysregulated genes into 4 regulated patterns. We analyzed gene list by Gene Set Enrichment Analysis (GSEA, Broad Institute). Gene ontology (GO) analysis identified top significantly enriched gene sets in category III including “positive regulation of growth” and “small molecule metabolic process” (Fig. 3B). Contractile fiber GO in category I included genes associated with sarcomere, such as Ankrd23, Fhl1 and Abra. The upregulation of four-and-a-half LIM domain protein 1 (Fhl1) and Ankyrin repeat domain-containing protein 23 (Ankrd23) by Ttn shRNA was suppressed by Yy1 (Fig. 3C). Importantly, Fhl1 and Ankrd23 interacted with Ttn protein, suggesting Yy1 modulates Ttn associated partners31,32. These results indicated Yy1 modulates Ttn partners and cardiac cell growth related gene expression which contributing to repair cardiac function.
Upregulation of Yy1 modulates cardiac structural contractile, cell proliferation and survival related gene expression. (A) Heat map representing color-coded expression level of 134 genes that significantly changed in Yy1 treated group (Ttn shRNA + Yy1) in comparison to Ttn shRNA group (Ttn shRNA + EGFP). Virus does, 0.2E + 13 vg/kg. Mice were harvest four weeks after transduction, n ≥ 3. (B) Top10 gene sets of GO of each category by GSEA, arranged by −log P value. (C) Quantitative real-time PCR analysis of Ankrd23, Fhl1, Abra expression in control, Ttn shRNA and Yy1 treated group, n ≥ 3.
Upregulation of Yy1 promotes cardiomyocyte cell cycle in dilated cardiomyopathy induced by Ttn shRNA
Because regulation of cell growth pathway was significantly enriched comparing Yy1 treated group to Ttn shRNA group by RNAseq analysis, we assessed cell cycle reentry genes including Ccnd1 and Ccnd2 by qPCR. Ccnd1 expression was significantly induced by ~1.36 fold (n = 12, P = 2.50E-02) in Ttn shRNA group compared to control shRNA group (n = 10). Ccnd2, another cyclin family member, was significantly induced by ~1.71 fold (n = 12, P = 1.92E-02) in Ttn shRNA group compared to control shRNA group (n = 10). Yy1 significantly enhanced additional Ccnd1 and Ccnd2 expression by ~ 1.33 fold (n = 11, P = 3.07E-03) and ~1.38 fold (n = 11, P = 3.58E-02) respectively compared to Ttn shRNA group (n = 12) (Fig. 4A). To locate Ccnd1 and Ccnd2 expression in heart tissues, we stained heart sections with Ccnd1 or Ccnd2 antibody together with an antibody recognizing cardiac Troponin I (cTn I) for cardiomyocytes (CM). Ccnd1 protein was predominantly detected in the nuclei of non-cardiomyocytes (non-CM) (Supplementary Fig. S5A). We observed a significant increase of non-CM positive for Ccnd1 (Ccnd1+) in Ttn shRNA group (1.16 ± 0.72%, n = 4, P = 2.38E-02) compared to control group (0.18 ± 0.09%, n = 5) (Fig. 4B). Importantly, a significant ~ 39.86 fold of Ccnd1 + CM (0.15 ± 0.04%, n = 4, P = 1.36E-02) in Ttn shRNA group was increased compared to those in control shRNA group (0.0038 ± 0.0026%, n = 5), suggesting that Ttn shRNA activates CM cell cycle reentry. In addition, Ccnd1 + CM was further increased by ~3.76 fold (0.56 ± 0.11%, n = 4, P = 7.02E-06) in Yy1 treated group compared to Ttn shRNA group. Ccnd2 protein was predominantly expressed and elevated in the nuclei of CM (Supplementary Fig. S5B). In comparison to control group (0.11 ± 0.03%, n = 5), we detected significantly ~ 37.20 fold more CM positive for Ccnd2 (Ccnd2+) in Ttn shRNA group (4.14 ± 0.52%, n = 4, P = 3.21E-07) (Fig. 4C). Yy1 significantly increased additional Ccnd2 + CM by~2.30 fold (9.53 ± 0.64%, n = 4, P = 3.48E-08) compared to Ttn shRNA group. We also observed a significant increase of Ccnd2 + non-CM in Ttn shRNA group (0.37 ± 0.14%, n = 4, P = 4.56E-03) compared to controls (0.05 ± 0.02%, n = 5), and Yy1 further significantly increased Ccnd2 + non-CM (0.65 ± 0.14%, n = 4, P = 1.36E-02) compared to Ttn shRNA group. We quantified protein levels of Ccnd1 and Ccnd2 in heart tissues by western blot analysis (Fig. 4D, Supplementary Fig. S6). Upon Ttn shRNA, we detected a significant ~ 3.01 fold (n = 4, P = 4.78E-03) and ~ 4.14 fold (n = 4, P = 7.46E-04) increase of Ccnd1 and Ccnd2 proteins compare to control shRNA (n = 4). In comparison to Ttn shRNA group (n = 4), additional ~ 1.98 fold (n = 4, P = 3.52E-04) and ~ 1.53 fold (n = 4, P = 7.45E-03) of Ccnd1 and Ccnd2 was increased in Yy1 group (Fig. 4E). We further examined how cyclin D was regulated in CM. Ccnd1 and Ccnd2 was assayed in isolated CM by qPCR as previous described33. Ccnd1 and Ccnd2 expression was increased by ~ 2.63 fold (2.71 ± 0.2%, n = 3, P = 7.35E-04) and ~ 1.93 fold (1.96 ± 0.33%, n = 3, P = 9.95E-04) in CM derived from Ttn shRNA group compared to control shRNA group (1.03 ± 0.27%, 1.02 ± 0.21%, n = 5), suggesting CM activated cell cycle reentry by upregulating Ccnd1 and Ccnd2 expression in response to Ttn insufficiency (Fig. 4F). Yy1 significantly enhanced additional Ccnd1 expression by ~ 1.56 fold (4.22 ± 0.62%, n = 3, P = 2.86E-03) compared to Ttn shRNA group (n = 3). Yy1 did not further increase Ccnd2 expression, suggesting Ccnd2 is not transcriptionally regulated by Yy1 in CM. To assess whether activated cardiac cell cycle advanced to G1/S phase, we assayed DNA synthesis by EdU incorporation. Most EdU was detected in non-CM because they do not exit from cell cycle (Supplementary Fig. S5C). We observed a comparable level of CM DNA positive for EdU (EdU + ) between control group (0.04±0.01%, n = 4, P = ns) and Ttn shRNA group (0.02 ± 0.004%, n = 4), suggesting CM cell cycle activated by endogenously upregulated genes including Ccnd1 and Ccnd2 does not advance to S phase (Fig. 4G). Upon Yy1 treatment, ~ 7.63 fold (0.18 ± 0.04%, P = 3.03E-05, n = 4) increase of EdU + CM was detected compared to Ttn shRNA group, indicating Yy1 promoted CM cell cycle from G1 to S phase. EdU incorporation also was increased by ~ 6.41 fold in non-CM of Yy1 group. To assess whether cardiac cell cycle further progressed to mitotic phase, we stained heart sections with M phase marker, phosphorylated histone (pH3) (Supplementary Fig S5D). We observed an elevated number of pH3 positive (pH3+) non-CM in Ttn shRNA group (0.04 ± 0.01%, n = 4, P = 6.78E-04) compared to control shRNA group (0.01 ± 0.003%, n = 5). Although Yy1 group showed an increased EdU incorporation in non-CM, upregulation of Yy1 did not further increase pH3 + signals (0.05 ± 0.01%, n = 4, P = ns) in non-CM of Yy1 group compared to Ttn shRNA group, suggesting upregulation of Yy1 does not increase non-CM mitosis. We did not observed pH3 + CM in the hearts of all three groups, indicating activated CM reaching to M phase are limited (Fig. 4H).
Upregulation of Yy1 promotes cardiac cell cycle in dilated cardiomyopathy induced by Ttn shRNA. (A) Quantitative real-time PCR analysis of Ccnd1 and Ccnd2 expression in mouse heart tissue from control, Ttn shRNA and Yy1 treated groups. Virus does, 0.2E + 13 vg/kg. Mice were harvest four weeks after transduction. n ≥ 10. (B) Paraffin sections (left) stained with DAPI (blue), cTnI (green) and Ccnd1 (red), representing Non-CM and CM with positive Ccnd1 signal (arrowed); quantification of Ccnd1 + Non-CM and CM (right) from control, Ttn shRNA and Yy1 treated groups, n ≥ 4. (C) Paraffin section (left) and quantification (right) of Ccnd2 + in Non-CM and CM respectively, n ≥ 4. (D) Western blot and quantitative analysis (E) of Yy1, Ccnd1 and Ccnd2 protein levels in mouse heart tissue, n = 4. Data were normalized by Lamin B1 antibody. Full-length blots were presented in Supplementary Fig. S6. (F) Quantitative real-time PCR analysis of Ccnd1 and Ccnd2 expression in isolated CM from control, Ttn shRNA and Yy1 treated groups four weeks after virus transduction, n ≥ 3. (G) Paraffin section (left) and quantification (right) of EdU + in Non-CM and CM respectively, n = 4. (H) Paraffin section (left) and quantification (right) of pH3 + in non-CM, n ≥ 4. In (B), (C), (G) and (H), magnification = 100×, scale bar = 10 µm. Data were shown as mean ± SD and were normalized to total nucleus number labelled by DAPI.
Upregulation of Ccnd1 and Ccnd2 suppresses DCM induced by Ttn shRNA
Ccnd1 and Ccnd2 was significantly enhanced by Yy1 in CM of mouse hearts. To assess whether enhancing of Cyclin D suppresses DCM caused by Ttn insufficiency, we co-introduced Ccnd1 or Ccnd2 (0.2E + 13 vg/kg) with Ttn shRNA (0.2E + 13 vg/kg). Cardiac performance was assessed by echocardiography. Cardiac function (FS) of mice treated with Ccnd1 or Ccnd2 was significantly improved compared to Ttn shRNA mice treated by EGFP. Ccnd1 or Ccnd2 overexpression significantly suppressed DCM induced by Ttn shRNA, suggesting cyclin D serves as a crucial downstream of Yy1 to enhance cardiac performance (Supplementary Table S5).
Discussion
Ttn truncating variants are enriched in DCM patients suggesting a causal effect of TTN variants on DCM3. TTN Haploinsufficiency caused by TTNtv is emerging as the disease mechanism. To assess whether Ttn insufficiency causes DCM, we used shRNA to modulate Ttn expression. By reducing ~ 50% of Ttn expression, we generated a mouse model demonstrating a severe DCM phenotype, including ventricular wall thinning, dilated ventricular chambers and impaired cardiac function. To develop potential therapy for suppressing DCM caused by Ttn insufficiency, we screened 10 genes involved in different pathways. As a transcriptional regulator of TTN during direct cardiac reprogramming, we included Yy1 for the rescue experiments. Ttn expression was not induced by Yy1 in DCM hearts caused by Ttn shRNA, suggesting that Yy1 does not inhibit DCM through Ttn gene regulation in mouse hearts. Recently, Yy1 was shown to promote both Nkx2.5 expression, cardiac progenitor cell commitment and maintenance during early embryo development34,35. Cardiomyocyte-specific ablation of Yy1 mediated by Myh6-cre resulted in perinatal death of mutant mice, suggesting Yy1 plays an important role in early and late cardiac lineage development36. Importantly, YY1 is shown to be upregulated in human idiopathic dilated cardiomyopathy (IDC) and heart failure37,38.
We detected over 35 fold increase of Ccnd1 and Ccnd2 in cardiomyocyte nuclei upon Ttn shRNA, indicating cardiomyocytes response to Ttn insufficiency by activating cell cycle reentry signals. We did not observed this reaction in another DCM model caused by Lmna insufficiency. Activated cardiomyocyte cell cycle induced by Ttn insufficiency does not advance to S phase. We found that upregulation of Yy1 promoted cardiac cell cycle reentry by further enhancing Ccnd1 and Ccnd2. Yy1 promoted cardiomyocyte cell cycle to S phase by a significant increase of EdU incorporation. However, we did not detect mitotic phase marker pH3 in cardiomyocytes, suggesting activated cardiomyocytes undergoing mitotic phase are limited. Importantly, Upregulation of Ccnd1 and Ccnd2 suppressed DCM caused by Ttn insufficiency. Taken together, Yy1 promotes cardiac cell cycle to facilitate to suppress Ttn shRNA-induced DCM.
Our recent study showed insufficiency of Mybpc3, another causal gene for sarcomeric cardiomyopathy, induces an extra round of cardiomyocyte cell cycle during neonatal stage19. It is now of great interest to know whether this reactivation of cardiac cell cycle is a common mechanism for sarcomere deficiency and enhancing of this process by Yy1 or other regulators is able to suppress cardiovascular disease related to sarcomere. Reactivation of cardiomyocyte cell cycle does not always lead to cell number increase. Cytokinesis is still rarely detected in adult hearts upon regenerative enhancement. Yy1 might promote hypertrophic growth coupled with cell cycle reentry as Ccnd2 is a mediator for Myc overexpression or exercise-induced hypertrophic growth in cardiomyocytes39. Recently, Yy1 is suggested to serve as a structural regulator between enhancer and promoter interactions and facilitates gene expression40. Apart from modulating cell growth, upregulation of Yy1 might indeed reinstate or promote enhancer - promoter interaction to restore gene regulatory network dysregulated by Ttn insufficiency.
Our selected candidates included many therapeutic targets for heart failure including Atp2a2, Bcl2, Sod2 and Sirt323,24,28,41. Most of them failed to protect DCM induced by Ttn shRNA, suggesting root causes and disease mechanisms should be taken into account for DCM prevention and treatment strategies. It is of great interest to know whether Yy1 is able to suppress DCM caused by other genes including LMNA, MYH7 and PLN. To translate our research, one concern is whether upregulation of Yy1 could cause any cardiac defects. Previous study showed overexpression of Yy1 induced a relative marginal hypertrophy cardiomyopathy only in male mice. Here, we specified the Yy1 expression by cardiac specific promoter, virus dose and postnatal transduction. In contrast to a previous study, no detectable impairment of cardiac structure and performance was observed after ~ 5 months of virus transduction37. Taken together, our findings provide a strong supporting evidence for translational research.
Materials and Methods
Animal protocols
All mice were maintained and studied using protocols approved by the Institutional Animal Care and Use Committee (IACUC) of National University of Singapore. Animal work was undertaken in accordance with Singapore National Advisory Committee for Laboratory Animal Research guidelines. Relevant national and institutional guidelines and regulations must be consulted before commencement of any animal work. All studies were conducted in male C57BL/6JINV (Jax) mice. For virus injection, 50 µl viruses were injected into thoracic cavity of 10 days old pups via insulin syringe, avoiding the heart and lungs. For EdU injection, EdU (Sigma, 900584) was dissolved in saline and 5 mg/kg EdU was delivered to mice by intraperitoneal injection for two weeks after AAV transduction. For heart harvesting, mouse was anesthetized by 2% isoflurane and the heart was exposed by opening chest. After that, 15% KCl was injected into inferior vena cava to achieve asystole at diastole, then the heart was rapidly isolated and flushed with D-PBS through LV to wash out blood. Half of the apex was isolated and immersed in RNALater (Qiagen, 76104) at room temperature for RNA extraction, while the other half was snap frozen in liquid nitrogen for protein extraction. The rest part of heart was fixed in 4% paraformaldehyde for 24 hours and subsequently embedded by paraffin.
Echocardiogram (Echo) and surface electrocardiogram (ECG)
Cardiac dimension and function of mice three and/or four weeks after virus transduction were achieved by echocardiography (VisualSonics, Vevo 2100, 40 Mhz-550S probe). All mice were shaved to expose chest area one day before experiment. During echo, 1.5% isoflurane with oxygen were applied to each mouse, and cine of 300 frames of both B mode and M mode (left parasternal long and short axes) were recorded when heart rate was around 450–500 bpm. Measurements were processed by Vevo®LAB (VisualSonics Inc.). LV tracings were averaged from at least 3 consecutive heart beats of M-mode. LVDD (LV diastolic dimensions), LVWT (LV posterior wall thickness), EF (ejection fraction) and FS (fractional shortening) were obtained from short axis images.
Cell culture and transfection
HEK293T cells were cultured at 37 °C with 5% CO2 and maintained in DMEM (Hyclone) supplemented with 10% FBS, 1 mM sodium pyruvate and 10 µg/ml gentamicin. Transfection of shRNA constructs and other plasmids was performed using PEI (Polysciences. Inc, 24765-2) according to manufacturer’s instructions.
Cardiomyocyte isolation
We adapted protocol from Ackers-Johnson et al. In brief, the heart was perfused with warm EDTA, perfusion buffer and collagenase buffer through LV when aorta was clamped, then heart tissue became softened and swollen. After that, heart tissue was teared into pieces and dissociated by pipetting. Cell suspension was then passed through 100 µm strainer and processed to three rounds of gravity settling. Cardiomyocytes, in the pellet, were harvested for RNA extraction.
Recombinant adeno-associated viruses 9 (rAAV9) production, purification and titration
Protocol was adapted from Grieger et al. In brief, rAAVs were produced on HEK293T cells by transient triple plasmid transfection, including rAAV viral vector with gene to be delivered, helper plasmids pAdΔF6 and plasmid pAAV2/9 (Penn Vector Core). Three days after transfection, viruses were collected from cell pellets and were purified by Optiprep density gradient medium (Sigma, D-1556). After concentrating by centrifugal filter (Milipore, UFC910096), viruses were aliquoted and stored at −80 °C. For virus titration, forward primer: gataaaagcagtctgggctttcaca and reverse primer: gagcccatataagcccaagctattg were designed to target at rAAV genome-containing particle, cTnT promotor region, and to determinate the physical titers by qPCR.
Lentiviral production and transduction
Lentiviral plasmids teto-HAND2, FUW-TetO-GATA4, teto-MEF2C, teto-TBX5, teto-MYOCD and teto-YY1were obtained from Addgene or by cloning. Lentiviruses were produced on HEK293T cells. Two days after transfection, viruses were harvested from supernatant and concentrated by centrifugal filter (Milipore, UFC910096). Lenti-XTM qRT-PCR Titration Kit was applied for titration. As for virus infection, human primary fibroblasts (ATCC PCS-201-010) were subjected to two rounds of overnight transduction at 50% confluency.
shRNA vector construction
Ttn shRNA candidates were designed to target 21 base-pair gene-specific regions (Invitrogen). The sequences of mouse Ttn shRNAs are as follows: shRNA-1: AGTACTTTCAGCTTAATGGTG (Exon 262); shRNA-2: TAAAGAAGCCGATTTCTTGGT (Exon 275); shRNA-3: ATTACTGGCACACTCAGTTGT (Exon 275); shRNA-4: ATAAGTTGGAGACAAGGAGCG (Exon 307). The sequence against LacZ, AAATCGCTGATTTGTGTAGTC, was used as control shRNA. The miR-155 backbone based shRNA cassette was inserted at 3 prime of AAV-cTnT-EGFP vector.
RNAseq library preparation and next generation sequencing
Total RNA from left ventricular tissue of male mice (n = 3 per group) was achieved to establish RNAseq library. RNA samples were pre-treated with Truseq Stranded Total RNA Library Prep kit (Illumina, RS-122–2201) to remove abundant cytoplasmic rRNA. The remaining intact RNA was fragmented using a chemical mix, followed by first- and second-strand cDNA synthesis using random hexamer primers. End-repaired fragments were ligated with a unique illumina adapter. All individually indexed samples were subsequently pooled together and multiplexed for sequencing. Libraries were sequenced using the Illumina Hiseq. 2000 sequencing system and paired-end 101 bp reads were generated for analysis. RNAseq data was deposited to NCBI. For selecting potential therapeutic genetic candidates, 3101 genes (log fold change <−0.59 or >0.7, FDR <0.05) were identified from DCM group (Ttn shRNA) compared to control group (Ctrl shRNA). For dissecting molecular mechanism of Yy1, 134 genes (P < 0.005 and FDR <0.2) were selected from Yy1 treated group (Ttn shRNA + Yy1) compared to Ttn shRNA group (Ttn shRNA + EGFP). Differentially expressed genes were uploaded to Morpheus for Hierarchical clustering and color-coded heat-map. Gene ontology (GO) of each category were analyzed by Gene Set Enrichment Analysis (GSEA, Broad Institute).
Quantitative real-time PCR (qPCR)
Transcription level were quantified by qPCR. cDNA was synthesized using Maxima First Strand kit (ThermoFisher, K1641) and qPCR was carried out by KAPA SYBR Fast qPCR Master Mix kit (KAPA Biosystems, KR0389). All qPCR primers are listed as follows:
Histological and immunostaining analysis
Heart samples were fixed in 4% paraformaldehyde for 24 hours, then embedded in paraffin and sectioned at 5 µm intervals. Paraffin samples were further treated with xylene (to remove paraffin), re-hydrated, and permeabilized in 0.1%(v/v) Triton-X100 in PBS. Hematoxylin and eosin (HE) was applied to observe myocyte architecture and Masson trichrome (MT) to identify cardiac fibrosis. Fibrosis was quantified by Image J. The percentage of fibrosis was calculated as the blue-stained areas divided by total ventricular area. As for immunostaining, boiled citric acid was used for Ccnd1, Ccnd2, EdU and pH3. Primary antibodies: Ccnd1 (Abcam, ab16663), Ccnd2 (Cell Signaling Technology, 3741), pH3 (Cell Signaling Technology, 9718) and cTnI (Abcam, ab8295). Other labeling dyes: DAPI for nucleus (ThermoFisher, D1306) and EdU Imaging kit (ThermoFisher, C10229). All the positive signals from three completed cross sections were counted for each heart sample, and data were normalized to total nucleus number.
Western blots
Frozen heart tissues were lysed in cold RIPA buffer with protease inhibitor (Sigma, 4693116001) and were homogenized with prechilled TissueLyser (Qiagen, 25/s, 2 mins, 3 cycles) by adding metal beads. After centrifuge, supernatants were collected to detect proteins with small molecular weight like Yy1, Ccnd1 and Ccnd2. 20 µg of each sample was loaded into SDS-page gel and further transfer to nitrocellulose membrane (0.2 µm, Biorad, 162-0112) for blotting. Primary antibody including: Yy1 (Thermofisher, PA5-29171), Ccnd1 (Abcam, ab16663), Ccnd2 (Cell Signaling Technology. Inc, 3741), and Lamin B1 (Abcam, ab16048). Secondary antibody including: Donkey anti-Rabbit IgG (H + L) Highly Cross-Adsorbed Secondary Antibody (Thermofisher, A16035).
Coomassie blue staining for Ttn protein
As for pellet fraction, protocol was adapted from Neagoe et al.42. Briefly, pellets were resuspended in solubilization buffer (1% SDS, 1% 2-mercaptoethanol, 10% glycerol, 8 µg/ml leupeptin, 4.3 mM Tris–HCl, pH 8.8, 4.3 mM EDTA) and samples were further incubated on ice for 10 mins followed by boiling (95 °C) for 3 mins. 35 µg protein from each sample was added with 6 µM of bromophenol blue (Sigma, 114391) and then loaded onto agarose-strengthened SDS-PAGE gel (3.6% polyacrylamide, 1% agarose with a laemmli buffer system). Protein bands were visualized by SimplyBlue SafeStain (Life technologies, LC6060) and band intensity were quantified using ImageLab (Bio-Rad).
Statistical analyses
Statistical analysis was achieved by Prism Graphpad 7.0. P value between two groups was performed by two-tailed, unpaired T-test with Weltch correction, and one-way ANOVA with Tukey’s multiple comparisons test for multiple groups. Quantitative data were shown as mean ± SD. ns, non-significant, P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001.
Data availability
The datasets generated during and/or analysed during the current study are available in the NCBI repository, https://www.ncbi.nlm.nih.gov/Traces/study/?acc=PRJNA565836.
References
Hershberger, R. E., Hedges, D. J. & Morales, A. Dilated cardiomyopathy: the complexity of a diverse genetic architecture. Nat Rev Cardiol 10, 531–547, https://doi.org/10.1038/nrcardio.2013.105 (2013).
Weintraub, R. G., Semsarian, C. & Macdonald, P. Dilated cardiomyopathy. Lancet 390, 400–414, https://doi.org/10.1016/S0140-6736(16)31713-5 (2017).
Herman, D. S. et al. Truncations of titin causing dilated cardiomyopathy. N Engl J Med 366, 619–628, https://doi.org/10.1056/NEJMoa1110186 (2012).
Fatkin, D. et al. Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease. N Engl J Med 341, 1715–1724, https://doi.org/10.1056/NEJM199912023412302 (1999).
Olson, T. M. et al. Inherited and de novo mutations in the cardiac actin gene cause hypertrophic cardiomyopathy. J Mol Cell Cardiol 32, 1687–1694, https://doi.org/10.1006/jmcc.2000.1204 (2000).
Schmitt, J. P. et al. Dilated cardiomyopathy and heart failure caused by a mutation in phospholamban. Science 299, 1410–1413, https://doi.org/10.1126/science.1081578 (2003).
McNally, E. M., Golbus, J. R. & Puckelwartz, M. J. Genetic mutations and mechanisms in dilated cardiomyopathy. J Clin Invest 123, 19–26, https://doi.org/10.1172/JCI62862 (2013).
Ware, J. S. et al. Shared Genetic Predisposition in Peripartum and Dilated Cardiomyopathies. N Engl J Med 374, 233–241, https://doi.org/10.1056/NEJMoa1505517 (2016).
Gerull, B. et al. Mutations of TTN, encoding the giant muscle filament titin, cause familial dilated cardiomyopathy. Nat Genet 30, 201–204, https://doi.org/10.1038/ng815 (2002).
Schafer, S. et al. Titin-truncating variants affect heart function in disease cohorts and the general population. Nat Genet 49, 46–53, https://doi.org/10.1038/ng.3719 (2017).
Pasumarthi, K. B. & Field, L. J. Cardiomyocyte cell cycle regulation. Circ Res 90, 1044–1054 (2002).
Bergmann, O. et al. Evidence for cardiomyocyte renewal in humans. Science 324, 98–102, https://doi.org/10.1126/science.1164680 (2009).
Senyo, S. E. et al. Mammalian heart renewal by pre-existing cardiomyocytes. Nature 493, 433–436, https://doi.org/10.1038/nature11682 (2013).
Morikawa, Y., Heallen, T., Leach, J., Xiao, Y. & Martin, J. F. Dystrophin-glycoprotein complex sequesters Yap to inhibit cardiomyocyte proliferation. Nature 547, 227–231, https://doi.org/10.1038/nature22979 (2017).
Mahmoud, A. I. et al. Meis1 regulates postnatal cardiomyocyte cell cycle arrest. Nature 497, 249–253, https://doi.org/10.1038/nature12054 (2013).
See, K. et al. Single cardiomyocyte nuclear transcriptomes reveal a lincRNA-regulated de-differentiation and cell cycle stress-response in vivo. Nat Commun 8, 225, https://doi.org/10.1038/s41467-017-00319-8 (2017).
Chen, J. et al. mir-17-92 cluster is required for and sufficient to induce cardiomyocyte proliferation in postnatal and adult hearts. Circ Res 112, 1557–1566, https://doi.org/10.1161/CIRCRESAHA.112.300658 (2013).
Lin, Z. et al. Cardiac-specific YAP activation improves cardiac function and survival in an experimental murine MI model. Circ Res 115, 354–363, https://doi.org/10.1161/CIRCRESAHA.115.303632 (2014).
Jiang, J. et al. Cardiac myosin binding protein C regulates postnatal myocyte cytokinesis. Proc Natl Acad Sci USA 112, 9046–9051, https://doi.org/10.1073/pnas.1511004112 (2015).
Jiang, J., Wakimoto, H., Seidman, J. G. & Seidman, C. E. Allele-specific silencing of mutant Myh6 transcripts in mice suppresses hypertrophic cardiomyopathy. Science 342, 111–114, https://doi.org/10.1126/science.1236921 (2013).
Wakimoto, H., Seidman, J. G., Foo, R. S. & Jiang, J. AAV9 Delivery of shRNA to the Mouse Heart. Curr Protoc Mol Biol 115, 23 16 21–29, https://doi.org/10.1002/cpmb.9 (2016).
Wolf, C. M. et al. Lamin A/C haploinsufficiency causes dilated cardiomyopathy and apoptosis-triggered cardiac conduction system disease. J Mol Cell Cardiol 44, 293–303, https://doi.org/10.1016/j.yjmcc.2007.11.008 (2008).
Nelson, B. R. et al. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 351, 271–275, https://doi.org/10.1126/science.aad4076 (2016).
Zangi, L. et al. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat Biotechnol 31, 898–907, https://doi.org/10.1038/nbt.2682 (2013).
Pasumarthi, K. B., Nakajima, H., Nakajima, H. O., Soonpaa, M. H. & Field, L. J. Targeted expression of cyclin D2 results in cardiomyocyte DNA synthesis and infarct regression in transgenic mice. Circ Res 96, 110–118, https://doi.org/10.1161/01.RES.0000152326.91223.4F (2005).
Van Remmen, H. et al. Knockout mice heterozygous for Sod2 show alterations in cardiac mitochondrial function and apoptosis. Am J Physiol Heart Circ Physiol 281, H1422–1432, https://doi.org/10.1152/ajpheart.2001.281.3.H1422 (2001).
Hafner, A. V. et al. Regulation of the mPTP by SIRT3-mediated deacetylation of CypD at lysine 166 suppresses age-related cardiac hypertrophy. Aging (Albany NY) 2, 914–923, https://doi.org/10.18632/aging.100252 (2010).
Chatterjee, S. et al. Viral gene transfer of the antiapoptotic factor Bcl-2 protects against chronic postischemic heart failure. Circulation 106, I212–217 (2002).
Nikolova, V. et al. Defects in nuclear structure and function promote dilated cardiomyopathy in lamin A/C-deficient mice. J Clin Invest 113, 357–369, https://doi.org/10.1172/JCI19448 (2004).
Crone, S. A. et al. ErbB2 is essential in the prevention of dilated cardiomyopathy. Nat Med 8, 459–465, https://doi.org/10.1038/nm0502-459 (2002).
Raskin, A. et al. A novel mechanism involving four-and-a-half LIM domain protein-1 and extracellular signal-regulated kinase-2 regulates titin phosphorylation and mechanics. J Biol Chem 287, 29273–29284, https://doi.org/10.1074/jbc.M112.372839 (2012).
Miller, M. K. et al. The muscle ankyrin repeat proteins: CARP, ankrd2/Arpp and DARP as a family of titin filament-based stress response molecules. J Mol Biol 333, 951–964 (2003).
Ackers-Johnson, M. et al. A Simplified, Langendorff-Free Method for Concomitant Isolation of Viable Cardiac Myocytes and Nonmyocytes From the Adult Mouse Heart. Circ Res 119, 909–920, https://doi.org/10.1161/CIRCRESAHA.116.309202 (2016).
Gregoire, S. et al. Essential and unexpected role of Yin Yang 1 to promote mesodermal cardiac differentiation. Circ Res 112, 900–910, https://doi.org/10.1161/CIRCRESAHA.113.259259 (2013).
Gregoire, S., Li, G., Sturzu, A. C., Schwartz, R. J. & Wu, S. M. YY1 Expression Is Sufficient for the Maintenance of Cardiac Progenitor Cell State. Stem Cells 35, 1913–1923, https://doi.org/10.1002/stem.2646 (2017).
Beketaev, I. et al. Critical role of YY1 in cardiac morphogenesis. Dev Dyn 244, 669–680, https://doi.org/10.1002/dvdy.24263 (2015).
Stauffer, B. L. et al. Transgenic over-expression of YY1 induces pathologic cardiac hypertrophy in a sex-specific manner. Biochem Biophys Res Commun 462, 131–137, https://doi.org/10.1016/j.bbrc.2015.04.106 (2015).
Sucharov, C. C., Mariner, P., Long, C., Bristow, M. & Leinwand, L. Yin Yang 1 is increased in human heart failure and represses the activity of the human alpha-myosin heavy chain promoter. J Biol Chem 278, 31233–31239, https://doi.org/10.1074/jbc.M301917200 (2003).
Zhong, W. et al. Hypertrophic growth in cardiac myocytes is mediated by Myc through a Cyclin D2-dependent pathway. EMBO J 25, 3869–3879, https://doi.org/10.1038/sj.emboj.7601252 (2006).
Weintraub, A. S. et al. YY1 Is a Structural Regulator of Enhancer-Promoter Loops. Cell 171, 1573–1588 e1528, https://doi.org/10.1016/j.cell.2017.11.008 (2017).
Kawase, Y. et al. Reversal of cardiac dysfunction after long-term expression of SERCA2a by gene transfer in a pre-clinical model of heart failure. J Am Coll Cardiol 51, 1112–1119, https://doi.org/10.1016/j.jacc.2007.12.014 (2008).
Neagoe, C., Opitz, C. A., Makarenko, I. & Linke, W. A. Gigantic variety: expression patterns of titin isoforms in striated muscles and consequences for myofibrillar passive stiffness. J Muscle Res Cell Motil 24, 175–189 (2003).
Kent, W. J. et al. The human genome browser at UCSC. Genome Res. 12(6), 996–1006 (2002 Jun).
Acknowledgements
We thank Zenia Tiang for RNAseq technical assistance and Yiqing Li for assistance with echocardiograph. This work was supported by grants of NMRC (J.J., R.F, NMRC/CIRG/1431/2015 and NMRC/OFIRG/0056/2017), MOE T1 (J.J, NUHS O-CRG 2016) and Startup (NUHS J.J.).
Author information
Authors and Affiliations
Contributions
J.J. wrote the main manuscript text; D.L. wrote figure legends and materials and methods, and prepared Figures 1–4, Supplementary Figures 1,2 and Supplementary Tables 1, 2, 3; W.C prepared Figure. 3 and Supplementary Figures 2, 3; J.X.W., P.S.C. and C.Y.T. prepared Figure. 4 and Supplementary Figures 1, 4, 5;.L.W.T. and R.F. did RNAseq and analysis.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liao, D., Chen, W., Tan, C.Y. et al. Upregulation of Yy1 Suppresses Dilated Cardiomyopathy caused by Ttn insufficiency. Sci Rep 9, 16330 (2019). https://doi.org/10.1038/s41598-019-52796-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-52796-0
This article is cited by
-
Systematic in vivo candidate evaluation uncovers therapeutic targets for LMNA dilated cardiomyopathy and risk of Lamin A toxicity
Journal of Translational Medicine (2023)
-
Transcriptome studies of inherited dilated cardiomyopathies
Mammalian Genome (2023)
-
Disrupting the LINC complex by AAV mediated gene transduction prevents progression of Lamin induced cardiomyopathy
Nature Communications (2021)
-
Network integration and modelling of dynamic drug responses at multi-omics levels
Communications Biology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.